SSR Linkage Maps and Identification of QTL Controlling Morpho-Phenological Traits in Two Iranian Wheat RIL Populations
Abstract
:1. Introduction
2. Materials and Methods
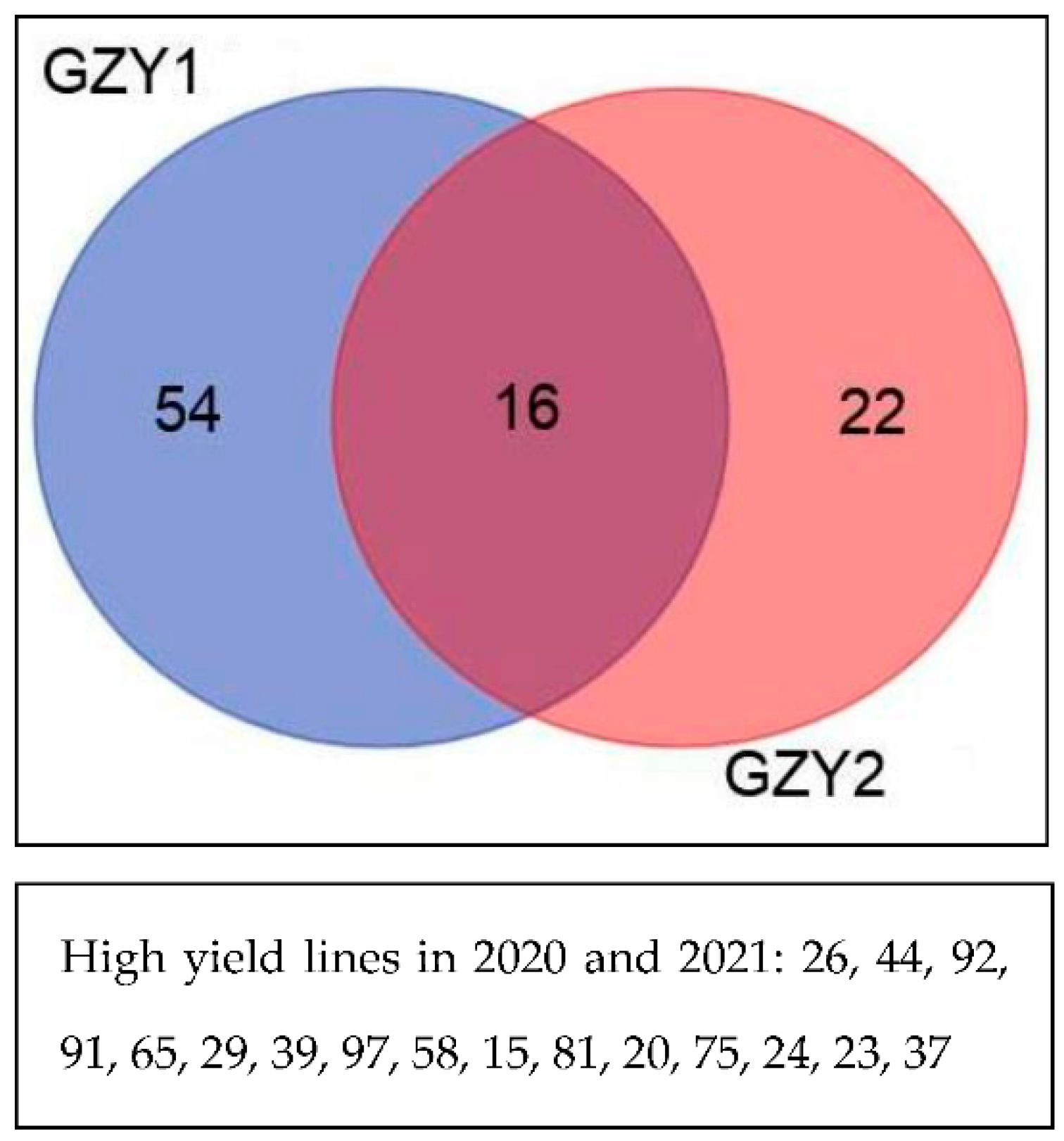
3. Results
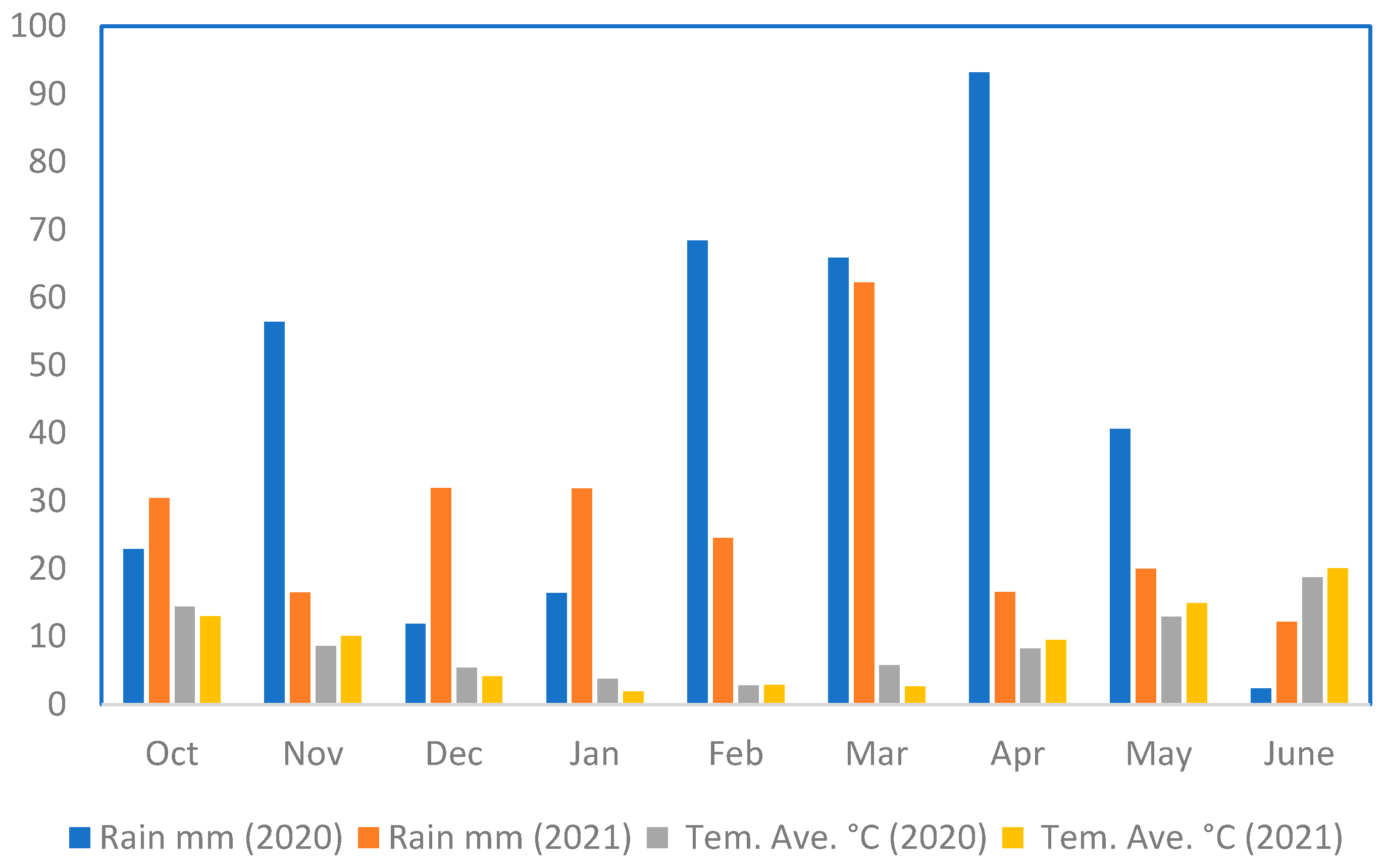
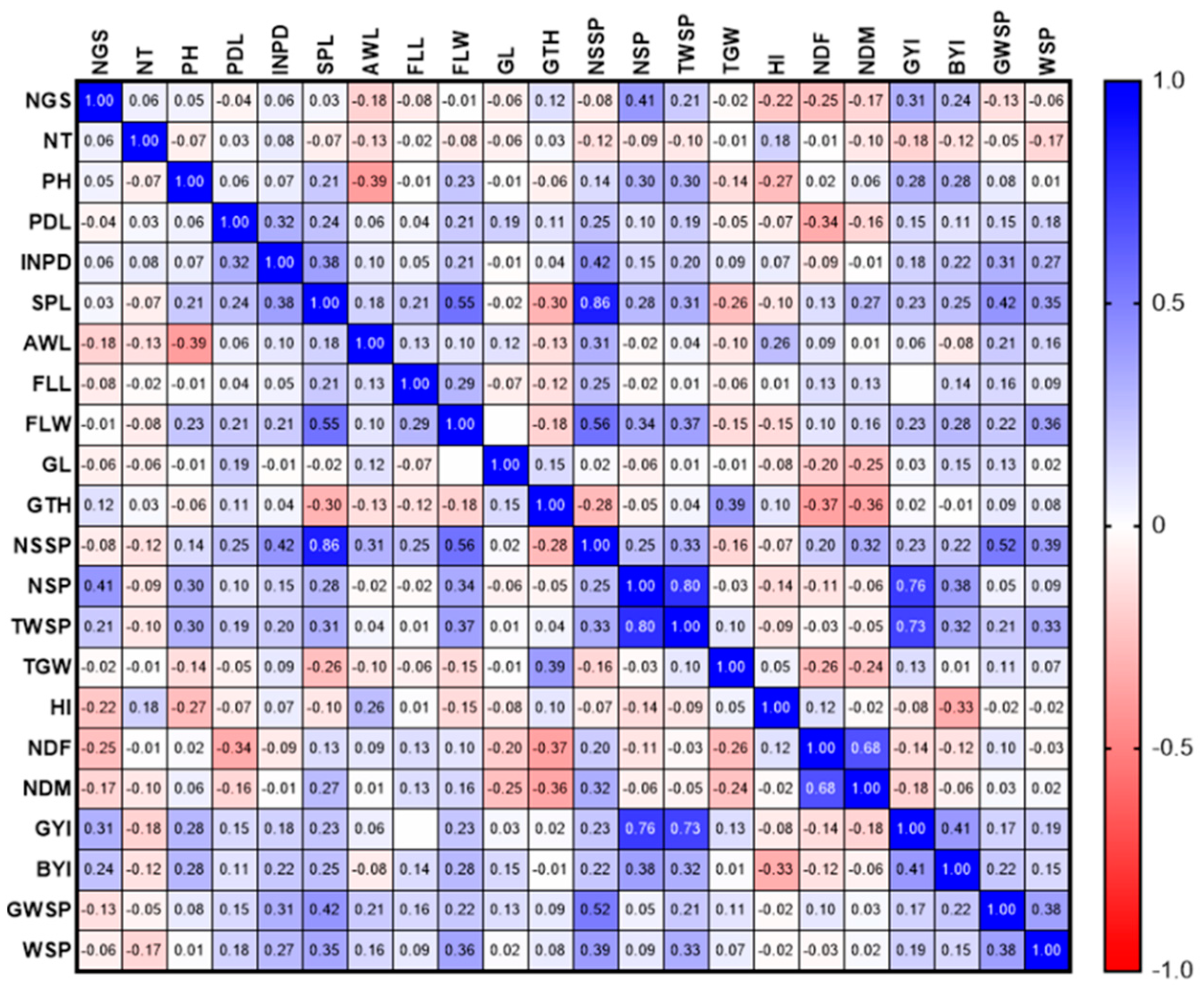
3.1. Gonbad Zagros RIL Population
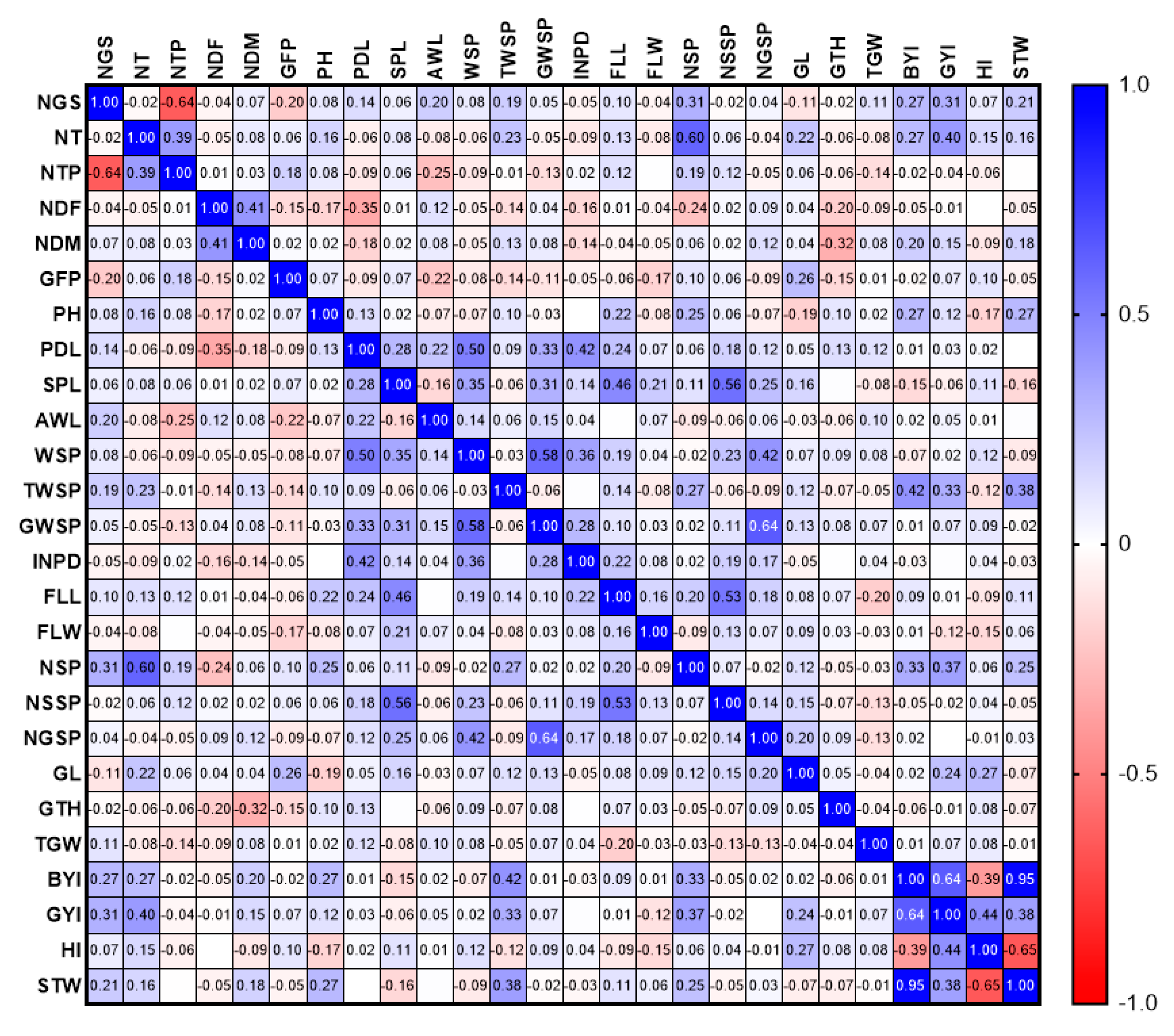
3.1.1. Phenotypic Evaluations
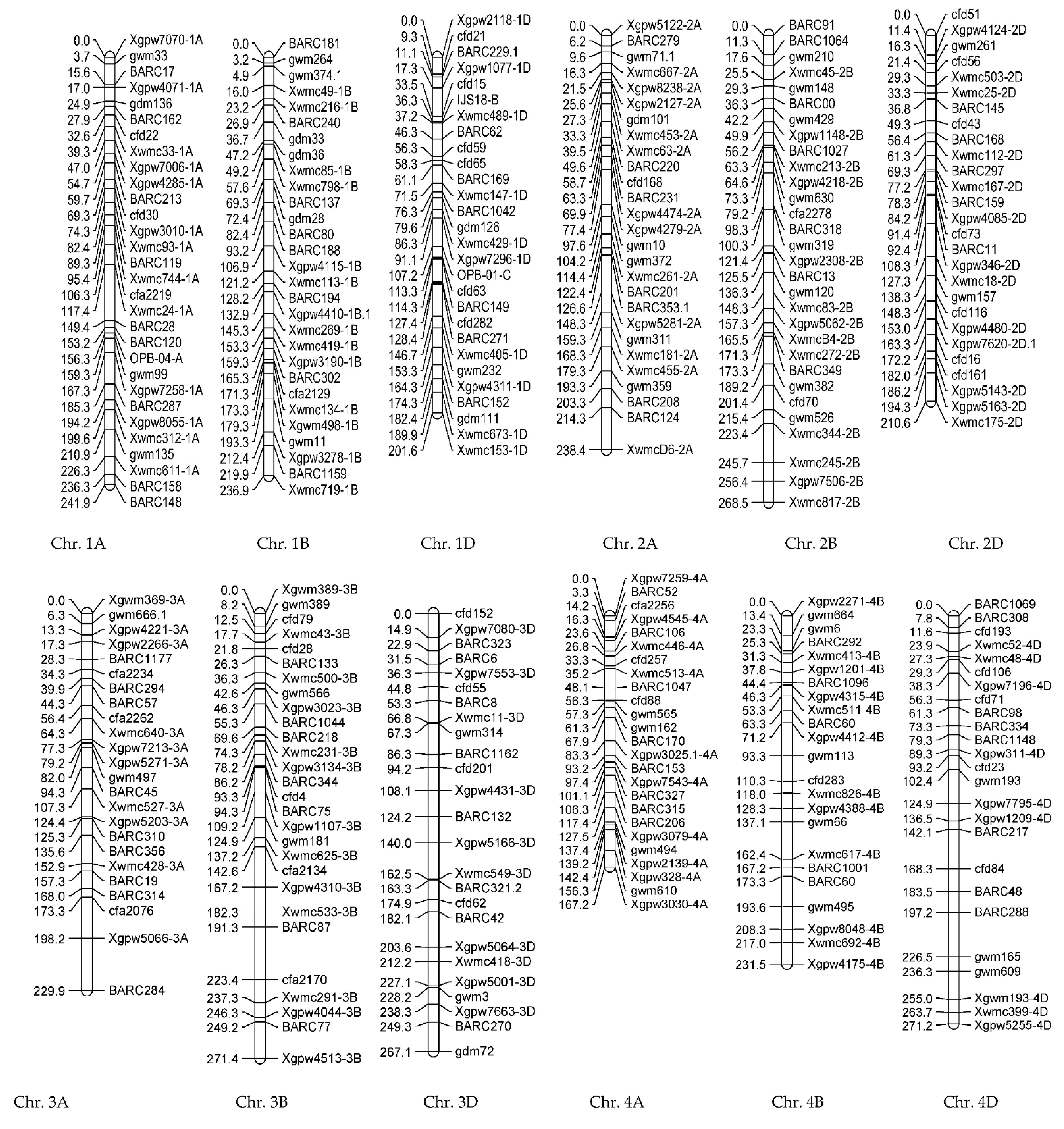
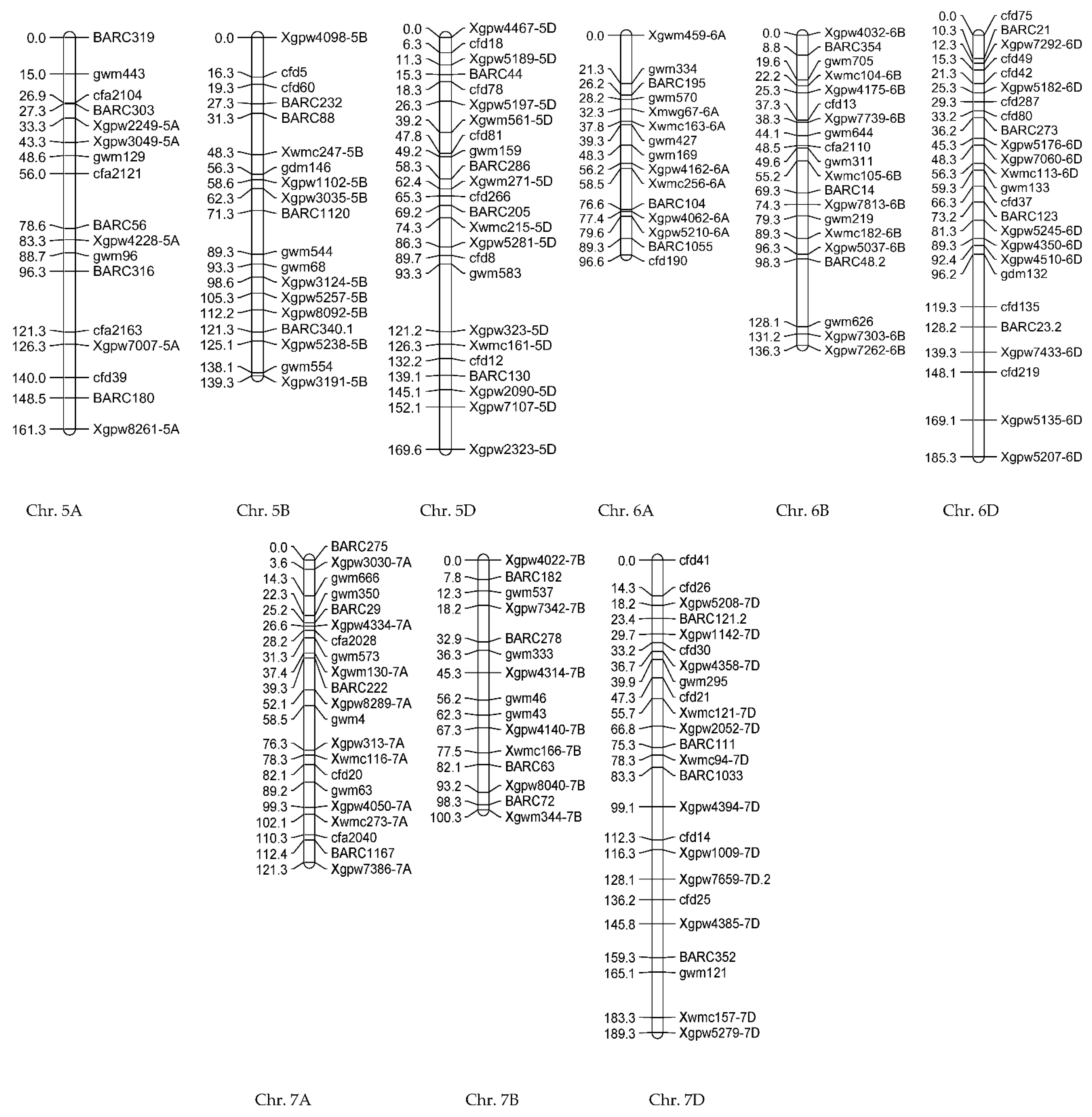
3.1.2. Genotypic Evaluations
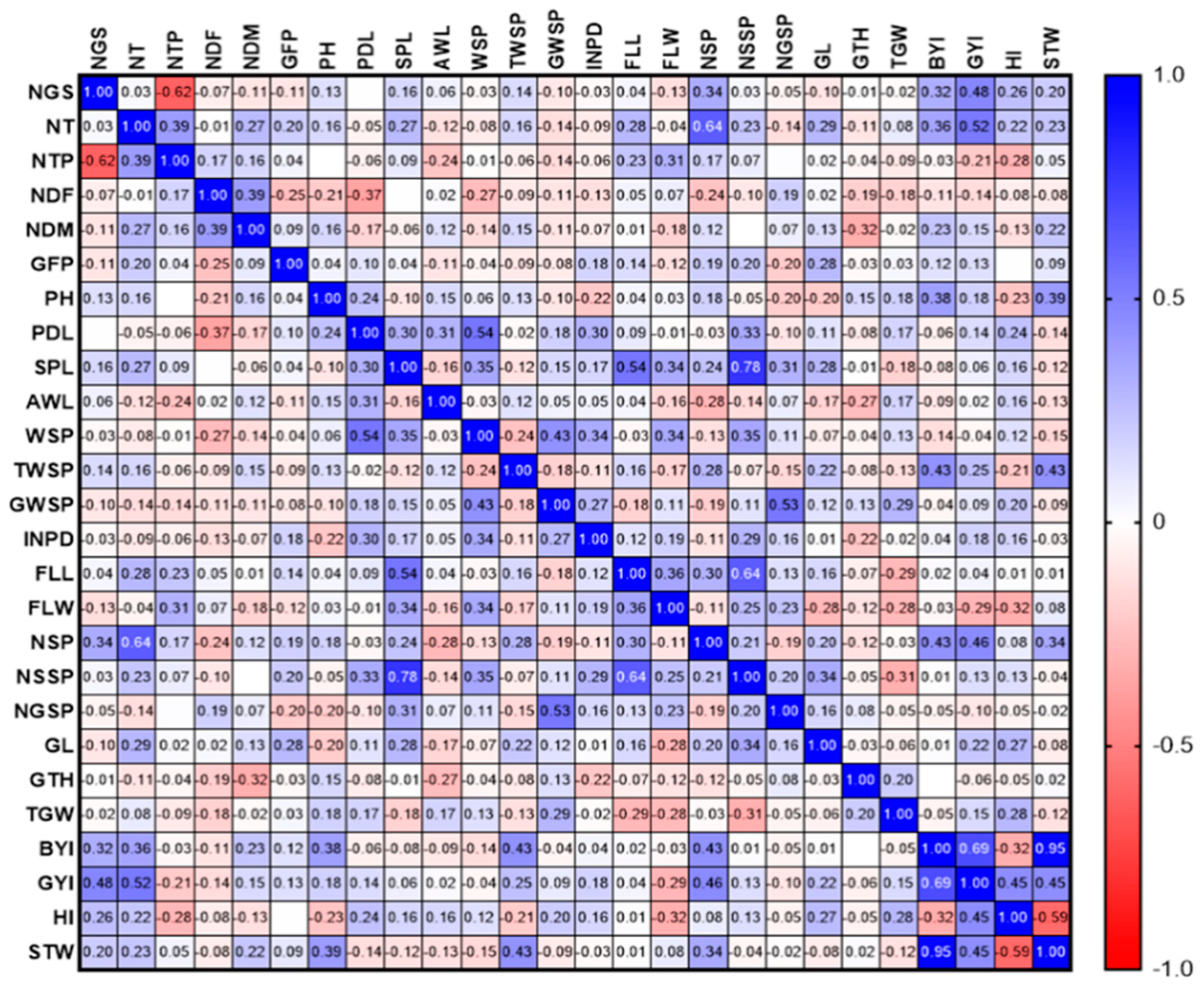
3.2. Gonbad Kohdasht RIL Population
3.2.1. Phenotypic Evaluations
3.2.2. Genotypic Evaluations
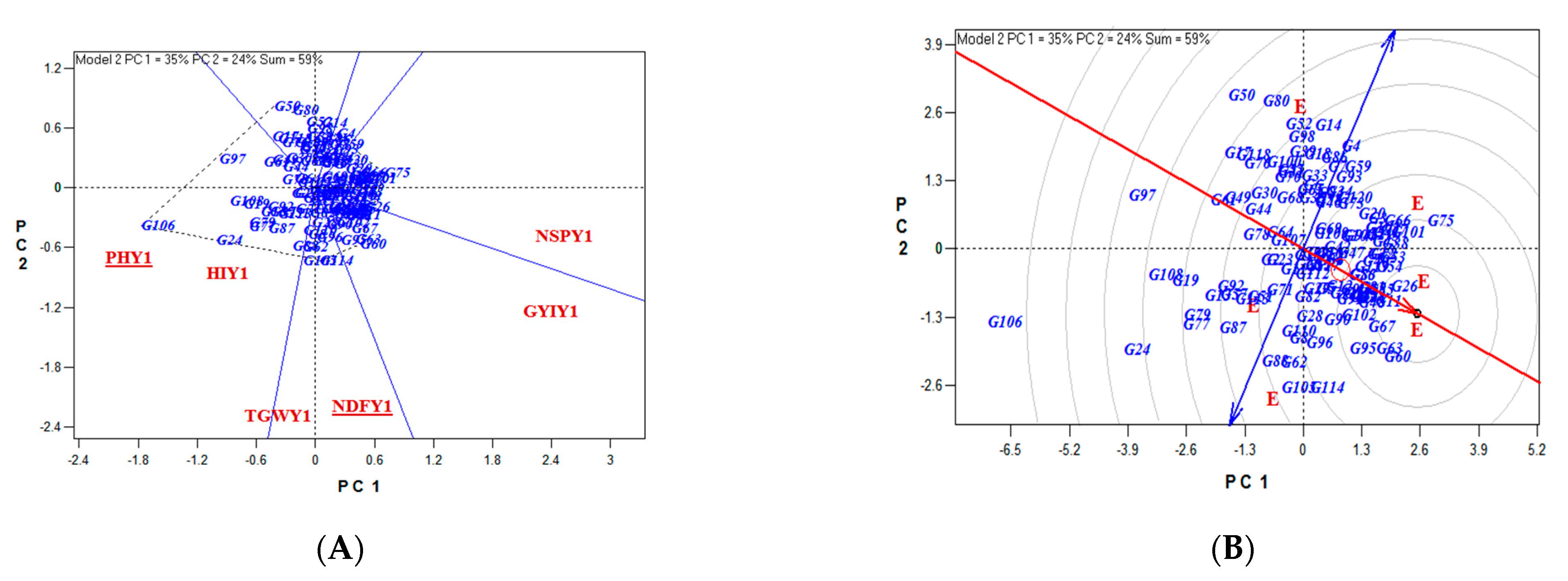
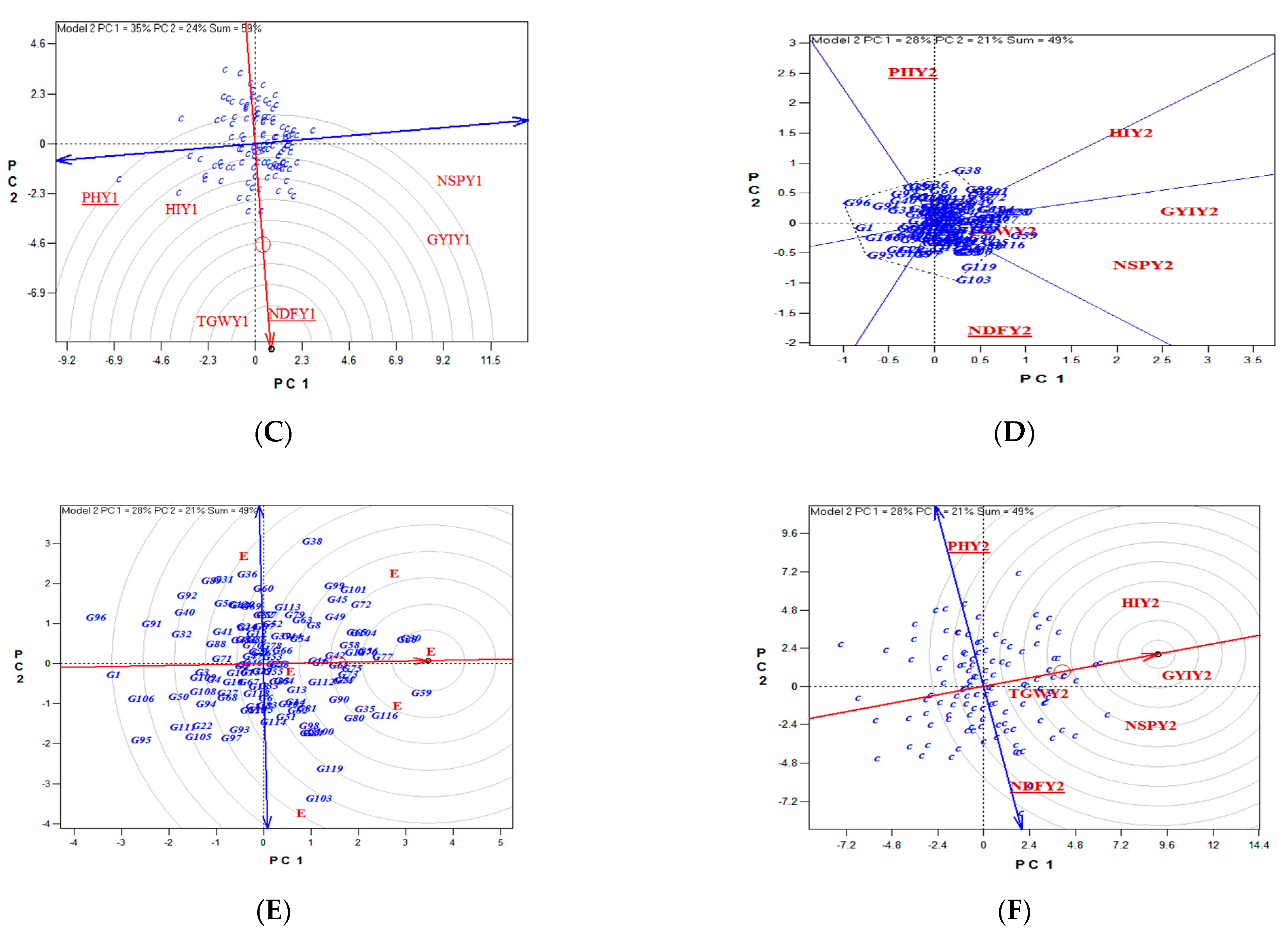
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hussain, W.; Baenziger, P.S.; Belamkar, V.; Guttieri, M.J.; Venegas, J.P.; Easterly, A.; Sallam, A.; Poland, J. Genotyping-by-sequencing derived high-density linkage map and its application to QTL mapping of flag leaf traits in bread wheat. Sci. Rep. 2017, 7, 16394. [Google Scholar] [CrossRef] [PubMed]
- Tomás, D.; Rodrigues, J.C.; Viegas, W.; Silva, M. Assessment of High Temperature Effects on Grain Yield and Composition in Bread Wheat Commercial Varieties. Agronomy 2020, 10, 499. [Google Scholar] [CrossRef] [Green Version]
- Gahlaut, V.; Jaiswal, V.; Singh, S.; Balyan, H.S.; Gupta, P.K. Multi Locus genome wide association mapping for yield and its contributing traits in hexaploid wheat under different water regimes. Sci. Rep. 2019, 9, 19486. [Google Scholar] [CrossRef]
- Hu, J.M.; Wang, X.Q.; Zhang, G.X.; Jiang, P.; Chen, W.Y.; Hao, Y.C.; Ma, X.; Xu, S.; Jia, J.; Kong, L.; et al. QTL mapping for yield-related traits in wheat based on four RIL populations. Theor. Appl. Genet. 2020, 133, 917–933. [Google Scholar] [CrossRef]
- Lin, Y.; Jiang, X.; Hu, H.; Zhou, K.; Wang, Q.; Yu, S.; Yang, X.; Wang, Z.; Wu, F.; Liu, S.; et al. QTL mapping for grain number per spikelet in wheat using a high-density genetic map. Crop J. 2021, 9, 1108–1114. [Google Scholar] [CrossRef]
- DeVita, P.; Taranto, T. Durum wheat (Triticumturgidum ssp. durum) breeding to meet the challenge of climate change. In Advances in Plant Breeding Strategies: Cereals; Al-Khayri, J.M., Jain, S.M., Johnson, D.V., Eds.; Springer: Berlin/Heidelberg, Germany, 2019; pp. 471–524. [Google Scholar] [CrossRef]
- Wang, P.; Zhou, G.L.; Yu, H.H.; Yu, S.B. Fine mapping a major QTL for flag leaf size and yield-related traits in rice. Theor. Appl. Genet. 2011, 123, 1319–1330. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.H.; Chen, Y.X.; Fu, L.; Zhou, S.; Chen, J.; Zhao, X.; Zhang, D.; Ouyang, S.; Wang, Z.; Li, D.; et al. QTL mapping of flag leaf traits in common wheat using an integrated high-density SSR and SNP genetic linkage map. Euphytica 2016, 208, 337–351. [Google Scholar] [CrossRef]
