Role of microRNAs in the Pathophysiology of Ulcerative Colitis
Abstract
1. Introduction
2. Overview of miRNA
3. miRNA Profile in UC
4. miRNAs in the Pathophysiology of UC
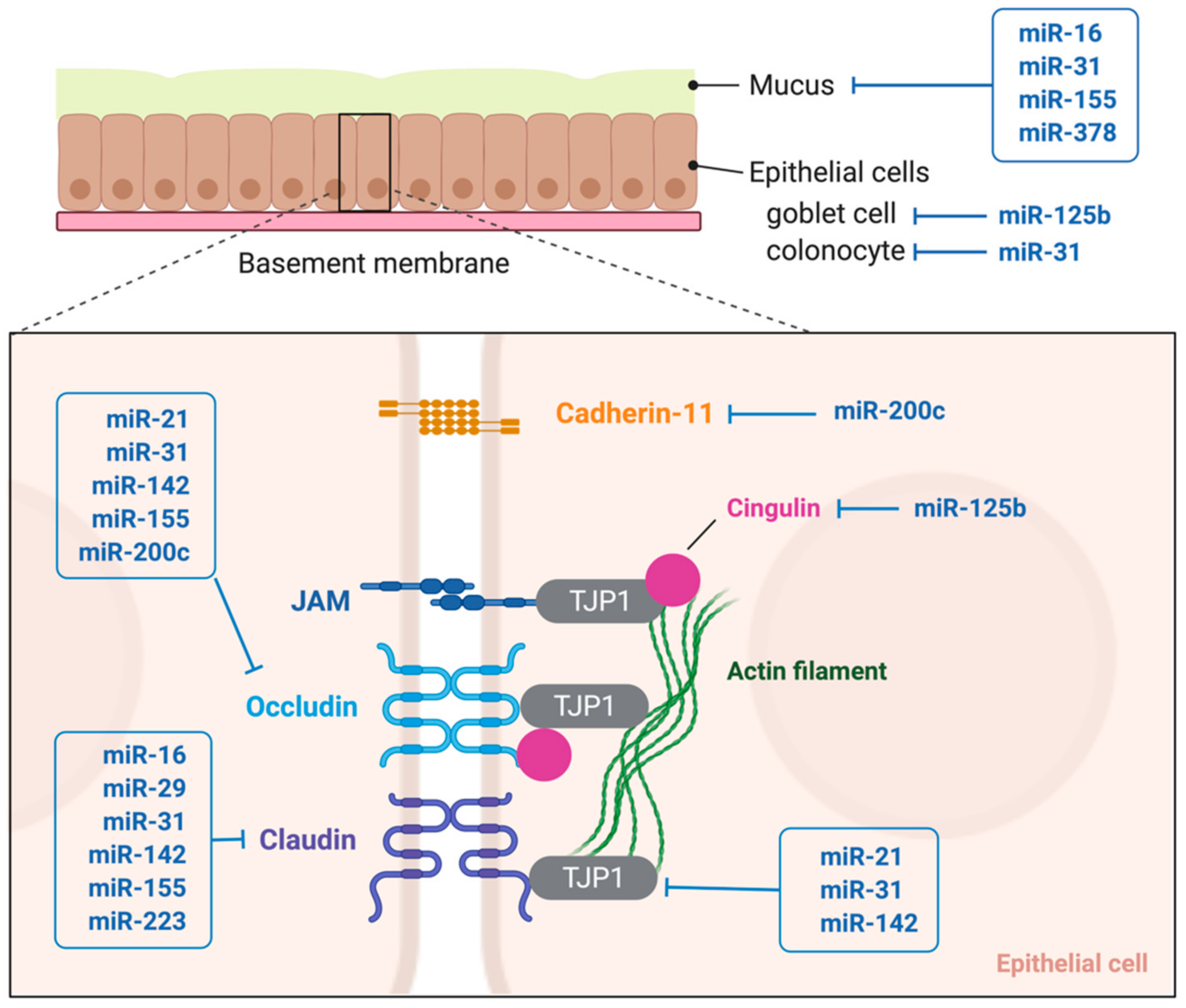
4.1. Mucus Barrier
4.2. Epithelial Permeability
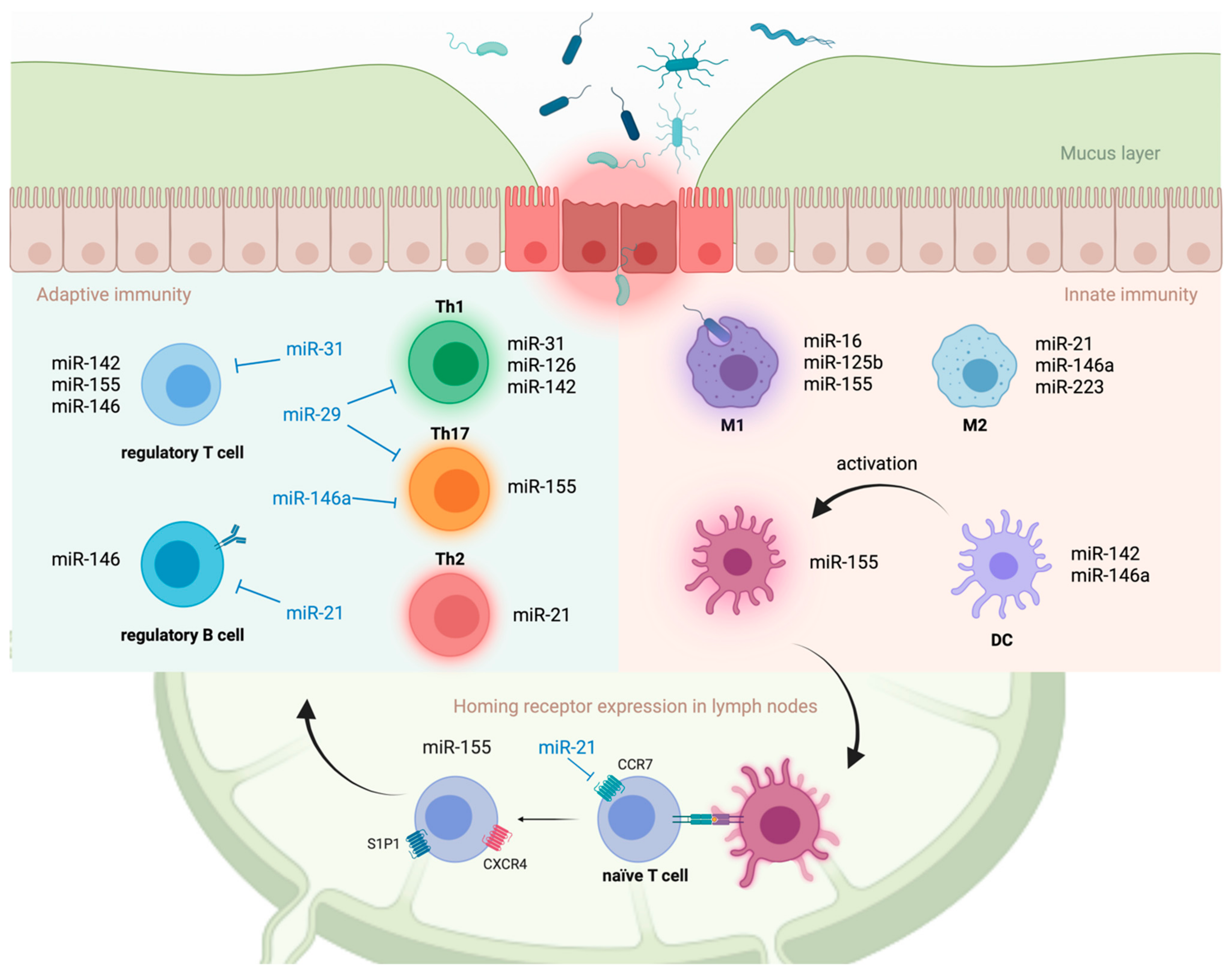
4.3. Innate Immune Cells
4.4. Adaptive Immune Cells
4.5. Intestinal T Cell Homing
4.6. Overall Effect of Individual miRNAs in the Development of Colitis
5. Potential of miRNAs as Therapeutic Targets in UC
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kobayashi, T.; Siegmund, B.; Le Berre, C.; Wei, S.C.; Ferrante, M.; Shen, B.; Bernstein, C.N.; Danese, S.; Peyrin-Biroulet, L.; Hibi, T. Ulcerative colitis. Nat. Rev. Dis. Primers 2020, 6, 74. [Google Scholar] [CrossRef]
- Sartor, R.B. Mechanisms of disease: Pathogenesis of Crohn’s disease and ulcerative colitis. Nat. Clin. Pract. Gastroenterol. Hepatol. 2006, 3, 390–407. [Google Scholar] [CrossRef]
- Khor, B.; Gardet, A.; Xavier, R.J. Genetics and pathogenesis of inflammatory bowel disease. Nature 2011, 474, 307–317. [Google Scholar] [CrossRef] [PubMed]
- Friedman, R.C.; Farh, K.K.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef]
- Bartel, D.P. Metazoan MicroRNAs. Cell 2018, 173, 20–51. [Google Scholar] [CrossRef]
- Esteller, M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011, 12, 861–874. [Google Scholar] [CrossRef]
- Kalla, R.; Ventham, N.T.; Kennedy, N.A.; Quintana, J.F.; Nimmo, E.R.; Buck, A.H.; Satsangi, J. MicroRNAs: New players in IBD. Gut 2015, 64, 504–517. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Liu, J.; Gao, Y.; Shen, L.; Li, S.; Chen, S. miRNA-Based Potential Biomarkers and New Molecular Insights in Ulcerative Colitis. Front. Pharmacol. 2021, 12, 707776. [Google Scholar] [CrossRef]
- Schaefer, J.S.; Attumi, T.; Opekun, A.R.; Abraham, B.; Hou, J.; Shelby, H.; Graham, D.Y.; Streckfus, C.; Klein, J.R. MicroRNA signatures differentiate Crohn’s disease from ulcerative colitis. BMC Immunol. 2015, 16, 5. [Google Scholar] [CrossRef]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Alles, J.; Fehlmann, T.; Fischer, U.; Backes, C.; Galata, V.; Minet, M.; Hart, M.; Abu-Halima, M.; Grasser, F.A.; Lenhof, H.P.; et al. An estimate of the total number of true human miRNAs. Nucleic Acids Res. 2019, 47, 3353–3364. [Google Scholar] [CrossRef]
- Gebert, L.F.R.; MacRae, I.J. Regulation of microRNA function in animals. Nat. Rev. Mol. Cell Biol. 2019, 20, 21–37. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Zikusoka, M.; Trindade, A.; Dassopoulos, T.; Harris, M.L.; Bayless, T.M.; Brant, S.R.; Chakravarti, S.; Kwon, J.H. MicroRNAs are differentially expressed in ulcerative colitis and alter expression of macrophage inflammatory peptide-2 alpha. Gastroenterology 2008, 135, 1624–1635.e24. [Google Scholar] [CrossRef] [PubMed]
- Ghafouri-Fard, S.; Eghtedarian, R.; Taheri, M. The crucial role of non-coding RNAs in the pathophysiology of inflammatory bowel disease. Biomed. Pharmacother. 2020, 129, 110507. [Google Scholar] [CrossRef]
- Iborra, M.; Bernuzzi, F.; Correale, C.; Vetrano, S.; Fiorino, G.; Beltran, B.; Marabita, F.; Locati, M.; Spinelli, A.; Nos, P.; et al. Identification of serum and tissue micro-RNA expression profiles in different stages of inflammatory bowel disease. Clin. Exp. Immunol. 2013, 173, 250–258. [Google Scholar] [CrossRef]
- Min, M.; Peng, L.; Yang, Y.; Guo, M.; Wang, W.; Sun, G. MicroRNA-155 is involved in the pathogenesis of ulcerative colitis by targeting FOXO3a. Inflamm. Bowel Dis. 2014, 20, 652–659. [Google Scholar] [CrossRef]
- Tian, T.; Zhou, Y.; Feng, X.; Ye, S.; Wang, H.; Wu, W.; Tan, W.; Yu, C.; Hu, J.; Zheng, R.; et al. MicroRNA-16 is putatively involved in the NF-kappaB pathway regulation in ulcerative colitis through adenosine A2a receptor (A2aAR) mRNA targeting. Sci. Rep. 2016, 6, 30824. [Google Scholar] [CrossRef]
- Coskun, M.; Bjerrum, J.T.; Seidelin, J.B.; Troelsen, J.T.; Olsen, J.; Nielsen, O.H. miR-20b, miR-98, miR-125b-1*, and let-7e* as new potential diagnostic biomarkers in ulcerative colitis. World J. Gastroenterol. 2013, 19, 4289–4299. [Google Scholar] [CrossRef]
- Takagi, T.; Naito, Y.; Mizushima, K.; Hirata, I.; Yagi, N.; Tomatsuri, N.; Ando, T.; Oyamada, Y.; Isozaki, Y.; Hongo, H.; et al. Increased expression of microRNA in the inflamed colonic mucosa of patients with active ulcerative colitis. J. Gastroenterol. Hepatol. 2010, 25 (Suppl. 1), S129–S133. [Google Scholar] [CrossRef]
