Development of SSR Markers Linked to Stress Responsive Genes along Tomato Chromosome 3 (Solanum lycopersicum L.)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. DNA Extraction
2.3. SSR Marker Development
2.4. Data Analysis
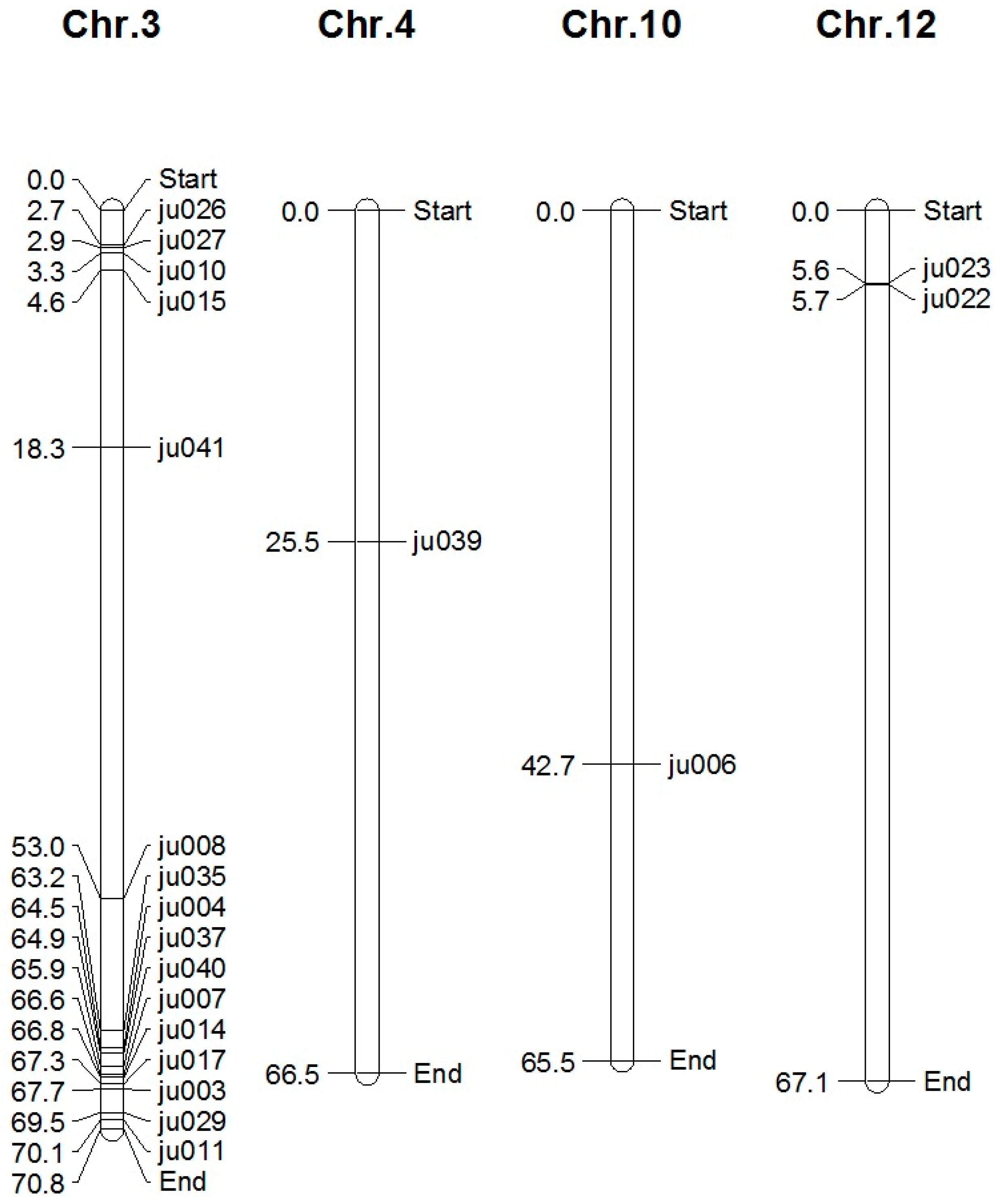
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- FAOSTAT 2020. Available online: http://faostat.fao.org (accessed on 1 June 2022).
