Abstract
Computing interactions among cells is computationally complex in multicellular molecular communication simulations, making the design of efficient algorithms crucial. We review simulation algorithms for computing interactions among cells in multicellular molecular communication simulations. The naïve algorithm, the Cell List algorithm, the Barnes–Hut algorithm, and a hybrid of the Cell List and Barnes–Hut algorithms are explored.
1. Introduction
Molecular communication is an emerging communication paradigm for bio-nanomachines (e.g., naturally occurring and artificially synthesized cells) [1,2,3]. In molecular communication, bio-nanomachines act as communication nodes and exchange information through biochemical and biophysical interactions. This form of communication is highly parallelizable, biocompatible, and energy-efficient. Applications of molecular communication and bio-nanomachines encompass in-body molecular sensing, targeted drug delivery, biological robotics, and 6G wireless molecular communication [4,5].
Computer simulation is promising in advancing research in molecular communication [6,7,8]. While wet laboratory experiments are essential, they can still be costly. Thus, simulation experiments, which can be conducted on personal computers, serve as an alternative approach. In previous work, we designed and implemented a multicellular molecular communication simulator [9,10]. The simulator was designed to be extensible, allowing users to design, model, and simulate their systems, such as cell-based drug delivery systems. Within the simulator, bio-nanomachines can be engineered from biological cells and exhibit cellular functions such as sensing the environment, sending and receiving molecules, migrating, dividing, and dying.
This article provides a comprehensive overview of algorithms for multicellular molecular communication simulations. In multicellular molecular communication simulations, computing interactions among cells is expensive, making the design of efficient algorithms crucial. Therefore, we review four algorithms, the naïve algorithm, the Cell List algorithm, the Barnes–Hut algorithm, and a hybrid of the Cell List and Barnes–Hut algorithms, as they vary in cost and accuracy depending on user needs. The Cell List algorithm is proposed in Ref. [9], the Barnes–Hut algorithm is proposed in Ref. [10], and a hybrid algorithm is designed in this study to simulate based on the Barnes–Hut algorithm.
The remaining sections of this article are organized as follows. Section 2 provides an overview of simulation algorithms, including the procedure to compute cell–cell interactions. We then provide detailed descriptions of the four algorithms: the naïve algorithm in Section 3, the Cell List algorithm in Section 4, the Barnes–Hut algorithm in Section 5, and a hybrid of the Cell List and Barnes–Hut algorithms in Section 6. Finally, in Section 7, we discuss future challenges and conclude this study.
2. Simulation Algorithms
In simulations, a two-dimensional simulation space is considered where the set of cells (or bio-nanomachines) interact through molecular communication and move collectively. Time is denoted as t, and it discretely advances by ∆t. For simplicity, cells do not divide, and the set is time-invariant.
In simulations, we assume that cells receive a force as a result of interactions with other cells. The magnitude of the force depends on cell–cell distance. Cells move based on the force in the simulation space; the velocity of a cell increases linearly as the force acting on it increases, with being the proportionality constant (i.e., we neglect inertia).
Let xi(t) be the position of cell i at time t, vi(t) be the velocity of cell i at time t, fi(t) be the force acting on cell i at time t, fij(t) be the force acting on cell i from cell j at time t, f(d) be the magnitude of the force between cells separated by a distance d, and dij be the distance between cell i and cell j at time t. Note that we use boldface to express a vector.
Under such assumptions, the position xi(t) of cell i at time t is given by the following equations:
Algorithm 1 outlines the simulation step. After initialization, it contains two for-loops: one for calculating interactions between cells (lines 2–4) and the other for updating cell positions (lines 5–7). In the first for-loop, we iteratively compute the forces acting on the cells. The ‘calcForce()’ function depends on the algorithm being used. In the second for-loop, we iteratively update the positions of all cells.
In the following, we describe four algorithms to compute ‘calcForce()’ in Algorithm 1: the naïve algorithm, the Cell List algorithm, the Barnes–Hut algorithm, and a hybrid of the Cell List and Barnes–Hut algorithms.