- Zahm, A.M.; Hand, N.J.; Tsoucas, D.M.; Le Guen, C.L.; Baldassano, R.N.; Friedman, J.R. Rectal microRNAs are perturbed in pediatric inflammatory bowel disease of the colon. J. Crohns Colitis 2014, 8, 1108–1117. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Wang, H.; Ye, S.; Guan, J.; Tan, W.; Cheng, S.; Wei, G.; Wu, W.; Wu, F.; Zhou, Y. Up-regulation of microRNA-126 may contribute to pathogenesis of ulcerative colitis via regulating NF-kappaB inhibitor IkappaBalpha. PLoS ONE 2012, 7, e52782. [Google Scholar] [CrossRef]
- Thorlacius-Ussing, G.; Schnack Nielsen, B.; Andersen, V.; Holmstrom, K.; Pedersen, A.E. Expression and Localization of miR-21 and miR-126 in Mucosal Tissue from Patients with Inflammatory Bowel Disease. Inflamm. Bowel Dis. 2017, 23, 739–752. [Google Scholar] [CrossRef] [PubMed]
- Koukos, G.; Polytarchou, C.; Kaplan, J.L.; Oikonomopoulos, A.; Ziring, D.; Hommes, D.W.; Wahed, R.; Kokkotou, E.; Pothoulakis, C.; Winter, H.S.; et al. A microRNA signature in pediatric ulcerative colitis: Deregulation of the miR-4284/CXCL5 pathway in the intestinal epithelium. Inflamm. Bowel Dis. 2015, 21, 996–1005. [Google Scholar] [CrossRef]
- Yang, Y.; Ma, Y.; Shi, C.; Chen, H.; Zhang, H.; Chen, N.; Zhang, P.; Wang, F.; Yang, J.; Yang, J.; et al. Overexpression of miR-21 in patients with ulcerative colitis impairs intestinal epithelial barrier function through targeting the Rho GTPase RhoB. Biochem. Biophys. Res. Commun. 2013, 434, 746–752. [Google Scholar] [CrossRef]
- Polytarchou, C.; Hommes, D.W.; Palumbo, T.; Hatziapostolou, M.; Koutsioumpa, M.; Koukos, G.; van der Meulen-de Jong, A.E.; Oikonomopoulos, A.; van Deen, W.K.; Vorvis, C.; et al. MicroRNA214 Is Associated with Progression of Ulcerative Colitis, and Inhibition Reduces Development of Colitis and Colitis-Associated Cancer in Mice. Gastroenterology 2015, 149, 981–992.e11. [Google Scholar] [CrossRef] [PubMed]
- Fasseu, M.; Treton, X.; Guichard, C.; Pedruzzi, E.; Cazals-Hatem, D.; Richard, C.; Aparicio, T.; Daniel, F.; Soule, J.C.; Moreau, R.; et al. Identification of restricted subsets of mature microRNA abnormally expressed in inactive colonic mucosa of patients with inflammatory bowel disease. PLoS ONE 2010, 5, e13160. [Google Scholar] [CrossRef]
- Van der Goten, J.; Vanhove, W.; Lemaire, K.; Van Lommel, L.; Machiels, K.; Wollants, W.J.; De Preter, V.; De Hertogh, G.; Ferrante, M.; Van Assche, G.; et al. Integrated miRNA and mRNA expression profiling in inflamed colon of patients with ulcerative colitis. PLoS ONE 2014, 9, e116117. [Google Scholar] [CrossRef] [PubMed]
- Fang, K.; Law, I.K.M.; Padua, D.; Sideri, A.; Huang, V.; Kevil, C.G.; Iliopoulos, D.; Pothoulakis, C. MicroRNA-31-3p Is Involved in Substance P (SP)-Associated Inflammation in Human Colonic Epithelial Cells and Experimental Colitis. Am. J. Pathol. 2018, 188, 586–599. [Google Scholar] [CrossRef]
- Lin, J.; Welker, N.C.; Zhao, Z.; Li, Y.; Zhang, J.; Reuss, S.A.; Zhang, X.; Lee, H.; Liu, Y.; Bronner, M.P. Novel specific microRNA biomarkers in idiopathic inflammatory bowel disease unrelated to disease activity. Mod. Pathol. 2014, 27, 602–608. [Google Scholar] [CrossRef]
- Valmiki, S.; Ahuja, V.; Paul, J. MicroRNA exhibit altered expression in the inflamed colonic mucosa of ulcerative colitis patients. World J. Gastroenterol. 2017, 23, 5324–5332. [Google Scholar] [CrossRef]
- Valmiki, S.; Ahuja, V.; Puri, N.; Paul, J. miR-125b and miR-223 Contribute to Inflammation by Targeting the Key Molecules of NFkappaB Pathway. Front. Med. 2019, 6, 313. [Google Scholar] [CrossRef]
- Han, J.; Li, Y.; Zhang, H.; Guo, J.; Wang, X.; Kang, Y.; Luo, Y.; Wu, M.; Zhang, X. MicroRNA-142-5p facilitates the pathogenesis of ulcerative colitis by regulating SOCS1. Int. J. Clin. Exp. Pathol. 2018, 11, 5735–5744. [Google Scholar]
- Beres, N.J.; Szabo, D.; Kocsis, D.; Szucs, D.; Kiss, Z.; Muller, K.E.; Lendvai, G.; Kiss, A.; Arato, A.; Sziksz, E.; et al. Role of Altered Expression of miR-146a, miR-155, and miR-122 in Pediatric Patients with Inflammatory Bowel Disease. Inflamm. Bowel Dis. 2016, 22, 327–335. [Google Scholar] [CrossRef]
- McKenna, L.B.; Schug, J.; Vourekas, A.; McKenna, J.B.; Bramswig, N.C.; Friedman, J.R.; Kaestner, K.H. MicroRNAs control intestinal epithelial differentiation, architecture, and barrier function. Gastroenterology 2010, 139, 1654–1664.e1. [Google Scholar] [CrossRef] [PubMed]
- Gantier, M.P.; Stunden, H.J.; McCoy, C.E.; Behlke, M.A.; Wang, D.; Kaparakis-Liaskos, M.; Sarvestani, S.T.; Yang, Y.H.; Xu, D.; Corr, S.C.; et al. A miR-19 regulon that controls NF-kappaB signaling. Nucleic Acids Res. 2012, 40, 8048–8058. [Google Scholar] [CrossRef] [PubMed]