- Arumuganathan, K.; Earle, E.D. Nuclear DNA content of some important plant species. Plant Mol. Biol. Rep. 1991, 9, 208–2018. [Google Scholar] [CrossRef]
- Rick, C.M. Biosystematic studies in Lycopersicon and closely related species of Solanum. In The Biology and Taxonomy of the Solanaceae; Hawkes, J.G., Lester, R.N., Skelding, A.D., Eds.; Academic Press: New York, NY, USA, 1979; pp. 667–677. [Google Scholar]
- Alsadon, A.A.; Sadder, M.T.; Wahb-Allah, M.A. Responsive gene screening and exploration of genotypes responses to salinity tolerance in tomato. Aust. J. Crop Sci. 2013, 7, 1383–1395. [Google Scholar]
- Sadder, M.T.; Alsadon, A.A.; Wahb-Allah, M.A. Transcriptomic analysis of tomato lines reveals putative stress-specific biomarkers. Turk. J. Agric. For. 2014, 38, 700–715. [Google Scholar] [CrossRef]
- Alsadon, A.A.; Ibrahim, A.A.; Wahb-Allah, M.A.; Ali, A.A.M.; Sadder, M.T. Tomato under salinity stress: Correlation between growth and yield components and responsive genes. Acta Hortic. 2015, 1081, 111–119. [Google Scholar] [CrossRef]
- Kissoudis, C.; Chowdhury, R.; van Heusden, S.; van de Wiel, C.; Finkers, R.; Visser, R.G.F.; Bai, Y.; van der Linden, G. Combined biotic and abiotic stress resistance in tomato. Euphytica 2015, 202, 317–332. [Google Scholar] [CrossRef]
- Kissoudis, C.; Sunarti, S.; van de Wiel, C.; Visser, R.G.F.; van der Linden, G.; Bai, Y. Responses to combined abiotic and biotic stress in tomato are governed by stress intensity and resistance mechanism. J. Exp. Bot. 2016, 67, 5119–5132. [Google Scholar] [CrossRef]
- Paul, A.; Rao, S.; Mathur, S. The α-crystallin domain containing genes: Identification, phylogeny and expression profiling in abiotic stress, phytohormone response and development in tomato (Solanum lycopersicum). Front. Plant Sci. 2016, 7, 426. [Google Scholar] [CrossRef]
- Reichardt, S.; Piepho, H.P.; Stintzi, A.; Schaller, A. Peptide signaling for drought-induced tomato flower drop. Science 2020, 367, 1482–1485. [Google Scholar] [CrossRef]
- Korir, N.K.; Diao, W.; Tao, R.; Li, X.; Kayesh, E.; Li, A.; Zhen, W.; Wang, S. Genetic diversity and relationships among different tomato varieties revealed by EST-SSR markers. Genet. Mol. Res. 2014, 13, 43–53. [Google Scholar] [CrossRef]
- Rick, C.M. Genetic relationship between self-incompatibility and floral traits in tomato species. Biol. Zent. Bl. 1982, 101, 185–198. [Google Scholar]
- Stevens, M.A.; Rick, C.M. Genetics and breeding. In The Tomato Crop; Atherton, J.G., Rudich, J., Eds.; Chapman and Hall: New York, NY, USA, 1986; pp. 35–109. [Google Scholar]
- Tanksley, S.D.; McCouch, S.R. Seed banks and molecular maps: Unlocking genetic potential from the wild. Science 1997, 277, 1063–1066. [Google Scholar] [CrossRef] [PubMed]
- Hay, A.; Craft, J.; Tsiantis, M. Plant hormones and homeoboxes: Bridging the gap? BioEssays 2004, 26, 395–404. [Google Scholar] [CrossRef]