| Algorithm 1 Simulation step |
|
3. Naïve Algorithm
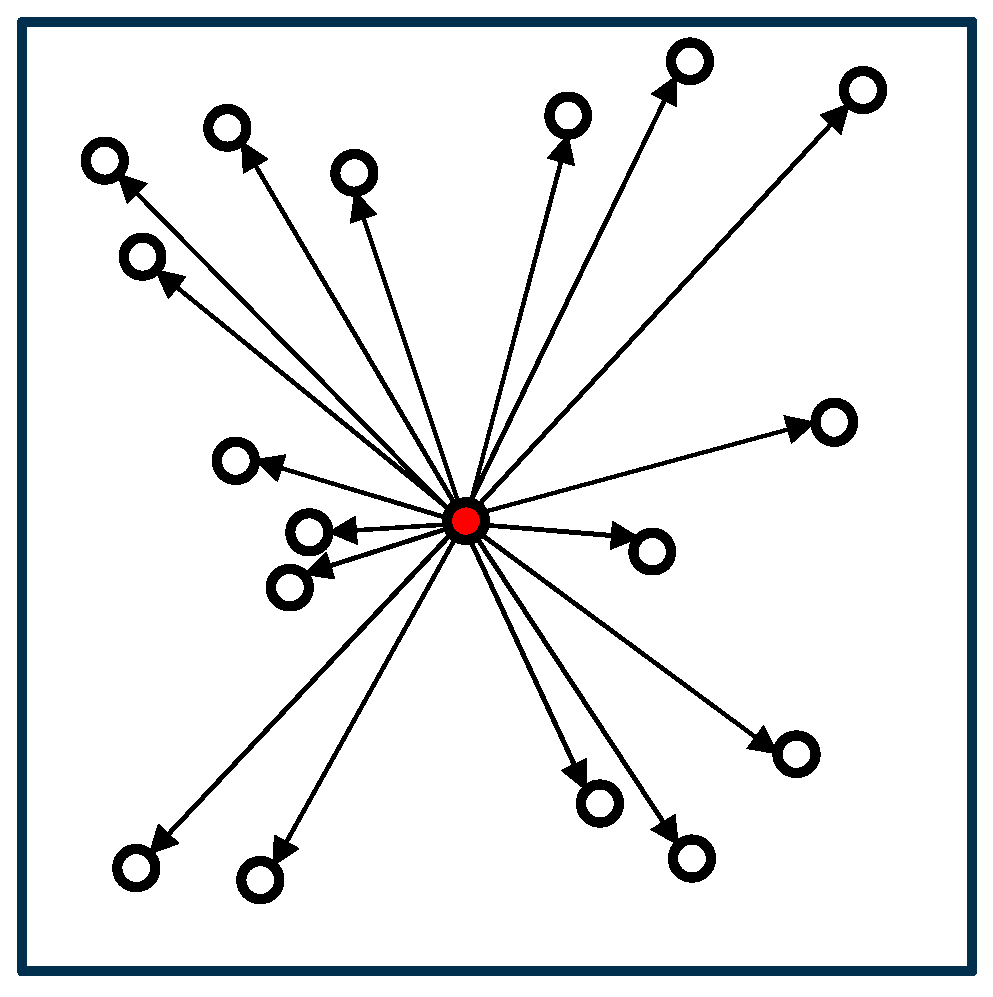
The naïve algorithm is the simplest algorithm for calculating the force acting on a cell by computing the interactions with all other cells. Figure 1 illustrates the computational process of the algorithm. In the figure, cells are represented as circles, and the cells exerting force on the red cell are depicted with arrows connecting them. While the naïve algorithm is the most accurate, its computational cost increases sharply as the number of cells increases.
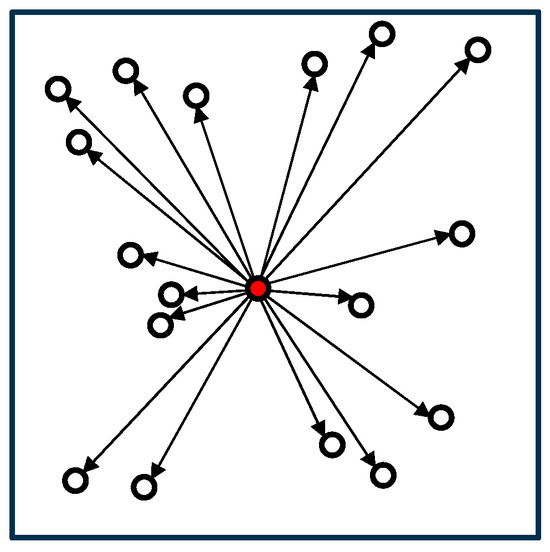
Figure 1.
Naïve algorithm.