- Liu, K.; Sun, X.X.; Ning, T.Y.; Duan, X.; Wang, Q.; Liu, T.; An, Y.; Guan, X.; Tian, J.; Cgen, J. Genetic dissection of wheat panicle traits using linkage analysis and a genome-wide association study. Theor. Appl. Genet. 2018, 131, 1073–1090. [Google Scholar] [CrossRef] [PubMed]
- Deng, Z.Y.; Cui, Y.; Han, Q.D.; Fang, W.; Li, J.; Tian, J. Discovery of consistent QTLs of wheat spike-related traits under nitrogen treatment at different development stages. Front. Plant Sci. 2017, 8, 2120. [Google Scholar] [CrossRef] [Green Version]
- Sakuma, S.; Golan, G.; Guo, Z.; Ogawa, T.; Tagiri, A.; Sugimoto, K.; Bernhardt, N.; Brassac, J.; Mascher, M.; Hensel, G.; et al. Unleashing floret fertility in wheat through the mutation of a homeobox gene. Proc. Natl. Acad. Sci. USA 2019, 116, 5182–5187. [Google Scholar] [CrossRef] [Green Version]
- Jia, H.Y.; Wan, H.S.; Yang, S.H.; Zgang, Z.; Kong, Z.; Xue, S.; Zhang, L.; Ma, Z. Genetic dissection of yieldrelated traits in a recombinant inbred line population created using a key breeding parent in China’s wheat breeding. Theor. Appl. Genet. 2013, 126, 2123–2139. [Google Scholar] [CrossRef]
- Noryan, M.; Hervan, I.M.; Sabouri, H.; Kojouri, F.D.; Mastinu, A. Drought Resistance Loci in Recombinant Lines of Iranian Oryza sativa L. in Germination Stage. BioTech 2021, 10, 26. [Google Scholar] [CrossRef]
- Ding, A.M.; Culi, F.; Li, J.; Zhao, C.H.; Wang, L.; Qi, X.L.; Bao, Y.G.; Li, X.F.; Wang, H.G. QTL Mapping for Grain Yield Conditioned on its Component Traits in Two RIL Populations of Bread Wheat. Cereal Res. Commun. 2013, 41, 45–53. [Google Scholar] [CrossRef]
- Rabiei, B.; Kordrostami, M.; Sabouri, A.; Sabouri, H. Identification of QTLs for yield related traits in Indica type rice using SSR and AFLP markers. Agric. Cons. Sci. 2015, 80, 91–99. [Google Scholar]
- Sabouri, A.; Reza Afshari, R.; Raiesi, T.; Babaei Raouf, H.; Nasiri, E.; Esfahani, M.; Kafi Ghasemi, A.; Kumar, A. Superior adaptation of aerobic rice under drought stress in Iran and validation test of linked SSR markers to major QTLs by MLM analysis across two years. Mol. Biol. Rep. 2018, 45, 1037–1053. [Google Scholar] [CrossRef]
- Shirmohammadli, S.; Sabouri, H.; Ahangar, L.; Ebadi, A.; Sajjadi, S. Genetic diversity and association analysis of rice genotypes for grain physical quality using iPBS, IRAP, and ISSR markers. J. Genet. Res. 2018, 4, 122–129. [Google Scholar] [CrossRef]
- Khapilina, O.; Turzhanova, A.; Danilova, A.; Tumenbayeva, A.; Shevtsov, V.; Kotukhov, Y.; Kalendar, R. Primer Binding Site (PBS) Profiling of Genetic Diversity of Natural Populations of Endemic Species Allium ledebourianum Schult. BioTech 2021, 10, 23. [Google Scholar] [CrossRef]
