Spectroscopic Studies of Interactions of Iron Oxide Nanoparticles with Ovalbumin Molecules †
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemical and Raw Materials
2.2. IONPs Synthesis and Suspension of IONP Preparation
2.3. Fluorescence Spectroscopy
2.4. Fourier Transform Infrared Spectroscopy (FTIR) Analysis
2.5. Statistical Analysis
3. Result and Discussion
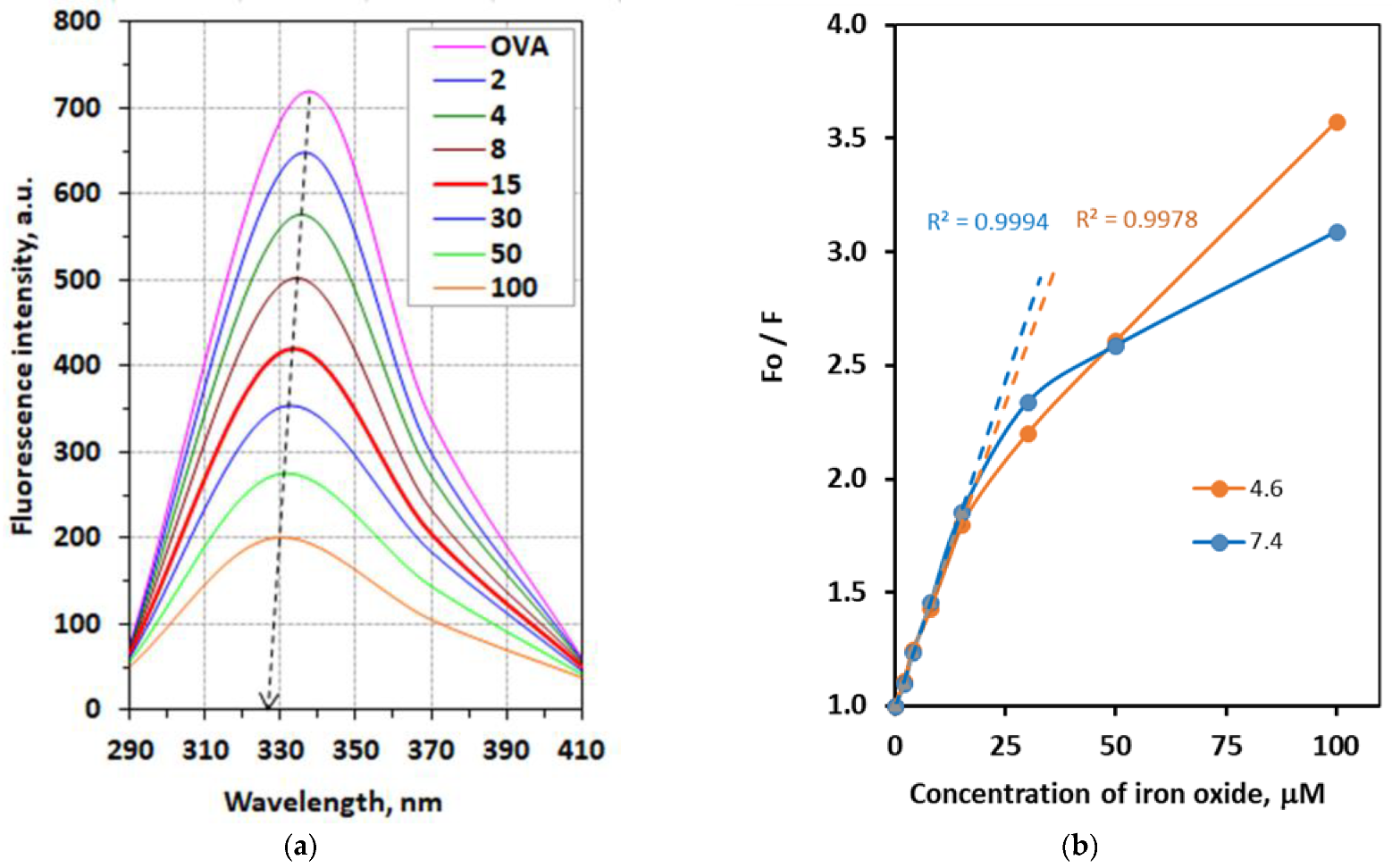
3.1. Fluorescence Spectroscopy
3.1.1. Fluorescence Quenching Mechanism
3.1.2. Stoichiometry, the Binding Constant, and Thermodynamic Parameters
3.2. Analysis of Protein Conformation
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dickinson, E. Stabilising Emulsion-Based Colloidal Structures with Mixed Food Ingredients. J. Sci. Food Agric. 2013, 93, 710–721. [Google Scholar] [CrossRef] [PubMed]
- Goralchuk, A.; Gubsky, S.; Omel’chenko, S.; Riabets, O.; Grinchenko, O.; Fedak, N.; Kotlyar, O.; Cheremska, T.; Skrynnik, V. Impact of Added Food Ingredients on Foaming and Texture of the Whipped Toppings: A Chemometric Analysis. Eur. Food Res. Technol. 2020, 246, 1955–1970. [Google Scholar] [CrossRef]
- Harper, W.J.; Hewitt, S.A.; Huffman, L.M. Model Food Systems and Protein Functionality. In Milk Proteins: From Expression to Food; Academic Press: Cambridge, MA, USA, 2019; pp. 573–598. [Google Scholar] [CrossRef]
- Sanguansri, P.; Augustin, M.A. Nanoscale Materials Development—A Food Industry Perspective. Trends Food Sci. Technol. 2006, 17, 547–556. [Google Scholar] [CrossRef]
- Tsykhanovska, I.; Evlash, V.; Alexandrov, A.; Lazarieva, T.; Svidlo, K.; Gontar, T.; Yurchenko, L.; Pavlotska, L. Substantiation of the Mechanism of Interaction between Biopolymers of Rye and wheat Flour and the Nanoparticles of the Magnetofood Food Additive in Order to Improve Moisture retaining Capacity of Dough. East.-Eur. J. Enterp. Technol. 2018, 2, 70–80. [Google Scholar] [CrossRef]
- Tsykhanovska, I.; Evlash, V.; Oleksandrov, O.; Gontar, T. Mechanism of Fat-Binding and Fat-Contenting of the Nanoparticles of a Food Supplement on the Basis of Double Oxide of Two- and Trivalent Iron. Ukr. Food J. 2018, 7, 702–715. [Google Scholar] [CrossRef]
- Tsykhanovska, I.; Evlash, V.; Blahyi, O. Mechanism of Water-Binding and Water-Retention of Food Additives Nanoparticles Based on Double Oxide of Two- and Trivalent Iron. Ukr. Food J. 2020, 9, 298–321. [Google Scholar] [CrossRef]
- Tsykhanovska, I.; Evlash, V.; Alexandrov, A.; Gontar, T. Dissolution Kinetics of Fe3O4 Nanoparticles in the Acid Media. Chem. Chem. Technol. 2019, 13, 170–184. [Google Scholar] [CrossRef]