- Muljo, S.A.; Ansel, K.M.; Kanellopoulou, C.; Livingston, D.M.; Rao, A.; Rajewsky, K. Aberrant T cell differentiation in the absence of Dicer. J. Exp. Med. 2005, 202, 261–269. [Google Scholar] [CrossRef] [PubMed]
- Chong, M.M.; Rasmussen, J.P.; Rudensky, A.Y.; Littman, D.R. The RNAseIII enzyme Drosha is critical in T cells for preventing lethal inflammatory disease. J. Exp. Med. 2008, 205, 2005–2017. [Google Scholar] [CrossRef]
- Nystrom, E.E.L.; Martinez-Abad, B.; Arike, L.; Birchenough, G.M.H.; Nonnecke, E.B.; Castillo, P.A.; Svensson, F.; Bevins, C.L.; Hansson, G.C.; Johansson, M.E.V. An intercrypt subpopulation of goblet cells is essential for colonic mucus barrier function. Science 2021, 372, 6539. [Google Scholar] [CrossRef]
- Van der Post, S.; Jabbar, K.S.; Birchenough, G.; Arike, L.; Akhtar, N.; Sjovall, H.; Johansson, M.E.V.; Hansson, G.C. Structural weakening of the colonic mucus barrier is an early event in ulcerative colitis pathogenesis. Gut 2019, 68, 2142–2151. [Google Scholar] [CrossRef]
- Ozaki, R.; Kobayashi, T.; Okabayashi, S.; Nakano, M.; Morinaga, S.; Hara, A.; Ohbu, M.; Matsuoka, K.; Toyonaga, T.; Saito, E.; et al. Histological Risk Factors to Predict Clinical Relapse in Ulcerative Colitis with Endoscopically Normal Mucosa. J. Crohns Colitis 2018, 12, 1288–1294. [Google Scholar] [CrossRef]
- Van der Sluis, M.; De Koning, B.A.; De Bruijn, A.C.; Velcich, A.; Meijerink, J.P.; Van Goudoever, J.B.; Buller, H.A.; Dekker, J.; Van Seuningen, I.; Renes, I.B.; et al. Muc2-deficient mice spontaneously develop colitis, indicating that MUC2 is critical for colonic protection. Gastroenterology 2006, 131, 117–129. [Google Scholar] [CrossRef]
- Olli, K.E.; Rapp, C.; O’Connell, L.; Collins, C.B.; McNamee, E.N.; Jensen, O.; Jedlicka, P.; Allison, K.C.; Goldberg, M.S.; Gerich, M.E.; et al. Muc5ac Expression Protects the Colonic Barrier in Experimental Colitis. Inflamm. Bowel Dis. 2020, 26, 1353–1367. [Google Scholar] [CrossRef] [PubMed]
- Parikh, K.; Antanaviciute, A.; Fawkner-Corbett, D.; Jagielowicz, M.; Aulicino, A.; Lagerholm, C.; Davis, S.; Kinchen, J.; Chen, H.H.; Alham, N.K.; et al. Colonic epithelial cell diversity in health and inflammatory bowel disease. Nature 2019, 567, 49–55. [Google Scholar] [CrossRef]
- Liu, Z.; Chen, X.; Wu, Q.; Song, J.; Wang, L.; Li, G. miR-125b inhibits goblet cell differentiation in allergic airway inflammation by targeting SPDEF. Eur. J. Pharmacol. 2016, 782, 14–20. [Google Scholar] [CrossRef]
- Gao, Y.; Yu, Z. MicroRNA16 inhibits interleukin13induced inflammatory cytokine secretion and mucus production in nasal epithelial cells by suppressing the IkappaB kinase beta/nuclear factorkappaB pathway. Mol. Med. Rep. 2018, 18, 4042–4050. [Google Scholar] [CrossRef] [PubMed]
- Gwiggner, M.; Martinez-Nunez, R.T.; Whiteoak, S.R.; Bondanese, V.P.; Claridge, A.; Collins, J.E.; Cummings, J.R.F.; Sanchez-Elsner, T. MicroRNA-31 and MicroRNA-155 Are Overexpressed in Ulcerative Colitis and Regulate IL-13 Signaling by Targeting Interleukin 13 Receptor alpha-1. Genes 2018, 9, 85. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Liu, P.; Lv, H.; Gao, Z.; Chang, W.; Xu, Y. miR-31 attenuates murine allergic rhinitis by suppressing interleukin-13-induced nasal epithelial inflammatory responses. Mol. Med. Rep. 2021, 23. [Google Scholar] [CrossRef]
- Liu, C.; Du, X.; Zhou, X.; Kolosov, V.P.; Perelman, J.M. MicroRNA-155 regulates lipopolysaccharide-induced mucin 5AC overproduction via a suppressor of cytokine signaling 1-mediated mechanism in human bronchial epithelial cells. Respir Physiol. Neurobiol. 2019, 264, 12–18. [Google Scholar] [CrossRef]
- Martini, E.; Krug, S.M.; Siegmund, B.; Neurath, M.F.; Becker, C. Mend Your Fences: The Epithelial Barrier and its Relationship with Mucosal Immunity in Inflammatory Bowel Disease. Cell. Mol. Gastroenterol. Hepatol. 2017, 4, 33–46. [Google Scholar] [CrossRef]
- Vivinus-Nebot, M.; Frin-Mathy, G.; Bzioueche, H.; Dainese, R.; Bernard, G.; Anty, R.; Filippi, J.; Saint-Paul, M.C.; Tulic, M.K.; Verhasselt, V.; et al. Functional bowel symptoms in quiescent inflammatory bowel diseases: Role of epithelial barrier disruption and low-grade inflammation. Gut 2014, 63, 744–752. [Google Scholar] [CrossRef]
- Chang, J.; Leong, R.W.; Wasinger, V.C.; Ip, M.; Yang, M.; Phan, T.G. Impaired Intestinal Permeability Contributes to Ongoing Bowel Symptoms in Patients with Inflammatory Bowel Disease and Mucosal Healing. Gastroenterology 2017, 153, 723–731.e1. [Google Scholar] [CrossRef] [PubMed]
- Khounlotham, M.; Kim, W.; Peatman, E.; Nava, P.; Medina-Contreras, O.; Addis, C.; Koch, S.; Fournier, B.; Nusrat, A.; Denning, T.L.; et al. Compromised intestinal epithelial barrier induces adaptive immune compensation that protects from colitis. Immunity 2012, 37, 563–573. [Google Scholar] [CrossRef]