- Tam, S.M.; Mhiri, C.; Vogelaar, A.; Kerkveld, M.; Pearce, S.R.; Grandbastien, M.A. Comparative analyses of genetic diversities within tomato and pepper collections detected by retrotransposon-based SSAP, AFLP and SSR. Theor. Appl. Genet. 2005, 110, 819–831. [Google Scholar] [CrossRef] [PubMed]
- Qaryouti, M.M.; Hamdan, H.H.; Edwan, M.A.; Hurani, O.M.; Al-Dabas, M.A. Evaluation and Characterization of Jordanian Tomato Landraces. Dirasat Agric. Sci. 2007, 34, 44–56. [Google Scholar]
- Song, Q.J.; Marek, L.F.; Shoemaker, R.C.; Lark, K.G.; Concibido, V.C.; Delannay, X.; Specht, J.E.; Cregan, P.B. A new integrated genetic linkage map of the soybean. Theor. Appl. Genet. 2004, 109, 122–128. [Google Scholar] [CrossRef]
- Temnykh, S.; DeClerck, G.; Lukashova, A.; Lipovich, L.; Cartinhour, S.; McCouch, S. Computational and experimental analysis of microsatellites in rice (Oryza sativa L.): Frequency, length variation, transposon associations, and genetic marker potential. Genome Res. 2001, 11, 1441–1452. [Google Scholar] [CrossRef] [PubMed]
- Caramante, M.; Rouphael, Y.; Corrado, G. The Genetic Diversity and Structure of Tomato Landraces from the Campania Region (Southern Italy) Uncovers a Distinct Population Identity. Agronomy 2021, 11, 564. [Google Scholar] [CrossRef]
- Al-Shammari, A.M.; Hamdi, G.J. Genetic diversity analysis and DNA fingerprinting of tomato breeding lines using SSR markers. J. Agric. Sci. 2021, 32, 1–7. [Google Scholar] [CrossRef]
- Pidigam, S.; Thuraga, V.; Munnam, S.B.; Amarapalli, G.; Kuraba, G.; Pandravada, S.R.; Nimmarajula, S.; Sudini, H.K. Genetic diversity, population structure and validation of SSR markers linked to Sw-5 and I-2 genes in tomato germplasm. Physiol. Mol. Biol. Plants 2021, 27, 695–1710. [Google Scholar] [CrossRef]
- Zhou, R.; Wu, Z.; Cao, X.; Jiang, F.L. Genetic diversity of cultivated and wild tomatoes revealed by morphological traits and SSR markers. Genet. Mol. Res. 2015, 14, 13868–13879. [Google Scholar] [CrossRef]
- Younis, A.; Ramzan, F.; Ramzan, Y.; Zulfiqar, F.; Ahsan, M.; Lim, K.B. Molecular markers improve abiotic stress tolerance in crops: A review. Plants 2020, 9, 1374. [Google Scholar] [CrossRef] [PubMed]
- Brake, M.H.; Al-Gharaibeh, M.A.; Hamasha, H.R.; Al-Sakarneh, N.S.; Alshomali, I.A.; Migdadi, H.M.; Qaryouti, M.M.; Haddad, N.J. Assessment of genetic variability among Jordanian tomato landrace using inter-simple sequence repeats markers. Jordan J. Biol. Sci. 2021, 14, 91–95. [Google Scholar]
- Jaccard, P. Nouvelles recherches sur la distribution florale. Bull. Société Vaud. Sci. Nat. 1908, 44, 223–270. [Google Scholar]
- Hammer, O.; Harper, D.A.T.; Ryan, P.D. PAST: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001, 4, 1–9. [Google Scholar]
- Park, S.D.E. The Excel Microsatellite Toolkit (Version 3.1). Ph.D. Thesis, Animal Genomics Laboratory, University College Dublin, Dublin, Ireland, 2001. [Google Scholar]
- Nei, M. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 1987, 89, 583–590. [Google Scholar] [CrossRef]
- Botstein, D.; White, R.L.; Skolnick, M.; Davis, R.W. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 1980, 32, 314–331. [Google Scholar]