In the naïve algorithm, the initialization process (line 1) in Algorithm 1 is unnecessary. Algorithm 2 presents the pseudocode for the ‘calcForce()’ function in the naïve algorithm. Since we calculate the force acting on cell i from cell j |N| times for both cells i and j, the time complexity of this algorithm is O(|N|2).
| Algorithm 2 calcForce(cell ) in the naïve algorithm |
|
4. Cell List Algorithm
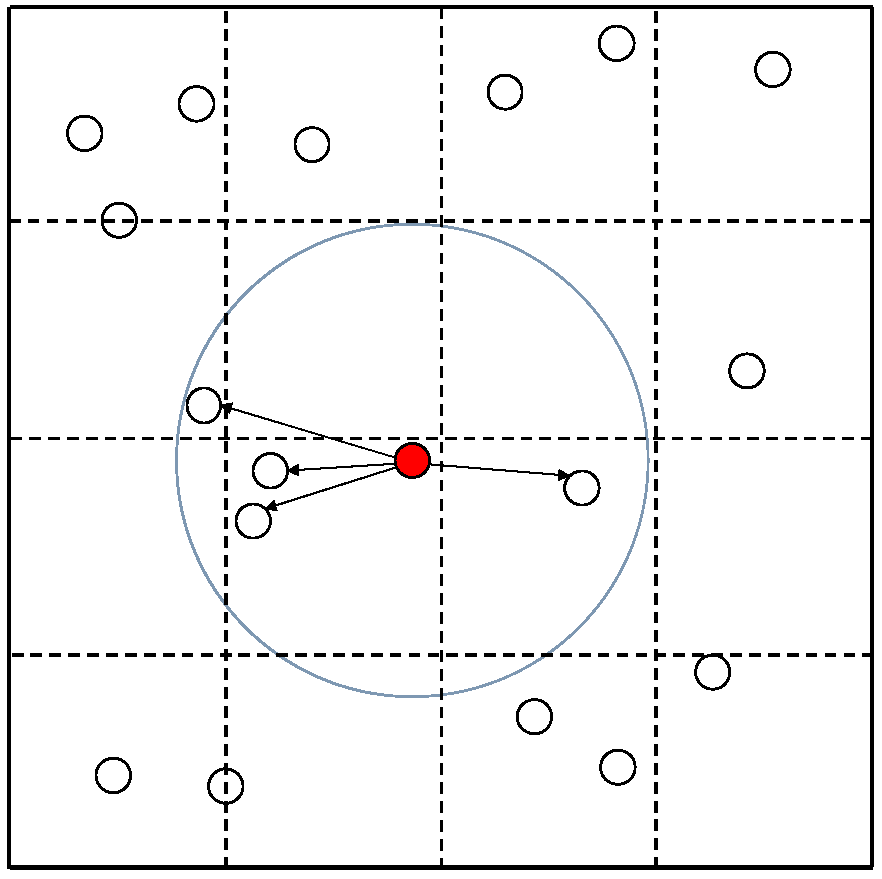
The Cell List algorithm accelerates simulations by limiting the number of cells that exert force on a cell. The Cell List algorithm is an accelerated algorithm that reduces computation time by limiting the calculation to nearby cells when determining the force acting on a cell. However, there is a trade-off between computation speed and accuracy, depending on the range considered. Figure 2 illustrates the computational process of the algorithm. In the Cell List algorithm, the simulation space is divided into grid units, with each grid maintaining the addresses of all cells within it. When calculating the force acting on the red cell, the algorithm searches for cells within a grid search range (yellow area) and then further limits the calculation to those cells within the red cell’s search radius (blue circle). The grid search range determines how many grid cells from the grid containing the red cell are included in the search area. For example, when the grid search range is w (a non-negative integer), the search area forms a square of grid cells. As shown in Figure 2, the grid search range is set to 1. The grid search range should be set to accommodate the search radius.
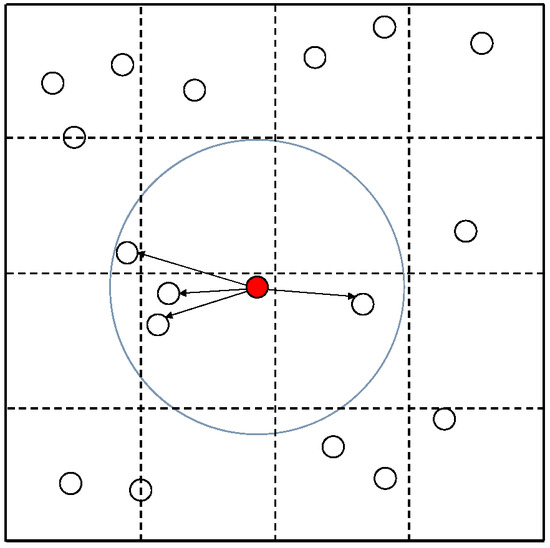
Figure 2.