- Sabouri, A.; Nasiri, E.; Esfahani, M.; Forghani, A. SSR marker-based study of the effects of genomic regions on Fe, Mn, Zn, and protein content in a rice diversity panel. J. Plant Biochem. Biotechnol. 2021, 30, 504–514. [Google Scholar] [CrossRef]
- Sabouri, A.; Alinezhad, F.; Mousanejad, S. Association analysis using SSR markers and identification of resistant aerobic and Iranian rice cultivars to blast disease. Eur. J. Plant Pathol. 2020, 158, 561–570. [Google Scholar] [CrossRef]
- Chopra, R.K.; Singh, K.; Shukla, S.; Kadam, S.; Singh, N.K. QTLs for cell membrane stability and flag leaf area under drought stress in a wheat RIL population. J. Plant Biochem. Biotechnol. 2019, 29, 276–286. [Google Scholar] [CrossRef]
- Liu, C.; Sukumaran, S.; Claverie, E.; Sansaloni, C.; Susanne Dreisigacker, S.; Reynolds, M. Genetic dissection of heat and drought stress QTLs in phenology-controlled synthetic-derived recombinant inbred lines in spring wheat. Mol. Breed. 2019, 39, 34. [Google Scholar] [CrossRef]
- Saghai, M.M.; Biyashev, R.M.; Yang, G.P.; Zhang, Q.; Allard, W. Extraordinarily polymorphic microsatellite DNA in barley: Species diversity, chromosomal locations, and population dynamics. Proc. Natl. Acad. Sci. USA 1994, 91, 5466–5470. [Google Scholar] [CrossRef] [Green Version]
- Xu, S.B.; Tao, Y.F.; Yang, Z.Q.; Chu, J.Y. A simple and rapid method used for silver staining and gel preservation. Hereditas 2002, 24, 335–336. [Google Scholar] [PubMed]
- Nelson, J. QGENE: Software for marker–based analysis and breeding. Mol. Breed. 1997, 3, 239–245. [Google Scholar] [CrossRef]
- Manly, K.F.; Olson, J.M. Overview of QTL mapping software and introduction to map manager QTL. Mamm. Genome 1999, 10, 327–334. [Google Scholar] [CrossRef]
- Kosambi, D.D. The estimation of map distances from recombination values. Ann. Eugen. 1944, 12, 172–175. [Google Scholar] [CrossRef]
- Lander, E.S.; Botstein, D. Mapping Mendelian factor underlying quantitative traits using RFLP linkage maps. Genetics 1989, 121, 185–199. [Google Scholar] [CrossRef]
- Zhang, Y.W.; Wen, Y.J.; Dunwell, J.M.; Zhang, Y.M. QTL. gCIMapping. GUI v2.0: An R software for detecting small-effect and linked QTLs for quantitative traits in bi-parental segregation populations. Comput. Struct. Biotechnol. J. 2020, 18, 59–65. [Google Scholar] [CrossRef]
- Haydar, F.M.A.; Ahamed, M.S.; Siddique, A.B.; Uddin, G.M.; Biswas, K.L.; Alam, M.F. Estimation of genetic variability, heritability and correlation for some quantitative traits in wheat (Triticumaestivum L.). J. Bio-Sci. 2020, 28, 81–86. [Google Scholar] [CrossRef]