- Kloosterman, A.D.; Budowle, B.; Daselaar, P. PCR amplification and detection of the human DIS80 VNTR locus. Amplification conditions, population genetics and application in forensic analysis. Int. J. Legal Med. 1993, 105, 257–264. [Google Scholar] [CrossRef]
- Li, Y.; Leveau, A.; Zhao, Q.; Feng, Q.; Lu, H.; Miao, J.; Osbourn, A. Subtelomeric assembly of a multi-gene pathway for antimicrobial defense compounds in cereals. Nat. Commun. 2021, 12, 1–13. [Google Scholar] [CrossRef]
- Cao, Y.; Li, K.; Li, Y.; Zhao, X.; Wang, L. MYB Transcription Factors as Regulators of Secondary Metabolism in Plants. Biology 2020, 9, 61. [Google Scholar] [CrossRef]
- Zhang, T.; Zhao, Y.; Wang, Y.; Liu, Z.; Gao, C. Comprehensive Analysis of MYB Gene Family and Their Expressions Under Abiotic Stresses and Hormone Treatments. Tamarix Hispida. Front. Plant Sci. 2018, 9, 1303. [Google Scholar] [CrossRef]
- He, C.; Teixeira da Silva, J.A.; Wang, H.; Si, C.; Zhang, M.; Zhang, X.; Li, M.; Tan, J.; Duan, J. Mining MYB transcription factors from the genomes of orchids (Phalaenopsis and Dendrobium) and characterization of an orchid R2R3-MYB gene involved in water-soluble polysaccharide biosynthesis. Sci. Rep. 2019, 9, 13818. [Google Scholar] [CrossRef] [PubMed]
- Zeng, H.; Xu, L.; Singh, A.; Wang, H.; Du, L.; Poovaiah, B.W. Involvement of calmodulin and calmodulin-like proteins in plant responses to abiotic stresses. Front. Plant Sci. 2015, 6, 600. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Nolan, T.M.; Jiang, H.; Yin, Y. AP2/ERF Transcription Factor Regulatory Networks in Hormone and Abiotic Stress Responses in Arabidopsis. Front. Plant Sci. 2019, 10, 228. [Google Scholar] [CrossRef]
- Chen, X.; Li, C.; Wang, H.; Guo, Z. WRKY transcription factors: Evolution, binding, and action. Phytopathol. Res. 2019, 1, 13. [Google Scholar] [CrossRef]
- Ng, A.; Xavier, R.J. Leucine-rich repeat (LRR) proteins: Integrators of pattern recognition and signaling in immunity. Autophagy 2011, 7, 1082–1084. [Google Scholar] [CrossRef]
- Hardie, D.G. Plant Protein Serine/Threonine Kinases: Classification and Functions. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999, 50, 97–131. [Google Scholar] [CrossRef]
- Xu, X.; Zhang, L.; Zhao, W.; Fu, L.; Han, Y.; Wang, K.; Yan, L.; Li, Y.; Zhang, X.H.; Min, D.H. Genome-wide analysis of the serine carboxypeptidase-like protein family in Triticum aestivum reveals TaSCPL184-6D is involved in abiotic stress response. BMC Genom. 2021, 22, 350. [Google Scholar] [CrossRef]
- Orozco-Nunnelly, D.A.; Muhammad, D.; Mezzich, R.; Lee, B.S.; Jayathilaka, L.; Kaufman, L.S.; Warpeha, K.M. Pirin1 (PRN1) Is a Multifunctional Protein that Regulates Quercetin, and Impacts Specific Light and UV Responses in the Seed-to-Seedling Transition of Arabidopsis thaliana. PLoS ONE 2014, 9, e93371. [Google Scholar] [CrossRef]
- Zhang, X.; Gonzalez-Carranza, Z.H.; Zhang, S.; Miao, Y.; Liu, C.J.; Roberts, J.A. F-Box proteins in plants. Annu. Plant Rev. 2019, 2, 307–328. [Google Scholar] [CrossRef]