Cell List algorithm.
Algorithm 1 shows the Cell List algorithm, including the initialization of the cells within the grids (line 1). Algorithm 3 presents the pseudocode for the ‘calcForce()’ function of the Cell List algorithm. We obtain the list of surrounding cells to be calculated using the ‘getCellList()’ function and stored in the ‘CellList’ list (line 1). We then calculate the interaction only for the cells in the ‘CellList’ list (lines 2–4). Algorithm 4 presents the pseudocode for the ‘getCellList()’ function of the Cell List algorithm. Based on the address of the input cell, the corresponding grid is identified (line 5), and a list of cells belonging to the surrounding grids is created based on the grid search range (lines 6–8). From this list, only the cells within the search radius are selected for inclusion in a new list (lines 9–11) and returned to the caller (line 16). Each grid is assigned a grid number for identification. Here, columns are denoted as ‘gridX’ and rows are denoted as ‘gridY’, defining the grid as ‘(gridX, gridY)’.
| Algorithm 3 calcForce(cell ) in the Cell List algorithm |
|
| Algorithm 4 getCellList(cell ) in the Cell List algorithm |
|
Since the Cell List algorithm’s processing speed and accuracy are influenced by parameters like search range and search radius, the time complexity can vary from O(|N|) to O(|N|2).
5. Barnes–Hut Algorithm