- Kandel, M.; Ghimire, S.K.; Ojha, B.R.; Shrestha, J. Genetic diversity for heat tolerant related traits in maize inbred lines. Agricultura 2018, 105, 23–34. [Google Scholar] [CrossRef]
- Kharel, L.; Ghimire, S.K.; Shrestha, J.; Kunwar, C.B.; Sharma, S. Evaluation of rice genotypes for its response to added fertility levels and induced drought tolerance during reproductive phase. J. AgriSearch 2018, 5, 3–18. [Google Scholar] [CrossRef] [Green Version]
- Dhami, N.B.; Kandel, M.; Gurung, S.B.; Shrestha, J. Agronomic performance and correlation analysis of finger millet genotypes (Elusinecoracana L.). Malays J. Sustain. Agric. 2018, 2, 16–18. [Google Scholar] [CrossRef]
- Ojha, R.; Sarkar, A.; Aryal, A.; Rahul, K.C.; Tiwari, S.; Poudel, M.; Pant, K.R.; Shrestha, J. Correlation and path coefficient analysis of wheat (Triticumaestivum L.) genotypes. Farming Manag. 2018, 3, 136–141. [Google Scholar] [CrossRef]
- Roy, A.; Kumar, A.; Singh, A.; Mandi, A.; Barman, M. Analysis of genetic diversity and correlation studies on grain yield and its component characters in bread wheat (Triticumaestivum L. emThell) genotypes. Pharma Innov. J. 2021, 10, 341–345. [Google Scholar]
- Fouad, H. Correlation, Path and Regression Analysis in Some Bread Wheat (Triticumaestivum L.) Genotypes under Normal Irrigation and Drought Conditions. Egypt. J. Agron. 2018, 40, 133–144. [Google Scholar]
- Kumar, S.; Attri, S.D.; Singh, K.K. Comparison of Lasso and stepwise regression technique for wheat yield prediction. J. Agrometeorol. 2019, 21, 188–192. [Google Scholar] [CrossRef]
- Mohamed, N.A. Some statistical procedures for evaluation of the relative contribution for yieldcomponents in wheat. Zagazig J. Agric. Res. 1999, 26, 281–290. [Google Scholar]
- Soleymanifard, A.; Naseri, R.; Meysam, M. The study genetic variation and factor analysis for agronomic traits of Durum wheat genotypes using cluster analysis and path analysis under drought stress condition in western of Iran. Res. J. Appl. Basic Sci. 2012, 3, 479–485. [Google Scholar]
- Nasri, R.; Kashani, A.; Paknejad, F.; Vazan, S.; Barary, M. Correlation, path analysis and stepwise regression in yield and yield components in wheat (Triticumaestivum L.) under the temperate climate of Ilam province, Iran. Indian Fundam Appl. Life Sci. 2014, 4, 188–198. [Google Scholar]
- Mirela, M.S.; Veselinka, Z.; Sofija, P.; Danica, M.; Borislav, B.; Desimir, K. Variability, correlation, path analysis and stepwise regression for yield components of different wheat genotypes. Genetika 2018, 50, 817–827. [Google Scholar] [CrossRef] [Green Version]