- Su, L.; Shen, L.; Clayburgh, D.R.; Nalle, S.C.; Sullivan, E.A.; Meddings, J.B.; Abraham, C.; Turner, J.R. Targeted epithelial tight junction dysfunction causes immune activation and contributes to development of experimental colitis. Gastroenterology 2009, 136, 551–563. [Google Scholar] [CrossRef]
- Peck, B.C.; Weiser, M.; Lee, S.E.; Gipson, G.R.; Iyer, V.B.; Sartor, R.B.; Herfarth, H.H.; Long, M.D.; Hansen, J.J.; Isaacs, K.L.; et al. MicroRNAs Classify Different Disease Behavior Phenotypes of Crohn’s Disease and May Have Prognostic Utility. Inflamm. Bowel Dis. 2015, 21, 2178–2187. [Google Scholar] [CrossRef]
- Pekow, J.R.; Kwon, J.H. MicroRNAs in inflammatory bowel disease. Inflamm. Bowel Dis. 2012, 18, 187–193. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Chao, K.; Ng, S.C.; Bai, A.H.; Yu, Q.; Yu, J.; Li, M.; Cui, Y.; Chen, M.; Hu, J.F.; et al. Pro-inflammatory miR-223 mediates the cross-talk between the IL23 pathway and the intestinal barrier in inflammatory bowel disease. Genome Biol. 2016, 17, 58. [Google Scholar] [CrossRef] [PubMed]
- Tili, E.; Michaille, J.J.; Piurowski, V.; Rigot, B.; Croce, C.M. MicroRNAs in intestinal barrier function, inflammatory bowel disease and related cancers-their effects and therapeutic potentials. Curr. Opin. Pharmacol. 2017, 37, 142–150. [Google Scholar] [CrossRef]
- Toyonaga, T.; Steinbach, E.C.; Keith, B.P.; Barrow, J.B.; Schaner, M.R.; Wolber, E.A.; Beasley, C.; Huling, J.; Wang, Y.; Allbritton, N.L.; et al. Decreased Colonic Activin Receptor-Like Kinase 1 Disrupts Epithelial Barrier Integrity in Patients With Crohn’s Disease. Cell. Mol. Gastroenterol. Hepatol. 2020, 10, 779–796. [Google Scholar] [CrossRef]
- Zhu, J.; Zhang, Y.; Zhang, W.; Zhang, W.; Fan, L.; Wang, L.; Liu, Y.; Liu, S.; Guo, Y.; Wang, Y.; et al. MicroRNA-142-5p contributes to Hashimoto’s thyroiditis by targeting CLDN1. J. Transl. Med. 2016, 14, 166. [Google Scholar] [CrossRef]
- Qin, W.; Ren, Q.; Liu, T.; Huang, Y.; Wang, J. MicroRNA-155 is a novel suppressor of ovarian cancer-initiating cells that targets CLDN1. FEBS Lett. 2013, 587, 1434–1439. [Google Scholar] [CrossRef]
- Geremia, A.; Biancheri, P.; Allan, P.; Corazza, G.R.; Di Sabatino, A. Innate and adaptive immunity in inflammatory bowel disease. Autoimmun. Rev. 2014, 13, 3–10. [Google Scholar] [CrossRef]
- Zigmond, E.; Bernshtein, B.; Friedlander, G.; Walker, C.R.; Yona, S.; Kim, K.W.; Brenner, O.; Krauthgamer, R.; Varol, C.; Muller, W.; et al. Macrophage-restricted interleukin-10 receptor deficiency, but not IL-10 deficiency, causes severe spontaneous colitis. Immunity 2014, 40, 720–733. [Google Scholar] [CrossRef] [PubMed]
- Nejad, C.; Stunden, H.J.; Gantier, M.P. A guide to miRNAs in inflammation and innate immune responses. FEBS J. 2018, 285, 3695–3716. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.D.; Xu, J.; Zhang, M.; Zhu, T.M.; Zhang, Y.H.; Sun, K. MicroRNA-21 inhibits lipopolysaccharide-induced acute lung injury by targeting nuclear factor-kappaB. Exp. Ther. Med. 2018, 16, 4616–4622. [Google Scholar] [CrossRef]
- Das, A.; Ganesh, K.; Khanna, S.; Sen, C.K.; Roy, S. Engulfment of apoptotic cells by macrophages: A role of microRNA-21 in the resolution of wound inflammation. J. Immunol. 2014, 192, 1120–1129. [Google Scholar] [CrossRef] [PubMed]
- Taganov, K.D.; Boldin, M.P.; Chang, K.J.; Baltimore, D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. USA 2006, 103, 12481–12486. [Google Scholar] [CrossRef]
- Li, S.; Yue, Y.; Xu, W.; Xiong, S. MicroRNA-146a represses mycobacteria-induced inflammatory response and facilitates bacterial replication via targeting IRAK-1 and TRAF-6. PLoS ONE 2013, 8, e81438. [Google Scholar] [CrossRef]
- Garo, L.P.; Ajay, A.K.; Fujiwara, M.; Gabriely, G.; Raheja, R.; Kuhn, C.; Kenyon, B.; Skillin, N.; Kadowaki-Saga, R.; Saxena, S.; et al. MicroRNA-146a limits tumorigenic inflammation in colorectal cancer. Nat. Commun. 2021, 12, 2419. [Google Scholar] [CrossRef]
- O’Connell, R.M.; Taganov, K.D.; Boldin, M.P.; Cheng, G.; Baltimore, D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc. Natl. Acad. Sci. USA 2007, 104, 1604–1609. [Google Scholar] [CrossRef]
- Ceppi, M.; Pereira, P.M.; Dunand-Sauthier, I.; Barras, E.; Reith, W.; Santos, M.A.; Pierre, P. MicroRNA-155 modulates the interleukin-1 signaling pathway in activated human monocyte-derived dendritic cells. Proc. Natl. Acad. Sci. USA 2009, 106, 2735–2740. [Google Scholar] [CrossRef]