The Barnes–Hut algorithm is commonly used for -body problems in physics [11]. In our previous work, we applied it to multicellular molecular communication simulations [10]. Unlike the Cell List algorithm, the Barnes–Hut algorithm allows a cell to receive forces from all other cells in multicellular molecular communication simulations, therefore providing improved accuracy. In the Barnes–Hut algorithm, distant cells were grouped and treated as a unified cell located at the center of mass of the cell group. The Barnes–Hut algorithm consists of the following three steps: Constructing the Barnes–Hut tree, computing centers of mass, and calculating cell–cell interaction forces.
5.1. Construction of the Barnes–Hut Tree
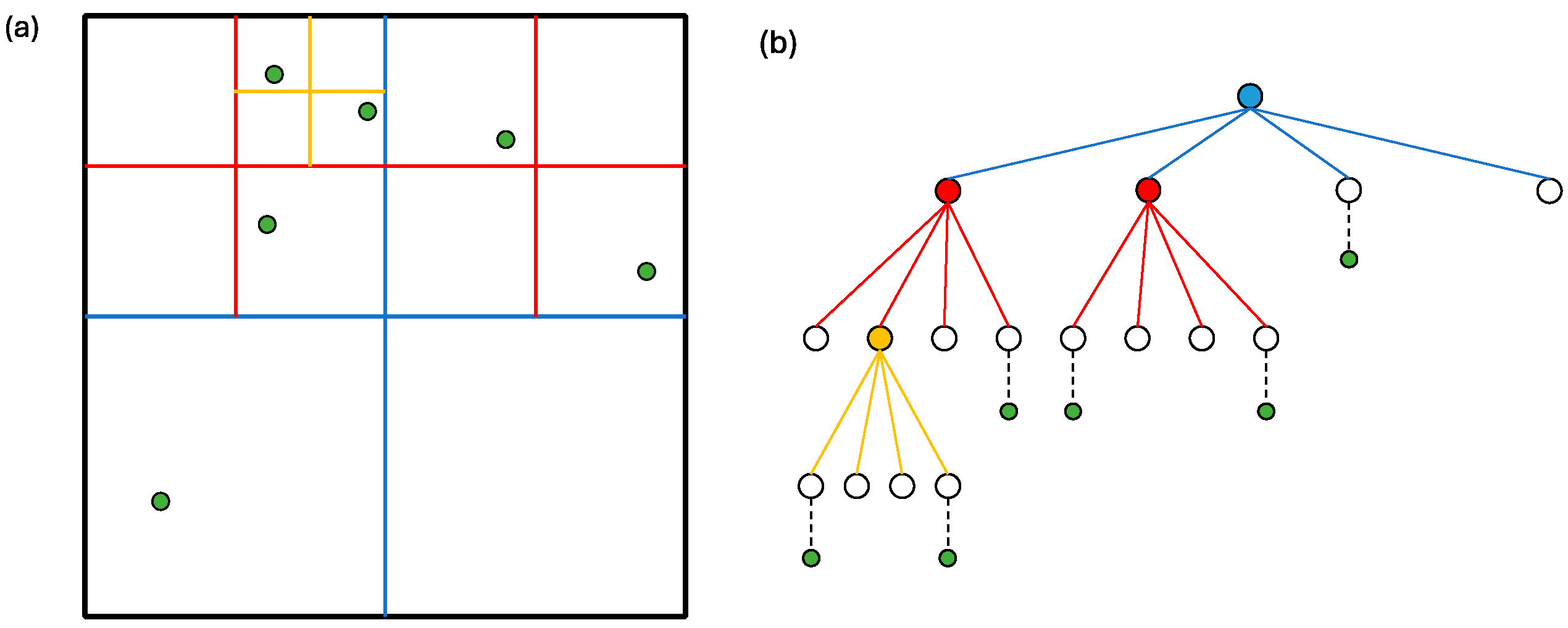
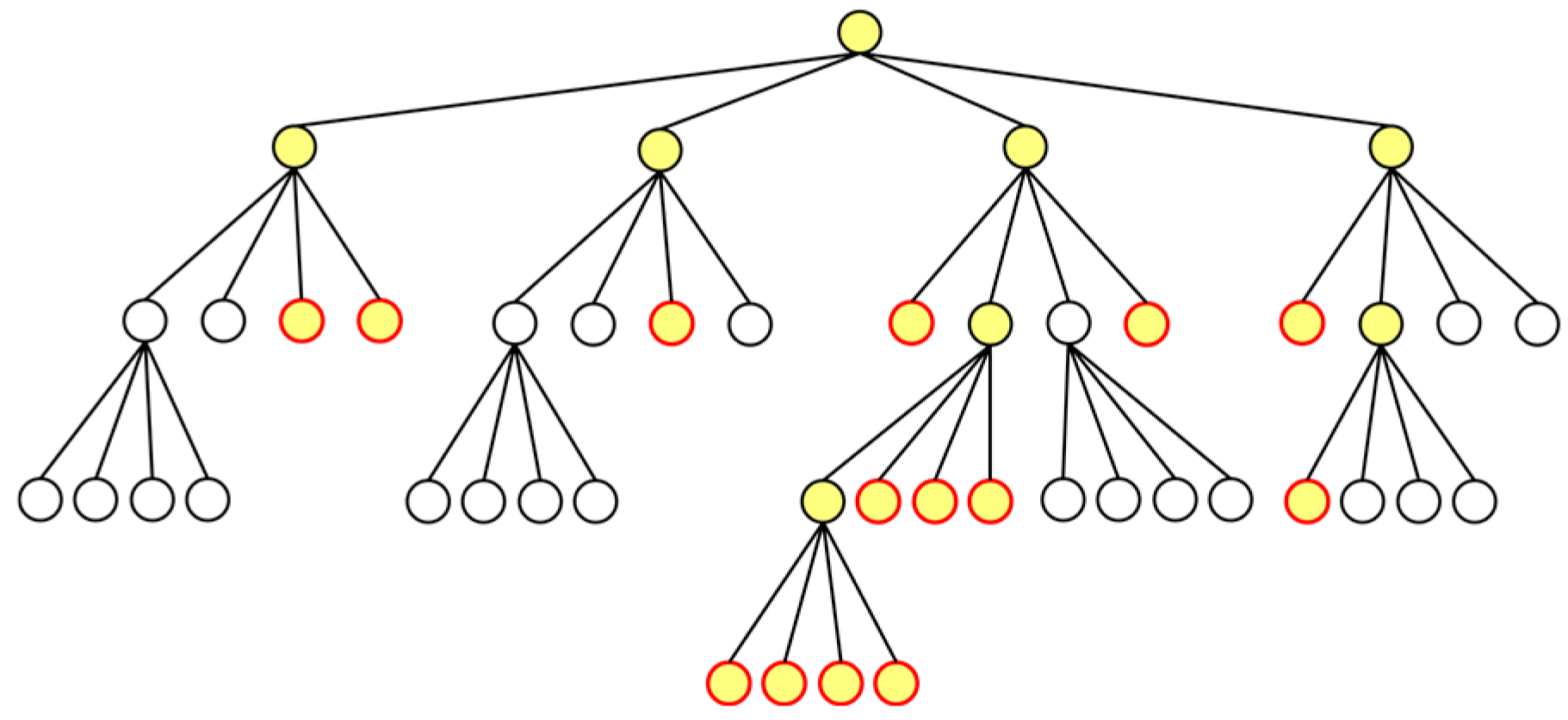
In the Barnes–Hut algorithm, the simulation space is divided into grids based on the distribution of cells and managed as a tree structure. This approach shares similarities with the Cell List algorithm, but the number and size of the grids in the Barnes–Hut algorithm are dynamic and updated at each step. Figure 3a illustrates how the simulation space is divided into grids using the Barnes–Hut algorithm. The grid division process continues until the number of cells in a grid is less than one, as depicted in Figure 3a,b. The figures also present the tree structure referred to as the Barnes–Hut tree, with nodes corresponding to these grids.