- Adilova, S.S.; Qulmamatova, D.E.; Baboev, S.K.; Bozorov, T.A.; Morgunov, A.I. Multivariate Cluster and Principle Component Analyses of Selected Yield Traits in Uzbek Bread Wheat Cultivars. Am. J. Plant. Sci. 2020, 11, 101205. [Google Scholar] [CrossRef]
- Marza, F.; Bai, G.H.; Carver, B.F.; Zhou, W.C. Quantitative trait loci for yield and related traits in the wheat population Ning7840 Clark. Theor. Appl. Genet. 2005, 112, 688–698. [Google Scholar] [CrossRef] [PubMed]
- Sourdille, P.; Cadalen, T.; Guyomarc’h, H.; Snape, J.W.; Perretant, M.R.; Charmet, G.; Boeuf, C.; Bernard, S.; Bernard, M. An update of the Courtot × Chinse spring intervarietal molecular marker linkage map for QTL detection of agronomic traits in wheat. Theor. Appl. Genet. 2003, 106, 530–538. [Google Scholar] [CrossRef]
- Keller, M.; Karutz, C.; Shmid, J.; Stamp, P.; Wnnzeler, M.; Keller, B.; Messmer, M. Quantitrative trait loci for lodging resistance in a segregating wheat × spelt population. Theor. Appl. Genet. 1999, 98, 1171–1182. [Google Scholar] [CrossRef]
- Borner, A.; Schumann, E.; Furste, A.; Coster, H.; Leithold, B.; Roder, M.S.; Weber, W.E. Mapping of quantitative trait loci determining agronomic important characters in hexaploidwheat (Triticumaestivum L.). Theor. Appl. Genet. 2002, 105, 921–936. [Google Scholar] [CrossRef]
- Sishen, L.; Jizeng, J.; Xyanyun, W. An interval genetic map and QTL analysis for height in wheat using a doubled-haploid population. Theor. Appl. Genet. 2007, 96, 933–940. [Google Scholar]
- Huang, X.Q.; Coster, H.; Ganal, M.W.; Röder, M.S. Advanced backcross QTL analysis for the identification of quantitative trait loci alleles from wild relatives of wheat (Triticumaestivum L.). Theor. Appl. Genet. 2003, 106, 1379–1389. [Google Scholar] [CrossRef]
- Kumar, N.; Kulwal, P.L.; Balyan, H.S.; Guta, P.K. QTL mapping for yield and yield contributing traits in two mapping populations of bread wheat. Mol. Breed. 2007, 19, 163–177. [Google Scholar] [CrossRef]
- Holland, J.B. Genetic architecture of complex traits in plants. Curr. Opin. Plant Biol. 2007, 10, 156–161. [Google Scholar] [CrossRef]
- Li, Z.K.; Yu, S.B.; Lafitte, H.R.; Huang, N.; Courtois, B.; Hittalmani, S.; Vijayakumar, C.H.M.; Liu, G.F.; Wang, G.C.; Shashidhar, H.E.; et al. QTL × environment interactions in rice. I. Heading date and plant height. Theor. Appl. Genet. 2003, 108, 141–153. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, Y.; Wang, Y.; Liu, Z.; Liu, C.; PEeng, N.B.; Tan, W.; Wang, A.D.; Shi, Y.; Sun, B.; et al. Stability of QTL Across Environments and QTL-by-Environment Interactions for Plant and Ear Height in Maize. Agric. Sci. China 2010, 9, 1400–1412. [Google Scholar] [CrossRef]





| Traits | Intercept | Coefficients | Std. Error | F | R2 | |
|---|---|---|---|---|---|---|
| b1 | b2 | |||||