- O’Connell, R.M.; Kahn, D.; Gibson, W.S.; Round, J.L.; Scholz, R.L.; Chaudhuri, A.A.; Kahn, M.E.; Rao, D.S.; Baltimore, D. MicroRNA-155 promotes autoimmune inflammation by enhancing inflammatory T cell development. Immunity 2010, 33, 607–619. [Google Scholar] [CrossRef]
- Zhang, N.; Fu, L.; Bu, Y.; Yao, Y.; Wang, Y. Downregulated expression of miR-223 promotes Toll-like receptor-activated inflammatory responses in macrophages by targeting RhoB. Mol. Immunol. 2017, 91, 42–48. [Google Scholar] [CrossRef] [PubMed]
- Choy, M.C.; Visvanathan, K.; De Cruz, P. An Overview of the Innate and Adaptive Immune System in Inflammatory Bowel Disease. Inflamm. Bowel Dis. 2017, 23, 2–13. [Google Scholar] [CrossRef]
- Haneklaus, M.; Gerlic, M.; Kurowska-Stolarska, M.; Rainey, A.A.; Pich, D.; McInnes, I.B.; Hammerschmidt, W.; O’Neill, L.A.; Masters, S.L. Cutting edge: miR-223 and EBV miR-BART15 regulate the NLRP3 inflammasome and IL-1beta production. J. Immunol. 2012, 189, 3795–3799. [Google Scholar] [CrossRef]
- Neudecker, V.; Haneklaus, M.; Jensen, O.; Khailova, L.; Masterson, J.C.; Tye, H.; Biette, K.; Jedlicka, P.; Brodsky, K.S.; Gerich, M.E.; et al. Myeloid-derived miR-223 regulates intestinal inflammation via repression of the NLRP3 inflammasome. J. Exp. Med. 2017, 214, 1737–1752. [Google Scholar] [CrossRef]
- Curtale, G.; Rubino, M.; Locati, M. MicroRNAs as Molecular Switches in Macrophage Activation. Front. Immunol. 2019, 10, 799. [Google Scholar] [CrossRef]
- Italiani, P.; Boraschi, D. From Monocytes to M1/M2 Macrophages: Phenotypical vs. Functional Differentiation. Front. Immunol. 2014, 5, 514. [Google Scholar] [CrossRef]
- Chaudhuri, A.A.; So, A.Y.; Sinha, N.; Gibson, W.S.; Taganov, K.D.; O’Connell, R.M.; Baltimore, D. MicroRNA-125b potentiates macrophage activation. J. Immunol. 2011, 187, 5062–5068. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Nunez, R.T.; Louafi, F.; Sanchez-Elsner, T. The interleukin 13 (IL-13) pathway in human macrophages is modulated by microRNA-155 via direct targeting of interleukin 13 receptor alpha1 (IL13Ralpha1). J. Biol. Chem. 2011, 286, 1786–1794. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Liu, X.J.; QunZhou; Xie, J.; Ma, T.T.; Meng, X.M.; Li, J. MiR-146a modulates macrophage polarization by inhibiting Notch1 pathway in RAW264.7 macrophages. Int. Immunopharmacol. 2016, 32, 46–54. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Duan, M.; Feng, Y.; Geng, L.; Li, X.; Zhang, W. Corrigendum to “MiR-146a modulates macrophage polarization in systemic juvenile idiopathic arthritis by targeting INHBA” [Mol. Immunol. 77 (2016) 205-212]. Mol. Immunol. 2017, 87, 329–330. [Google Scholar] [CrossRef] [PubMed]
- Ying, W.; Tseng, A.; Chang, R.C.; Morin, A.; Brehm, T.; Triff, K.; Nair, V.; Zhuang, G.; Song, H.; Kanameni, S.; et al. MicroRNA-223 is a crucial mediator of PPARgamma-regulated alternative macrophage activation. J. Clin. Investig. 2015, 125, 4149–4159. [Google Scholar] [CrossRef]
- Heller, F.; Florian, P.; Bojarski, C.; Richter, J.; Christ, M.; Hillenbrand, B.; Mankertz, J.; Gitter, A.H.; Burgel, N.; Fromm, M.; et al. Interleukin-13 is the key effector Th2 cytokine in ulcerative colitis that affects epithelial tight junctions, apoptosis, and cell restitution. Gastroenterology 2005, 129, 550–564. [Google Scholar] [CrossRef]
- Lu, T.X.; Munitz, A.; Rothenberg, M.E. MicroRNA-21 is up-regulated in allergic airway inflammation and regulates IL-12p35 expression. J. Immunol. 2009, 182, 4994–5002. [Google Scholar] [CrossRef]
- Lu, T.X.; Hartner, J.; Lim, E.J.; Fabry, V.; Mingler, M.K.; Cole, E.T.; Orkin, S.H.; Aronow, B.J.; Rothenberg, M.E. MicroRNA-21 limits in vivo immune response-mediated activation of the IL-12/IFN-gamma pathway, Th1 polarization, and the severity of delayed-type hypersensitivity. J. Immunol. 2011, 187, 3362–3373. [Google Scholar] [CrossRef]
- Meisgen, F.; Xu, N.; Wei, T.; Janson, P.C.; Obad, S.; Broom, O.; Nagy, N.; Kauppinen, S.; Kemeny, L.; Stahle, M.; et al. MiR-21 is up-regulated in psoriasis and suppresses T cell apoptosis. Exp. Dermatol. 2012, 21, 312–314. [Google Scholar] [CrossRef]
- Ruan, Q.; Wang, P.; Wang, T.; Qi, J.; Wei, M.; Wang, S.; Fan, T.; Johnson, D.; Wan, X.; Shi, W.; et al. MicroRNA-21 regulates T-cell apoptosis by directly targeting the tumor suppressor gene Tipe2. Cell Death Dis. 2014, 5, e1095. [Google Scholar] [CrossRef] [PubMed]
