In-Line Monitoring of Downstream Purification Processes for VSV Based SARS-CoV-2 Vaccine Using a Novel Technique
Abstract
:1. Introduction
2. Materials and Methods
2.1. Virus Upstream Production
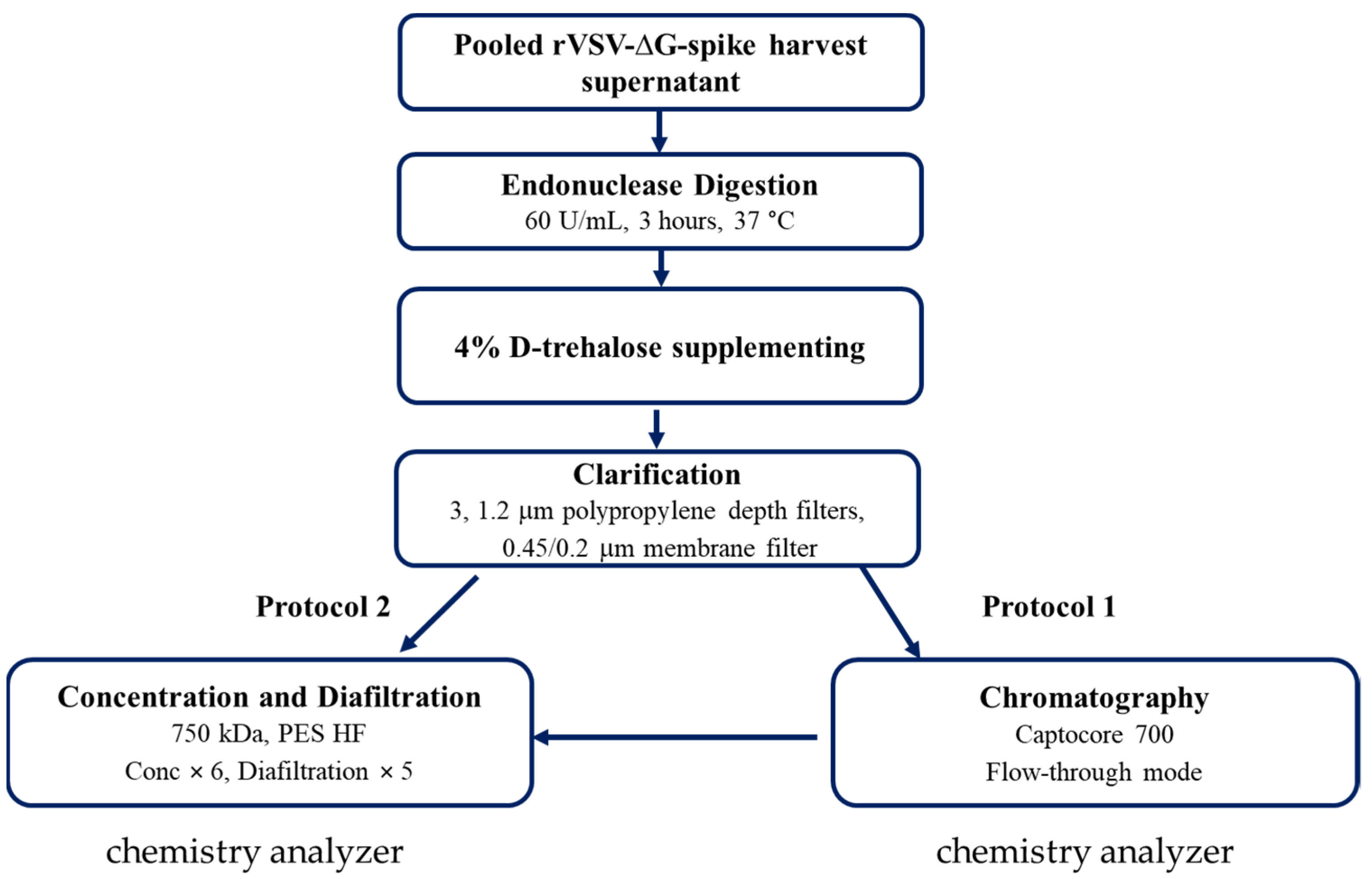
2.2. Virus Purification
2.3. Chemistry Analyzer: Cobas Integra 400 Plus
2.4. Host Cell Protein (HCP) Analysis
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Guan, W.-J.; Ni, Z.-Y.; Hu, Y.; Liang, W.-H.; Ou, C.-Q.; He, J.-X.; Liu, L.; Shan, H.; Lei, C.-L.; Hui, D.S.C.; et al. Clinical Characteristics of Coronavirus Disease 2019 in China. N. Engl. J. Med. 2020, 382, 1708–1720. [Google Scholar] [CrossRef] [PubMed]
- WHO. The COVID-19 Candidate Vaccine Landscape and Tracker. 2021. Available online: https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines (accessed on 1 November 2021).