| NSP | 1854.461 | 43.909 ** | 1044.419 | 160.974 ** | 0.577 | |
| TGW | −1397.54 | 44.152 ** | 83.025 ** | 1018.535 | 88.166 ** | 0.594 |
| Traits | Intercept | Coefficients | Std. Error | F | R2 | ||||
|---|---|---|---|---|---|---|---|---|---|
| b1 | b2 | b3 | b4 | b5 | |||||
| NT | 3682.314 | 6.156 ** | 1005.028 | 18.641 ** | 0.276 | ||||
| NGS | 1678.096 | 6.016 ** | 9.014 ** | 850.764 | 23.197 ** | 0.491 | |||
| STW | 946.991 | 5.345 ** | 8.019 ** | 0.108 * | 806.549 | 19.343 ** | 0.552 | ||
| FLW | 2121.728 | 5.144 ** | 7.257 ** | 0.121 ** | −46,073.598 * | 759.584 | 18.104 ** | 0.612 | |
| PD | −1864.79 | 5.400 ** | 7.249 ** | 0.124 ** | −56,296.787 ** | 1591.834 ** | 680.978 | 20.466 ** | 0.695 |
| Trait | Chr | Position (cM) | Additive Effect | LOD | Left_Marker | Right_Marker | R2 (%) | |
|---|---|---|---|---|---|---|---|---|
| NGS | qNGS-1A | 1A | 102.29 | −17.440 | 3.42 | Xwmc744-1A | cfa2219 | 22.15 |
| qNGS-4B | 4B | 109.25 | 14.8282 | 3.15 | gwm113 | cfd283 | 16.01 | |
| PDL | qPDL-1B | 1B | 4.85 | 1.764 | 3.11 | gwm374.1 | gwm374.1 | 13.775 |
| qPDL-4A | 4A | 97.36 | 1.5644 | 2.73 | Xgpw7543-4A | Xgpw7543-4A | 10.835 | |
| qPDL-4B | 4B | 193.57 | −1.663 | 2.76 | gwm495 | gwm495 | 12.245 | |
| FLL | qFLL-1Da | 1D | 70.51 | 1.2198 | 3.86 | BARC169 | Xwmc147-1D | 17.625 |
| qFLL-1Db | 1D | 161.52 | −1.2523 | 3.17 | gwm232 | Xgpw4311-1D | 18.575 | |
| GL | qGL-4D | 4D | 196.25 | 0.6967 | 2.79 | BARC48 | BARC288 | 27.985 |
| GW | qGW-2A | 2A | 49.62 | 0.1408 | 3.31 | BARC220 | BARC220 | 26.035 |
| NDF | qNDF-6D | 6D | 0 | 2.52 | 2.97 | Xgpw7292-6D | Xgpw7292-6D | 30.635 |
| TGW | qTGW-1D | 1A | 177.28 | 2.6711 | 3.10 | Xgpw7258-1A | BARC287 | 28.495 |
| WSP | qWSP-2B | 1B | 74.358 | 0.3048 | 2.59 | gdm28 | BARC80 | 22.91 |
| Trait | Chr | Position (cM) | Additive Effect | LOD | Left_Marker | Right_Marker | R2 (%) | |
|---|---|---|---|---|---|---|---|---|
| NT | qNT-2B | 2B | 37.23 | 104.097 | 3.12 | BARC00 | gwm429 | 27.645 |
| PH | qPH-1A | 1A | 82.36 | −6.064 | 2.99 | Xwmc93-1A | Xwmc93-1A | 16.635 |
| qPH-3A | 3A | 53.56 | −4.763 | 2.93 | BARC57 | cfa2262 | 10.265 | |
| PDL | qPDL-1Ba | 1B | 49.22 | 2.292 | 2.59 | Xwmc85-1B | Xwmc85-1B | 16.94 |
| qPDL-1Bb | 1B | 160.32 | −2.180 | 2.68 | Xgpw3190-1B | BARC302 | 15.325 | |
| SPL | qSPL-7B | 7B | 127.75 | 0.777 | 2.92 | Xwmc335-7B | gwm302 | 26.06 |
| WSP | qWSP-2B | 2B | 73.25 | 0.224 | 3.45 | gwm630 | gwm630 | 19.585 |
| qWSP-3A | 3A | 15.65 | −0.202 | 3.94 | Xgpw4221-3A | Xgpw2266-3A | 15.925 | |
| TWSP | qTWSP-6A | 6A | 12.36 | 200.093 | 4.04 | BARC171 | BARC171 | 31.365 |
| GWSP | qGWSP-7A | 7A | 121.47 | −0.195 | 2.54 | cfa2257 | cfa2257 | 21.36 |
| GW | qGW-7B | 7B | 209.45 | 0.216 | 2.12 | gwm611 | Xwmc792-7B | 21.58 |