- Van der Heide, V.; Mohnle, P.; Rink, J.; Briegel, J.; Kreth, S. Down-regulation of MicroRNA-31 in CD4+ T Cells Contributes to Immunosuppression in Human Sepsis by Promoting TH2 Skewing. Anesthesiology 2016, 124, 908–922. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Ke, F.; Liu, Z.; Bai, J.; Liu, J.; Yan, S.; Xu, Z.; Lou, F.; Wang, H.; Zhu, H.; et al. MicroRNA-31 negatively regulates peripherally derived regulatory T-cell generation by repressing retinoic acid-inducible protein 3. Nat. Commun. 2015, 6, 7639. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, A.; Vigorito, E.; Clare, S.; Warren, M.V.; Couttet, P.; Soond, D.R.; van Dongen, S.; Grocock, R.J.; Das, P.P.; Miska, E.A.; et al. Requirement of bic/microRNA-155 for normal immune function. Science 2007, 316, 608–611. [Google Scholar] [CrossRef] [PubMed]
- Thai, T.H.; Calado, D.P.; Casola, S.; Ansel, K.M.; Xiao, C.; Xue, Y.; Murphy, A.; Frendewey, D.; Valenzuela, D.; Kutok, J.L.; et al. Regulation of the germinal center response by microRNA-155. Science 2007, 316, 604–608. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.R.; Zhu, H.F.; Zhu, Y. Knockout of MicroRNA-155 Ameliorates the Th17/Th9 Immune Response and Promotes Wound Healing. Curr. Med. Sci. 2019, 39, 954–964. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.F.; Thai, T.H.; Calado, D.P.; Chaudhry, A.; Kubo, M.; Tanaka, K.; Loeb, G.B.; Lee, H.; Yoshimura, A.; Rajewsky, K.; et al. Foxp3-dependent microRNA155 confers competitive fitness to regulatory T cells by targeting SOCS1 protein. Immunity 2009, 30, 80–91. [Google Scholar] [CrossRef] [PubMed]
- Brain, O.; Owens, B.M.; Pichulik, T.; Allan, P.; Khatamzas, E.; Leslie, A.; Steevels, T.; Sharma, S.; Mayer, A.; Catuneanu, A.M.; et al. The intracellular sensor NOD2 induces microRNA-29 expression in human dendritic cells to limit IL-23 release. Immunity 2013, 39, 521–536. [Google Scholar] [CrossRef]
- Steiner, D.F.; Thomas, M.F.; Hu, J.K.; Yang, Z.; Babiarz, J.E.; Allen, C.D.; Matloubian, M.; Blelloch, R.; Ansel, K.M. MicroRNA-29 regulates T-box transcription factors and interferon-gamma production in helper T cells. Immunity 2011, 35, 169–181. [Google Scholar] [CrossRef]
- Li, B.; Wang, X.; Choi, I.Y.; Wang, Y.C.; Liu, S.; Pham, A.T.; Moon, H.; Smith, D.J.; Rao, D.S.; Boldin, M.P.; et al. miR-146a modulates autoreactive Th17 cell differentiation and regulates organ-specific autoimmunity. J. Clin. Investig. 2017, 127, 3702–3716. [Google Scholar] [CrossRef]
- Lu, L.F.; Boldin, M.P.; Chaudhry, A.; Lin, L.L.; Taganov, K.D.; Hanada, T.; Yoshimura, A.; Baltimore, D.; Rudensky, A.Y. Function of miR-146a in controlling Treg cell-mediated regulation of Th1 responses. Cell 2010, 142, 914–929. [Google Scholar] [CrossRef]
- Catalan, D.; Mansilla, M.A.; Ferrier, A.; Soto, L.; Oleinika, K.; Aguillon, J.C.; Aravena, O. Immunosuppressive Mechanisms of Regulatory B Cells. Front. Immunol. 2021, 12, 611795. [Google Scholar] [CrossRef]
- Mizoguchi, A.; Mizoguchi, E.; Takedatsu, H.; Blumberg, R.S.; Bhan, A.K. Chronic intestinal inflammatory condition generates IL-10-producing regulatory B cell subset characterized by CD1d upregulation. Immunity 2002, 16, 219–230. [Google Scholar] [CrossRef]
- Wang, S.; Qin, C. Interleukin 35 Rescues Regulatory B Cell Function, but the Effect Is Dysregulated in Ulcerative Colitis. DNA Cell Biol. 2017, 36, 413–421. [Google Scholar] [CrossRef]
- Wang, X.; Zhu, Y.; Zhang, M.; Wang, H.; Jiang, Y.; Gao, P. Ulcerative Colitis Is Characterized by a Decrease in Regulatory B Cells. J. Crohns Colitis. 2016, 10, 1212–1223. [Google Scholar] [CrossRef]
- Wang, H.; Xu, W.; Shao, Q.; Ding, Q. miR-21 silencing ameliorates experimental autoimmune encephalomyelitis by promoting the differentiation of IL-10-producing B cells. Oncotarget 2017, 8, 94069–94079. [Google Scholar] [CrossRef]
- Xiao, S.; Brooks, C.R.; Zhu, C.; Wu, C.; Sweere, J.M.; Petecka, S.; Yeste, A.; Quintana, F.J.; Ichimura, T.; Sobel, R.A.; et al. Defect in regulatory B-cell function and development of systemic autoimmunity in T-cell Ig mucin 1 (Tim-1) mucin domain-mutant mice. Proc. Natl. Acad. Sci. USA 2012, 109, 12105–12110. [Google Scholar] [CrossRef]
- Amrouche, L.; You, S.; Sauvaget, V.; Manda, V.; Lamarthee, B.; Desbuissons, G.; Tinel, C.; Rabant, M.; Nguyen, C.; Isnard, P.; et al. MicroRNA-146a-deficient mice develop immune complex glomerulonephritis. Sci. Rep. 2019, 9, 15597. [Google Scholar] [CrossRef]
- Habtezion, A.; Nguyen, L.P.; Hadeiba, H.; Butcher, E.C. Leukocyte Trafficking to the Small Intestine and Colon. Gastroenterology 2016, 150, 340–354. [Google Scholar] [CrossRef]
- Matloubian, M.; Lo, C.G.; Cinamon, G.; Lesneski, M.J.; Xu, Y.; Brinkmann, V.; Allende, M.L.; Proia, R.L.; Cyster, J.G. Lymphocyte egress from thymus and peripheral lymphoid organs is dependent on S1P receptor 1. Nature 2004, 427, 355–360. [Google Scholar] [CrossRef]