- ClinicalTrials.gov. Evaluate the Safety, Immunogenicity and Potential Efficacy of an rVSV-SARS-CoV-2-S Vaccine, NCT04608305. 2021. Available online: https://clinicaltrials.gov/ct2/results?cond=&term=NCT04608305&cntry=&state=&city=&dist= (accessed on 1 November 2021).
- ClinicalTrials.gov. Phase 2b/3 Trial of VSV-ΔG SARS-CoV-2 Vaccine (BRILIFE) Against Approved Comparator Vaccine. (BRILIFE002), NCT04990466. 2021. Available online: https://clinicaltrials.gov/ct2/show/NCT04990466?term=IIBR100&draw=2&rank=1 (accessed on 1 November 2021).
- Yahalom-Ronen, Y.; Tamir, H.; Melamed, S.; Politi, B.; Shifman, O.; Achdout, H.; Vitner, E.B.; Israeli, O.; Milrot, E.; Stein, D.; et al. A single dose of recombinant VSV-∆G-spike vaccine provides protection against SARS-CoV-2 challenge. Nat. Commun. 2020, 11, 6402. [Google Scholar] [CrossRef] [PubMed]
- Cino, J.; Mirro, R.; Kedzierski, S. An Update on the Advantages of Fibra-Cel® Disks for Cell Culture. 2011. Available online: https://www.eppendorf.com/uploads/media/Application_bioprocess_shakers_incubators_Application-Note-Boo.pdf (accessed on 24 October 2021).
- Wolf, M.W.; Reichl, U. Downstream processing of cell culture-derived virus particles. Expert Rev. Vaccines 2011, 10, 1451–1475. [Google Scholar] [CrossRef] [PubMed]
- Nestola, P.; Peixoto, C.; Silva, R.R.J.S.; Alves, P.M.; Mota, J.P.B.; Carrondo, M.J.T. Improved virus purification processes for vaccines and gene therapy. Biotechnol. Bioeng. 2015, 112, 843–857. [Google Scholar] [CrossRef] [PubMed]
- Makovitzki, A.; Lerer, E.; Kafri, Y.; Adar, Y.; Cherry, L.; Lupu, E.; Monash, A.; Levy, R.; Israeli, O.; Dor, E.; et al. Evaluation of a Downstream Process for the Recovery and Concentration of a Cell-Culture-Derived rVSV-Spike COVID-19 Vaccine Candidate. Vaccine 2021, in press. [Google Scholar] [CrossRef]
- Lerer, E.; Oren, Z.; Kafri, Y.; Adar, Y.; Cherry, L.; Lupu, E.; Monash, A.; Levy, R.; Dor, E.; Epstein, E.; et al. Highly Efficient Purification of Recombinant VSV-∆G-spike Vaccine Against SARS-CoV-2 By Flow-through Chromatography. BioTech 2021, 10, 22. [Google Scholar] [CrossRef]
- Mandenius, C.-F.; Titchener-Hooker, N.J. Measurement, Monitoring, Modelling and Control of Bioprocesses; Springer: Berlin/Heidelberg, Germany, 2013; ISBN 978-3-642-36838-7. [Google Scholar]