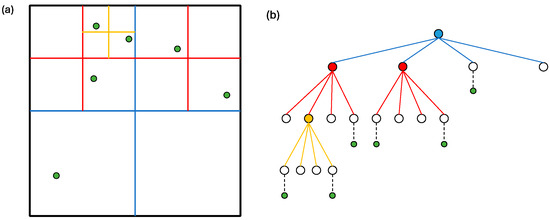
Figure 3.
(a) Example simulation space divided into grids; (b) quad-tree representation where nodes correspond to the grids in (a).
In general, for an -dimensional space, the partitioning is . Since we focus on a two-dimensional space, it is divided into four () partitions, resulting in a quad-tree. Hence, we consider a quad-tree in the rest of this section.
5.2. Computation of Centers of Mass
After constructing the quad-tree, the center of mass for each node is calculated. Each node corresponds to a grid in the simulation space, and the center of mass is determined based on the positions of all cells within that grid. As shown in Figure 3b, leaf nodes either have no cells or contain only one cell. In the former case, nodes without cells are not assigned a center of mass (or the mass of the center of mass was set to 0). In the latter case, nodes containing a single cell had their cell positions as their center of mass.
5.3. Computation of Cell–Cell Interaction Forces
In the Barnes–Hut algorithm, distant groups of cells are calculated using centers of mass, while calculations between cells that are not part of such groups are performed directly between individual cells. A group of cells in this context refers to all the cells contained within a specific grid. Additionally, the center-of-mass points associated with each grid are used to determine distances. In the algorithm, nodes in the quad-tree are traversed in a depth-first manner starting from the root node, and calculations are performed using the center of mass associated with the corresponding distant grid node. Since the cells within the grid are collectively accounted for by the center of mass, there is no need to search for subsequent descendant nodes during this process. For leaf nodes containing cells, the center of mass is also utilized in the calculations. As mentioned earlier, when a node has only one cell, the cell position is treated as a center of mass.
In the algorithm, we use the indicator given below to determine whether a node is sufficiently distant from the cell:
where is a threshold, is the grid size, and is the distance from the cell to the node (i.e., the center of mass). When the given expression evaluates to true, the force is computed by utilizing the center of mass. Algorithm 5 presents the pseudocode for the ‘getCellList()’ function of the Barnes Hut algorithm.
| Algorithm 5 calcForce(cell , node ) in the Barnes Hut algorithm |
|
6. Hybrid Algorithm
We designed a hybrid algorithm combining the Barnes–Hut algorithm and the Cell List algorithm to further accelerate the original Barnes–Hut algorithm. The original Barnes–Hut algorithm computes interactions with all cells distributed in the simulation space. In cases where the impact of interactions decreases sharply with increasing distance between cells, the original Barnes–Hut algorithm becomes inefficient. In the hybrid algorithm, we introduce the search radius used in the Cell List algorithm and exclude cells beyond a certain distance from the computation of cell–cell interactions.
The hybrid algorithm consists of the following four steps.
- Construction of the tree structure
- Calculation of centers of mass
- Coloring of the tree
- Calculation of interactions using the colored tree
The first two steps are the same as those in the original Barnes–Hut algorithm, and the last two steps are different, whereby the hybrid algorithm creates and uses a colored tree. In the following, we describe the last two steps in detail.