| GL | qGL-1Da | 1D | 67.69 | −2.630 | 2.99 | BARC169 | Xwmc147-1D | 6.875 |
| qGL-3Aa | 3A | 76.32 | −3.134 | 3.39 | Xwmc640-3A | Xgpw7213-3A | 9.765 | |
| qGL-1Db | 1D | 37.23 | −1.828 | 2.77 | Xwmc489-1D | Xwmc489-1D | 3.32 | |
| qGL-3Ab | 3A | 44.28 | 2.393 | 3.97 | BARC57 | BARC57 | 5.69 | |
| qGL-7D | 7D | 78.32 | 3.345 | 5.16 | Xgpw4385-7D | gdm145 | 11.12 | |
| TGW | qTGW-1D | 1D | 70.51 | 2.041 | 2.59 | BARC169 | Xwmc147-1D | 22.54 |
| HI | qHI-7D | 7D | 35.22 | 6.22 | 3.18 | cfd41 | Xgpw2160-7D | 30.235 |
| NFSP | qNFSP-5D | 5D | 44.24 | −0.28 | 2.07 | BARC143 | Xgpw7238-5D | 18.765 |
| NDF | qNDF-4A | 4A | 18.09 | −2.603 | 3.37 | Xgpw4545-4A | BARC106 | 26.095 |
| NDM | qNDM-6B | 6B | 174.36 | −1.452 | 2.65 | gwm626 | gwm626 | 25.99 |
| GFP | qGFP-5D | 5D | 138.29 | −1.966 | 2.89 | Xwmc264-5D | cfd7 | 23.185 |
| Traits | Intercept | Coefficients | Std. Error | F | R2 (%) | |
|---|---|---|---|---|---|---|
| b1 | b2 | |||||
| NSP | 2671.308 | 37.511 ** | 1142.479 | 108.576 ** | 0.679 | |
| PD | −557.581 | 37.939 ** | 1201.858 ** | 1093.953 | 65.065 ** | 0.726 |
| Traits Entered in Mode | Intercept | Coefficients | Std. Error | F | R2 (%) | ||
|---|---|---|---|---|---|---|---|
| b1 | b2 | b3 | |||||
| NT | 4584.416 | 4.059 ** | 1022.691 | 21.971 ** | 0.157 | ||
| NGS | 3179.422 | 4.136 ** | 5.922 ** | 963.953 | 20.274 ** | 0.257 | |
| GL | 2183.118 | 3.682 ** | 6.310 ** | 78.775 * | 943.137 | 16.193 ** | 0.295 |
| Trait | QTL | Chr | Position (cM) | Additive Effect | LOD | Left_Marker | Right_Marker | R2 (%) |
|---|---|---|---|---|---|---|---|---|
| NGS | qNGS-6B | 6B | 33.26 | −8.0386 | 2.76 | Xgpw4175-6B | cfd13 | 18.91 |
| FLL | qFLL-1Db | 3A | 40.26 | 0.6082 | 2.89 | BARC284 | Xwmc264-3A | 18.05 |
| GL | qGL-3D | 3D | 110.25 | 0.2817 | 2.70 | Xgwm645-3D | Xgpw7114-3D | 23.92 |
| GW | qGW-2A | 2A | 144.17 | −0.0598 | 2.56 | gwm71.1 | BARC208 | 9.90 |
| NSSP | qNSSP-2A | 2A | 86.25 | 0.3751 | 2.88 | BARC201 | BARC201 | 11.07 |
| NSP | qNSP-1B | 1B | 45.23 | 8.8226 | 3.07 | Xgpw4134 | Xgpw4134 | 8.72 |
| qNSP-2A | 2A | 119.54 | −8.4177 | 2.68 | Xwmc261-2A | Xwmc261-2A | 7.94 | |
| qNSP-5D | 5D | 1.80 | −10.8852 | 2.77 | Xgpw4467-5D | cfd18 | 13.28 | |
| HI | qHI-1A | 1A | 108.75 | −3.8254 | 3.15 | gwm135 | Xwmc24-1A | 21.35 |
| GYI | qGYI-1B | 1B | 0 | −526.736 | 3.23 | BARC181 | BARC181 | 9.71 |
| qGYI-5B | 5B | 102.39 | −747.969 | 3.51 | Xgpw3124-5B | Xgpw5257-5B | 19.5 | |
| BYI | qBYI-1B | 1B | 32.26 | 1579.43 | 3.03 | Xgpw3122-1B | Xgpw3122-1B | 8.23 |
| qBYI-4A | 4A | 74.59 | −1627.6 | 3.04 | Xwmc173-4A.1 | Xwmc173-4A.1 | 8.74 | |
| qBYI-5D | 5D | 130.18 | −2353.14 | 4.05 | Xwmc161-5D | cfd12 | 18.28 | |
| GWSP | qGWSP-2B | 2B | 1.57 | 0.0966 | 5.71 | BARC1027 | gwm614 | 13.35 |