- Flickinger, M.C. Downstream Industrial Biotechnology: Recovery and Purification; John Wiley & Sons: Hoboken, NJ, USA, 2013; ISBN 978-1-118-13124-4. [Google Scholar]
- Negrete, A.; Pai, A.; Shiloach, J. Use of hollow fiber tangential flow filtration for the recovery and concentration of HIV virus-like particles produced in insect cells. J. Virol. Methods 2014, 195, 240–246. [Google Scholar] [CrossRef] [PubMed]
- Yuk, I.H.; Nishihara, J.; Walker, D.J.; Huang, E.; Gunawan, F.; Subramanian, J.; Pynn, A.F.J.; Yu, X.C.; Zhu-Shimoni, J.; Vanderlaan, M.; et al. More similar than different: Host cell protein production using three null CHO cell lines. Biotechnol. Bioeng. 2015, 112, 2068–2083. [Google Scholar] [CrossRef] [PubMed]

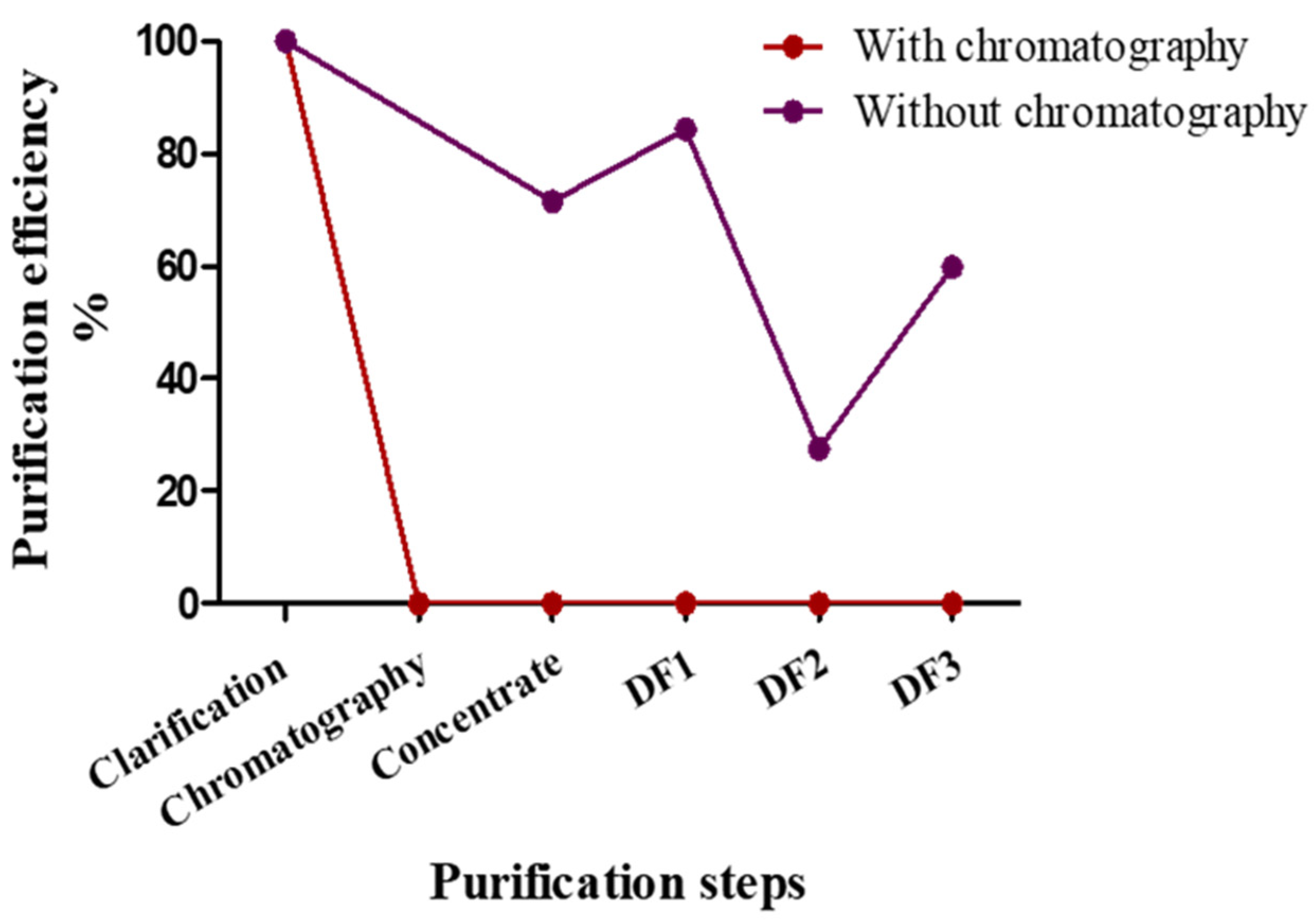
| Purification Step | Purification Efficiency % | |
|---|---|---|
| With chromatography | clarification | 100 |
| Chromatography | 0.6 | |
| DF1 | 0.2 | |
| DF3 | 0.2 | |
| Without chromatography | clarification | 100 |
| DF1 | 52 | |
| DF3 | 58 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Makovitzki, A.; Jayson, A.; Oren, Z.; Lerer, E.; Kafri, Y.; Dor, E.; Cherry, L.; Tzadok, H.; Levin, L.; Hazan, O.; et al. In-Line Monitoring of Downstream Purification Processes for VSV Based SARS-CoV-2 Vaccine Using a Novel Technique. BioTech 2021, 10, 25. https://doi.org/10.3390/biotech10040025
Makovitzki A, Jayson A, Oren Z, Lerer E, Kafri Y, Dor E, Cherry L, Tzadok H, Levin L, Hazan O, et al. In-Line Monitoring of Downstream Purification Processes for VSV Based SARS-CoV-2 Vaccine Using a Novel Technique. BioTech. 2021; 10(4):25. https://doi.org/10.3390/biotech10040025
Chicago/Turabian StyleMakovitzki, Arik, Avital Jayson, Ziv Oren, Elad Lerer, Yaron Kafri, Eyal Dor, Lilach Cherry, Hanan Tzadok, Lilach Levin, Ophir Hazan, and et al. 2021. "In-Line Monitoring of Downstream Purification Processes for VSV Based SARS-CoV-2 Vaccine Using a Novel Technique" BioTech 10, no. 4: 25. https://doi.org/10.3390/biotech10040025
APA StyleMakovitzki, A., Jayson, A., Oren, Z., Lerer, E., Kafri, Y., Dor, E., Cherry, L., Tzadok, H., Levin, L., Hazan, O., Simon, I., Tal, A., Girshengorn, M., & Rosen, O. (2021). In-Line Monitoring of Downstream Purification Processes for VSV Based SARS-CoV-2 Vaccine Using a Novel Technique. BioTech, 10(4), 25. https://doi.org/10.3390/biotech10040025