6.1. Coloring Tree
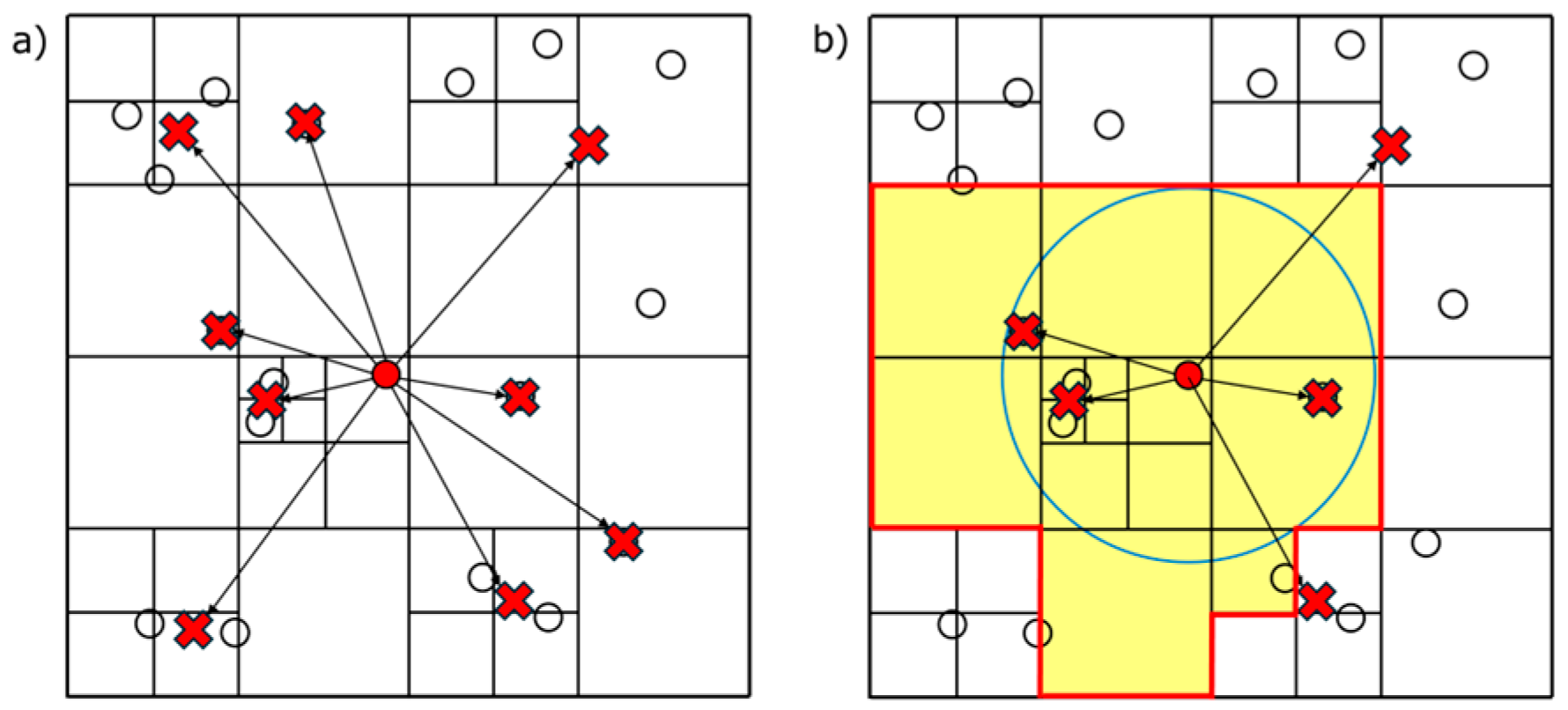
After constructing a tree and calculating the centers of mass, the Hybrid algorithm colors the tree. Figure 4a shows cell–cell interactions considered in the original Barnes–Hut algorithm. As an example, Figure 4b shows those in the hybrid algorithm; cells within the grids and the search radius are considered. Figure 5 shows the colored tree to compute the cell–cell interactions shown in Figure 4b.
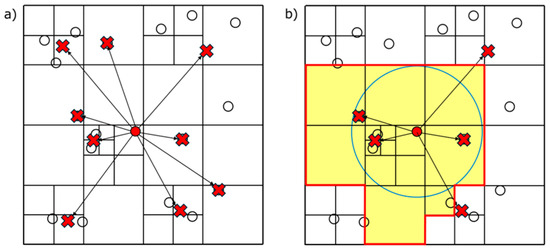
Figure 4.
(a) Barnes–Hut algorithm and (b) hybrid algorithm.
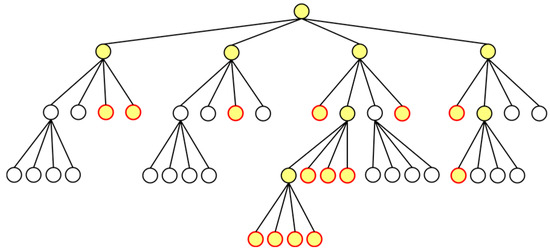
Figure 5.
Colored quad-tree.