- Feagan, B.G.; Rutgeerts, P.; Sands, B.E.; Hanauer, S.; Colombel, J.F.; Sandborn, W.J.; Van Assche, G.; Axler, J.; Kim, H.J.; Danese, S.; et al. Vedolizumab as induction and maintenance therapy for ulcerative colitis. N. Engl. J. Med. 2013, 369, 699–710. [Google Scholar] [CrossRef]
- Vermeire, S.; Sandborn, W.J.; Danese, S.; Hebuterne, X.; Salzberg, B.A.; Klopocka, M.; Tarabar, D.; Vanasek, T.; Gregus, M.; Hellstern, P.A.; et al. Anti-MAdCAM antibody (PF-00547659) for ulcerative colitis (TURANDOT): A phase 2, randomised, double-blind, placebo-controlled trial. Lancet 2017, 390, 135–144. [Google Scholar] [CrossRef]
- Sandborn, W.J.; Peyrin-Biroulet, L.; Zhang, J.; Chiorean, M.; Vermeire, S.; Lee, S.D.; Kuhbacher, T.; Yacyshyn, B.; Cabell, C.H.; Naik, S.U.; et al. Efficacy and Safety of Etrasimod in a Phase 2 Randomized Trial of Patients with Ulcerative Colitis. Gastroenterology 2020, 158, 550–561. [Google Scholar] [CrossRef]
- Sandborn, W.J.; Feagan, B.G.; D’Haens, G.; Wolf, D.C.; Jovanovic, I.; Hanauer, S.B.; Ghosh, S.; Petersen, A.; Hua, S.Y.; Lee, J.H.; et al. Ozanimod as Induction and Maintenance Therapy for Ulcerative Colitis. N. Engl. J. Med. 2021, 385, 1280–1291. [Google Scholar] [CrossRef]
- Smigielska-Czepiel, K.; van den Berg, A.; Jellema, P.; Slezak-Prochazka, I.; Maat, H.; van den Bos, H.; van der Lei, R.J.; Kluiver, J.; Brouwer, E.; Boots, A.M.; et al. Dual role of miR-21 in CD4+ T-cells: Activation-induced miR-21 supports survival of memory T-cells and regulates CCR7 expression in naive T-cells. PLoS ONE 2013, 8, e76217. [Google Scholar] [CrossRef] [PubMed]
- Zitzer, N.C.; Snyder, K.; Meng, X.; Taylor, P.A.; Efebera, Y.A.; Devine, S.M.; Blazar, B.R.; Garzon, R.; Ranganathan, P. MicroRNA-155 Modulates Acute Graft-versus-Host Disease by Impacting T Cell Expansion, Migration, and Effector Function. J. Immunol. 2018, 200, 4170–4179. [Google Scholar] [CrossRef] [PubMed]
- Ranganathan, P.; Heaphy, C.E.; Costinean, S.; Stauffer, N.; Na, C.; Hamadani, M.; Santhanam, R.; Mao, C.; Taylor, P.A.; Sandhu, S.; et al. Regulation of acute graft-versus-host disease by microRNA-155. Blood 2012, 119, 4786–4797. [Google Scholar] [CrossRef] [PubMed]
- Park, E.J.; Prajuabjinda, O.; Soe, Z.Y.; Darkwah, S.; Appiah, M.G.; Kawamoto, E.; Momose, F.; Shiku, H.; Shimaoka, M. Exosomal regulation of lymphocyte homing to the gut. Blood Adv. 2019, 3, 1–11. [Google Scholar] [CrossRef]
- Chen, Y.; Shan, T.; Qu, H.; Chen, Y.; Wang, N.; Xia, J. Inhibition of miR-16 Ameliorates Inflammatory Bowel Disease by Modulating Bcl-2 in Mouse Models. J. Surg. Res. 2020, 253, 185–192. [Google Scholar] [CrossRef]
- Shi, C.; Liang, Y.; Yang, J.; Xia, Y.; Chen, H.; Han, H.; Yang, Y.; Wu, W.; Gao, R.; Qin, H. MicroRNA-21 knockout improve the survival rate in DSS induced fatal colitis through protecting against inflammation and tissue injury. PLoS ONE 2013, 8, e66814. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Xu, J.; Li, Y.; Zhao, R.; Du, S.; Lv, C.; Wu, W.; Liu, R.; Sheng, X.; Song, Y.; et al. MicroRNA-31 Reduces Inflammatory Signaling and Promotes Regeneration in Colon Epithelium, and Delivery of Mimics in Microspheres Reduces Colitis in Mice. Gastroenterology 2019, 156, 2281–2296. [Google Scholar] [CrossRef]
- Chu, F.; Hu, Y.; Zhou, Y.; Guo, M.; Lu, J.; Zheng, W.; Xu, H.; Zhao, J.; Xu, L. MicroRNA-126 deficiency enhanced the activation and function of CD4(+) T cells by elevating IRS-1 pathway. Clin. Exp. Immunol. 2018, 191, 166–179. [Google Scholar] [CrossRef]
- Nata, T.; Fujiya, M.; Ueno, N.; Moriichi, K.; Konishi, H.; Tanabe, H.; Ohtake, T.; Ikuta, K.; Kohgo, Y. MicroRNA-146b improves intestinal injury in mouse colitis by activating nuclear factor-kappaB and improving epithelial barrier function. J. Gene Med. 2013, 15, 249–260. [Google Scholar] [CrossRef]
- Singh, U.P.; Murphy, A.E.; Enos, R.T.; Shamran, H.A.; Singh, N.P.; Guan, H.; Hegde, V.L.; Fan, D.; Price, R.L.; Taub, D.D.; et al. miR-155 deficiency protects mice from experimental colitis by reducing T helper type 1/type 17 responses. Immunology 2014, 143, 478–489. [Google Scholar] [CrossRef]
- Rawat, M.; Nighot, M.; Al-Sadi, R.; Gupta, Y.; Viszwapriya, D.; Yochum, G.; Koltun, W.; Ma, T.Y. IL1B Increases Intestinal Tight Junction Permeability by Up-regulation of MIR200C-3p, Which Degrades Occludin mRNA. Gastroenterology 2020, 159, 1375–1389. [Google Scholar] [CrossRef] [PubMed]
- Titze-de-Almeida, S.S.; Soto-Sanchez, C.; Fernandez, E.; Koprich, J.B.; Brotchie, J.M.; Titze-de-Almeida, R. The Promise and Challenges of Developing miRNA-Based Therapeutics for Parkinson’s Disease. Cells 2020, 9, 841. [Google Scholar] [CrossRef] [PubMed]