- de Dantas, M.D.A.; de Tenório, H.A.; Lopes, T.I.B.; Pereira, H.J.V.; Marsaioli, A.J.; Figueiredo, I.M.; Santos, J.C.C. Interactions of Tetracyclines with Ovalbumin, the Main Allergen Protein from Egg White: Spectroscopic and Electrophoretic Studies. Int. J. Biol. Macromol. 2017, 102, 505–514. [Google Scholar] [CrossRef] [PubMed]
- Byler, D.M.; Susi, H. Examination of the Secondary Structure of Proteins by Deconvolved FTIR Spectra. Biopolymers 1986, 25, 469–487. [Google Scholar] [CrossRef] [PubMed]
- Stein, P.E.; Leslie, A.G.W.; Finch, J.T.; Carrell, R.W. Crystal Structure of Uncleaved Ovalbumin at 1·95 Å Resolution. J. Mol. Biol. 1991, 221, 941–959. [Google Scholar] [CrossRef]
- Midoux, P.; Wahl, P.; Auchet, J.-C.; Monsigny, M. Fluorescence Quenching of Tryptophan by Trifluoroacetamide. Biochim. Biophys. Acta-Gen. Subj. 1984, 801, 16–25. [Google Scholar] [CrossRef]
- Tang, L.; Li, S.; Bi, H.; Gao, X. Interaction of Cyanidin-3-O-Glucoside with Three Proteins. Food Chem. 2016, 196, 550–559. [Google Scholar] [CrossRef] [PubMed]
- Lakowicz, J.R. Principles of Fluorescence Spectroscopy; Springer: Berlin/Heidelberg, Germany, 2006. [Google Scholar] [CrossRef]
- Zhang, G.; Ma, Y.; Wang, L.; Zhang, Y.; Zhou, J. Multispectroscopic Studies on the Interaction of Maltol, a Food Additive, with Bovine Serum Albumin. Food Chem. 2012, 133, 264–270. [Google Scholar] [CrossRef] [PubMed]
- Shu, Y.; Xue, W.; Xu, X.; Jia, Z.; Yao, X.; Liu, S.; Liu, L. Interaction of Erucic Acid with Bovine Serum Albumin Using a Multi-Spectroscopic Method and Molecular Docking Technique. Food Chem. 2015, 173, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, M.; Mukhopadhyay, S. Structural and Dynamical Insights into the Molten-Globule Form of Ovalbumin. J. Phys. Chem. B 2012, 116, 520–531. [Google Scholar] [CrossRef] [PubMed]
- Ross, P.D.; Subramanian, S. Thermodynamics of Protein Association Reactions: Forces Contributing to Stability. Biochemistry 1981, 20, 3096–3102. [Google Scholar] [CrossRef] [PubMed]
- Zolghadri, S.; Saboury, A.A.; Amin, E.; Moosavi-Movahedi, A.A. A Spectroscopic Study on the Interaction between Ferric Oxide Nanoparticles and Human Hemoglobin. J. Iran. Chem. Soc. 2010, 7, S145–S153. [Google Scholar] [CrossRef]