| qGWSP-6D | 6D | 141.22 | −0.0853 | 5.09 | Xgpw7433-6D | cfd219 | 10.41 | |
| WSP | qWSP-2B | 1A | 108.75 | −0.3722 | 3.07 | gwm135 | Xwmc24-1A | 12.81 |
| qWSP-2B | 5A | 37.26 | 0.331 | 2.71 | Xgpw2249-5A | Xgpw3049-5A | 10.13 | |
| qWSP-2B | 7B | 39.86 | 0.3526 | 2.51 | gwm333 | Xgpw4314-7B | 11.49 | |
| qWSP-2B | 2B | 83.26 | −0.2552 | 3.06 | Xgpw7641-2B | Xgpw7641-2B | 6.02 | |
| qWSP-2B | 2D | 103.26 | 0.2436 | 2.59 | XwmcD6-2D | XwmcD6-2D | 5.48 |
| Trait | Chr | Position (cM) | Additive Effect | LOD | Left_Marker | Right_Marker | R2 (%) | |
|---|---|---|---|---|---|---|---|---|
| NSG | qNSG-2A | 2A | 62.23 | 22.2059 | 3.23 | BARC279 | BARC279 | 13.67 |
| NT | qNT-5B | 5B | 64.30 | 0.413 | 4.05 | Xgpw3035-5B | BARC1120 | 21.16 |
| PL | qPL-2A | 2A | 28.82 | 1.0326 | 2.47 | BARC220 | Xgpw5177-2A | 15.83 |
| AWL | qOL-4D | 4D | 53.65 | 0.3434 | 2.44 | Xgpw4132-4D | Xgpw7795-4D | 12.79 |
| qOL-6B | 6B | 17.61 | −0.3967 | 2.91 | BARC354 | gwm705 | 17.06 | |
| STW | qSTW-6B | 6B | 42.16 | −94.138 | 2.37 | Xgpw7739-6B | gwm644 | 18.35 |
| NTS | qNTS-2D | 2D | 43.05 | 77.9566 | 4.13 | Xgpw332-2D | cfd233 | 23.87 |
| NSP | qNSP-6D | 6D | 57.25 | 0.6214 | 2.71 | Xwmc113-6D | gwm133 | 19.53 |
| qNSP-4A | 4A | 97.96 | 0.4065 | 2.44 | BARC206 | Xgpw7051-4A | 8.36 | |
| NGSP | qNGSP-1D | 1D | 49.9 | 3.145 | 3.52 | Xgpw7082-1D | Xgpw7109-1D | 15.25 |
| qNGSP-2A | 2A | 173.50 | −2.5984 | 2.69 | Xgwm382-2A | Xgwm356-2A | 10.41 | |
| qNGSP-4B | 4B | 8.43 | 2.9969 | 3.12 | gwm538 | BARC60 | 13.85 | |
| GL | qGL-1D | 1D | 49.9 | 1.3773 | 3.89 | Xgpw7082-1D | Xgpw7109-1D | 21.63 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sabouri, H.; Alegh, S.M.; Sahranavard, N.; Sanchouli, S. SSR Linkage Maps and Identification of QTL Controlling Morpho-Phenological Traits in Two Iranian Wheat RIL Populations. BioTech 2022, 11, 32. https://doi.org/10.3390/biotech11030032
Sabouri H, Alegh SM, Sahranavard N, Sanchouli S. SSR Linkage Maps and Identification of QTL Controlling Morpho-Phenological Traits in Two Iranian Wheat RIL Populations. BioTech. 2022; 11(3):32. https://doi.org/10.3390/biotech11030032
Chicago/Turabian StyleSabouri, Hossein, Sharifeh Mohammad Alegh, Narges Sahranavard, and Somayyeh Sanchouli. 2022. "SSR Linkage Maps and Identification of QTL Controlling Morpho-Phenological Traits in Two Iranian Wheat RIL Populations" BioTech 11, no. 3: 32. https://doi.org/10.3390/biotech11030032
APA StyleSabouri, H., Alegh, S. M., Sahranavard, N., & Sanchouli, S. (2022). SSR Linkage Maps and Identification of QTL Controlling Morpho-Phenological Traits in Two Iranian Wheat RIL Populations. BioTech, 11(3), 32. https://doi.org/10.3390/biotech11030032