- Naidu, S.; Magee, P.; Garofalo, M. MiRNA-based therapeutic intervention of cancer. J. Hematol. Oncol. 2015, 8, 68. [Google Scholar] [CrossRef] [PubMed]
- Pang, J.K.S.; Phua, Q.H.; Soh, B.S. Applications of miRNAs in cardiac development, disease progression and regeneration. Stem Cell Res. Ther. 2019, 10, 336. [Google Scholar] [CrossRef]
- Suri, K.; Bubier, J.A.; Wiles, M.V.; Shultz, L.D.; Amiji, M.M.; Hosur, V. Role of MicroRNA in Inflammatory Bowel Disease: Clinical Evidence and the Development of Preclinical Animal Models. Cells 2021, 10, 2204. [Google Scholar] [CrossRef]
- Rupaimoole, R.; Slack, F.J. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017, 16, 203–222. [Google Scholar] [CrossRef]
- Van Zandwijk, N.; Pavlakis, N.; Kao, S.C.; Linton, A.; Boyer, M.J.; Clarke, S.; Huynh, Y.; Chrzanowska, A.; Fulham, M.J.; Bailey, D.L.; et al. Safety and activity of microRNA-loaded minicells in patients with recurrent malignant pleural mesothelioma: A first-in-man, phase 1, open-label, dose-escalation study. Lancet Oncol. 2017, 18, 1386–1396. [Google Scholar] [CrossRef]
- Gallant-Behm, C.L.; Piper, J.; Lynch, J.M.; Seto, A.G.; Hong, S.J.; Mustoe, T.A.; Maari, C.; Pestano, L.A.; Dalby, C.M.; Jackson, A.L.; et al. A MicroRNA-29 Mimic (Remlarsen) Represses Extracellular Matrix Expression and Fibroplasia in the Skin. J. Investig. Dermatol. 2019, 139, 1073–1081. [Google Scholar] [CrossRef]
- Seto, A.G.; Beatty, X.; Lynch, J.M.; Hermreck, M.; Tetzlaff, M.; Duvic, M.; Jackson, A.L. Cobomarsen, an oligonucleotide inhibitor of miR-155, co-ordinately regulates multiple survival pathways to reduce cellular proliferation and survival in cutaneous T-cell lymphoma. Br. J. Haematol. 2018, 183, 428–444. [Google Scholar] [CrossRef] [PubMed]

| miRNA | Expression | Inflammation on Samples | Detection Method |
|---|---|---|---|
| miR-16 | Elevated | Inflamed [13,16,17] | Microarray [13,16], PCR [17] |
| miR-21 | Elevated | Inflamed [13,16,18,19,20,21,22], Uninflamed [13], Unspecified [9,23,24,25] | Microarray [13,16,18,19,23], PCR [9,13,20,21,22,24,25] |
| miR-29a/b | Elevated | Inflamed [13,16,26,27], Uninflamed [13,26] | Microarray [13,16,27], PCR [13,26] |
| miR-31 | Elevated | Inflamed [26,28], Unspecified [9,29] | smRNA-Seq [29], PCR [9,26,28,29] |
| miR-125b | Elevated | Inflamed [18,30,31], Uninflamed [18] | Microarray [18], PCR [18,30,31] |
| miR-126 | Elevated | Inflamed [13,16,21,22,26], Uninflamed [13,26] | Microarray [13,16], PCR [13,21,22,26] |
| miR-142 | Elevated | Inflamed [16,20,32] | Microarray [16,32], PCR [20,32] |
| miR-146a/b | Elevated | Inflamed [16,20,33], Unspecified [23,25,29] | smRNA-Seq [29], Microarray [16,23], PCR [20,25,29,33] |
| miR-155 | Elevated | Inflamed [16,19,30,33] | Microarray [16,19], PCR [16,30,33] |
| miR-223 | Elevated | Inflamed [22,26,27,30,31], Uninflamed [30], Unspecified [23] | Microarray [23,27], PCR [22,26,30,31] |
| miR-192 | Reduced | Inflamed [13,20,27] | Microarray [13,27], PCR [13,20] |
| miR-200a/b/c | Reduced | Inflamed [16,20,27] | Microarray [16,27], PCR [20,27] |
| miR-378 | Reduced | Inflamed [16,27], Unspecified [23] | Microarray [16,23,27] |
| miRNA | Expression in UC | miRNA Modulation in Mice | Result (Degree of Colitis) |
|---|---|---|---|
| miR-16 | Elevated | Inhibited | Ameliorated [115] |
| miR-21 | Elevated | Deficient | Ameliorated [116] |
| miR-29a/b | Elevated | Deficient | Deteriorated [94] |
| miR-31 | Elevated | Deficient | Deteriorated [117] |
| miR-126 | Elevated | Deficient | Deteriorated [118] |
| miR-146a/b | Elevated | Deficient (miR-146a) Overexpressed (miR-146b) | Deteriorated [68] Ameliorated [119] |
| miR-155 | Elevated | Deficient | Ameliorated [120] |
| miR-223 | Elevated | Overexpressed | Ameliorated [75] |
| miR-200a/b/c | Reduced | Inhibited (miR-200c) | Ameliorated [121] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Toyonaga, T.; Saruta, M. Role of microRNAs in the Pathophysiology of Ulcerative Colitis. Immuno 2021, 1, 558-573. https://doi.org/10.3390/immuno1040039
Toyonaga T, Saruta M. Role of microRNAs in the Pathophysiology of Ulcerative Colitis. Immuno. 2021; 1(4):558-573. https://doi.org/10.3390/immuno1040039
Chicago/Turabian StyleToyonaga, Takahiko, and Masayuki Saruta. 2021. "Role of microRNAs in the Pathophysiology of Ulcerative Colitis" Immuno 1, no. 4: 558-573. https://doi.org/10.3390/immuno1040039
APA StyleToyonaga, T., & Saruta, M. (2021). Role of microRNAs in the Pathophysiology of Ulcerative Colitis. Immuno, 1(4), 558-573. https://doi.org/10.3390/immuno1040039