- Xue, J.J.; Chen, Q.Y. The Interaction between Ionic Liquids Modified Magnetic Nanoparticles and Bovine Serum Albumin and the Cytotoxicity to HepG-2 Cells. Spectrochim. Acta-Part A Mol. Biomol. Spectrosc. 2014, 120, 161–166. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Wu, P.; Zhu, Z.; Wang, Y. Interaction of γ-Fe2O3 Nanoparticles with Fibrinogen. Spectrochim. Acta-Part A Mol. Biomol. Spectrosc. 2015, 151, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Ngarize, S.; Herman, H.; Adams, A.; Howell, N. Comparison of Changes in the Secondary Structure of Unheated, Heated, and High-Pressure-Treated β-Lactoglobulin and Ovalbumin Proteins Using Fourier Transform Raman Spectroscopy and Self-Deconvolution. J. Agric. Food Chem. 2004, 52, 6470–6477. [Google Scholar] [CrossRef] [PubMed]
| pH | Temperature, K | Stern–Volmer Constant | ||
|---|---|---|---|---|
| KSV·104, L·mol−1 | R2 | Kq·1012 L·mol−1·s−1 | ||
| 4.6 | 296 | 5.30 ± 0.27 | 0.9900 | 5.29 |
| 311 | 5.16 ± 0.27 | 0.9887 | 5.15 | |
| 7.4 | 296 | 5.72 ± 0.08 | 0.9994 | 5.72 |
| 311 | 5.63 ± 0.13 | 0.9984 | 5.63 | |
| pH | Temperature, K | Binding Parameters | Thermodynamic Properties | Preferential Interaction | ||||
|---|---|---|---|---|---|---|---|---|
| Kb·104 L·mol−1 | n | R2 | Δ G, kJ·mol−1 | ΔH, kJ·mol−1 | ΔS, J·mol−1 | |||
| 4.6 | 296 | 4.1 ± 0.2 | 0.76 ± 0.04 | 0.9892 | −26.2 ± 0.1 | −6.8 | 65.0 | Electrostatic forces |
| 311 | 3.6 ± 0.4 | 0.76 ± 0.04 | 0.9875 | −27.1 ± 0.1 | ||||
| 7.4 | 296 | 4.0 ± 0.2 | 0.74 ± 0.07 | 0.9840 | −26.2 ± 0.3 | −2.2 | 81.0 | |
| 311 | 3.8 ± 0.4 | 0.78 ± 0.09 | 0.9858 | −27.3 ± 0.3 | ||||
| Amide I Components | Frequency Range, cm−1 | Free OVA, % | OVA-IONPs, % |
|---|---|---|---|
| β-sheet | 1610–1640 | 32.95 | 25.21 |
| random coil | 1641–1649 | 7.02 | 21.14 |
| α-helix | 1650–1660 | 43.78 | 18.38 |
| β-turn | 1660–1680 | 16.28 | 34.15 |
| β-anti | 1680–1692 | - | 1.12 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsykhanovska, I.; Stabnikova, O.; Gubsky, S. Spectroscopic Studies of Interactions of Iron Oxide Nanoparticles with Ovalbumin Molecules. Mater. Proc. 2022, 9, 2. https://doi.org/10.3390/materproc2022009002
Tsykhanovska I, Stabnikova O, Gubsky S. Spectroscopic Studies of Interactions of Iron Oxide Nanoparticles with Ovalbumin Molecules. Materials Proceedings. 2022; 9(1):2. https://doi.org/10.3390/materproc2022009002
Chicago/Turabian StyleTsykhanovska, Irina, Olena Stabnikova, and Sergey Gubsky. 2022. "Spectroscopic Studies of Interactions of Iron Oxide Nanoparticles with Ovalbumin Molecules" Materials Proceedings 9, no. 1: 2. https://doi.org/10.3390/materproc2022009002
APA StyleTsykhanovska, I., Stabnikova, O., & Gubsky, S. (2022). Spectroscopic Studies of Interactions of Iron Oxide Nanoparticles with Ovalbumin Molecules. Materials Proceedings, 9(1), 2. https://doi.org/10.3390/materproc2022009002