To color a tree, we use the functions ‘colorTree()’ and ‘colorNode()’ shown in Algorithms 6 and 7, respectively. By executing ‘colorTree()’ with cell i and the root node as arguments, grids within the search radius centered on cell i are searched. When a grid is found, ‘colorNode()’ is called to color the node. ‘colorNode()’ colors the node passed as an argument, and then recursively calls itself with the parent node as an argument, thereby coloring all nodes containing the leaf nodes.
| Algorithm 6 colorTree(cell , node ) |
|
| Algorithm 7 colorNode(node ) in Algorithm 6 |
|
6.2. Calculating Interactions Using Colored Tree
The basic calculation of interactions follows the Barnes–Hut algorithm in Algorithm 2. In exploring child nodes at line 11 of Algorithm 2, we check whether the node is colored, and continue to explore only if the node is colored. Specifically, we insert a conditional statement right before line 11 of Algorithm 2 to check if the child node to be explored is colored.
7. Conclusions
Computing interactions among cells is computationally expensive in multicellular molecular communication simulations, making the design of efficient algorithms crucial. In this article, we provide an overview of simulation algorithms for computing interactions among cells. We explain the naïve algorithm, the Cell List algorithm, and the Barnes–Hut algorithm. We also designed a hybrid of the Cell List and Barnes–Hut algorithms. Currently, we are developing a multicellular molecular communication simulator [9,10] in which the four algorithms are incorporated, enabling users to select the algorithm best suited to their simulations. In addition, we are implementing parallel computations using multicore computers to further accelerate molecular communication simulations.
Author Contributions
Conceptualization, S.I. and T.N.; methodology, S.I.; writing—original draft preparation, S.I. and T.N. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by JSPS KAKENHI Grant Number JP21H04876.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Nakano, T.; Eckford, A.; Haraguchi, T. Molecular Communication; Cambridge University Press: Cambridge, UK, 2024. [Google Scholar]
- Farsad, N.; Yilmaz, H.B.; Eckford, A.; Chae, C.-B.; Guo, W. A comprehensive survey of recent advancements in molecular communication. IEEE Commun. Surv. Tutor. 2016, 18, 1887–1919. [Google Scholar] [CrossRef]
- Bi, D.; Almpanis, A.; Noel, A.; Deng, Y.; Schober, R. A survey of molecular communication in cell biology: Establishing a new hierarchy for interdisciplinary applications. IEEE Commun. Surv. Tutor. 2021, 23, 1494–1545. [Google Scholar] [CrossRef]
- Akyildiz, I.F.; Kak, A.; Nie, S. 6G and beyond: The future of wireless communications systems. IEEE Access 2020, 8, 133995–134030. [Google Scholar] [CrossRef]
- Guo, W.; Abbaszadeh, M.; Lin, L.; Charmet, J.; Thomas, P.; Wei, Z.; Li, B.; Zhao, C. Molecular physical layer for 6G in wave-denied environments. IEEE Commun. Mag. 2021, 59, 33–39. [Google Scholar] [CrossRef]
- Noel, A.; Cheung, K.C.; Schober, R.; Makrakis, D.; Hafid, A. Simulating with accord: Actor-based communication via reaction-diffusion. Nano Commun. Netw. 2017, 11, 44–75. [Google Scholar] [CrossRef]
- Llatser, I.; Demiray, D.; Cabellos-Aparicio, A.; Altilar, D.T.; Alarcón, E. N3sim: Simulation framework for diffusion-based molecular communication nanonetworks. Simul. Model. Pract. Theory 2014, 42, 210–222. [Google Scholar] [CrossRef]
- Yilmaz, H.B.; Chae, C.-B. Simulation study of molecular communication systems with an absorbing receiver: Modulation and isi mitigation techniques. Simul. Model. Pract. Theory 2014, 49, 136–150. [Google Scholar] [CrossRef]
- Saiki, T.; Nakano, T. Design and implementation of a multicellular molecular communication simulator. In Proceedings of the 2022 Joint 12th International Conference on Soft Computing and Intelligent Systems and 23rd International Symposium on Advanced Intelligent Systems (SCISISIS), Ise, Japan, 29 November–2 December 2022. [Google Scholar]
- Imanaka, S.; Saiki, T.; Nakano, T. Accelerating multicellular molecular communication simulations using the Barnes-Hut algorithm. In Proceedings of the 2024 IEEE 4th International Conference on Electronic Communications, Internet of Things and Big Data (ICIEB 2024), Taipei, Taiwan, 19–21 April 2024. [Google Scholar]
- Barnes, J.; Hut, P. A hierarchical o(n log n) force-calculation algorithm. Nature 1986, 324, 446–449. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).