- Sheehy, R.E.; Kramer, M.; Hiatt, W.R. Reduction of polygalacturonase activity in tomato fruit by antisense RNA. Proc. Natl. Acad. Sci. USA 1988, 85, 8805–8809. [Google Scholar] [CrossRef]
- Makhadmeh, I.M.; Thabet, S.G.; Ali, M.; Alabbadi, B.; Albalasmeh, A.; Alqudah, A.M. Exploring genetic variation among Jordanian Solanum Lycopersicon L. landraces and their performance under salt stress using SSR markers. J. Genet. Eng. Biotechnol. 2022, 20, 45. [Google Scholar] [CrossRef] [PubMed]
- Al-Qadumii, L.W.; Sadder, M.T.; Migdadi, H. Assessment of in silico BAC-based simple sequence repeat (SSR) marker development for tomato (Solanum lycopersicum L.). Afr. J. Biotechnol. 2012, 11, 13938–13946. [Google Scholar] [CrossRef]
- Al Shaye, N.; Migdadi, H.; Charbaji, A.; Alsayegh, S.; Daoud, S.; AL-Anazi, W.; Alghamdi, S. Genetic variation among Saudi tomato (Solanum lycopersicum L.) landraces studied using SDS-PAGE and SRAP markers. Saudi J. Biol. Sci. 2018, 25, 1007–1015. [Google Scholar] [CrossRef] [PubMed]
- Alzahib, R.H.; Migdadi, H.M.; Ghamdi, A.A.A.; Alwahibi, M.S.; Afzal, M.; Elharty, E.H.; Alghamdi, S.S. Exploring Genetic Variability among and within Hail Tomato Landraces Based on Sequence-Related Amplified Polymorphism Markers. Diversity 2021, 13, 135. [Google Scholar] [CrossRef]
- Henareh, M.; Dursun, A.; Abdollahi-Mandoulakani, B.; Haliloğlu, K. Assessment of genetic diversity in tomato landraces using ISSR markers. Genetika 2016, 48, 25–35. [Google Scholar] [CrossRef]
- Li, Q.; Su, X.; Ma, H.; Du, K.; Yang, M.; Chen, B.; Fu, S.; Fu, T.; Xiang, C.; Zhao, Q.; et al. Development of genic SSR marker resources from RNA-seq data in Camellia japonica and their application in the genus Camellia. Sci. Rep. 2021, 11, 9919. [Google Scholar] [CrossRef]
- Zheng, Z.; Zhang, N.; Huang, Z.; Zeng, Q.; Huang, Y.; Qi, Y. Genome survey sequencing and characterization of simple sequence repeat (SSR) markers in Platostoma palustre (Blume) A.J.Paton (Chinese mesona). Sci. Rep. 2022, 12, 355. [Google Scholar] [CrossRef]



| Accession Number | Growth Type | Plant Size | Foliage Density | Growth Habit |
|---|---|---|---|---|
| 951 | Determinate | Large | Dense | Erect |
| 952 | Determinate | Med.–large | Dense | Prostrate branched |
| 955 | Determinate | Med.–large | Intermediate | Prostrate branched |
| 956 | Determinate | Small | Intermediate | Erect |
| 958 | Determinate | Medium | Intermediate | Erect less branched |
| 959 | Determinate | Med.–large | Intermediate | Half erect branched |
| 960 | Determinate | Small | Sparse | Erect less branched |
| 961 | Determinate | Large | Dense | Prostrate |
| 963 | Determinate | Large | Dense | Erect branched |
| 964 | Semi-determinate | Large | Dense | Erect |
| 969 | Determinate | Medium | Intermediate | Prostrate branched |
| 970 | Determinate | Med.–large | Dense | Erect branched |
| 971A | Semi-determinate | Medium | Intermediate | Erect |
| 971B | Determinate | Small-med. | Sparse | Prostrate branched |
| 972 | Determinate | Large | Dense | Erect branched |
| 973 | Determinate | Medium | Intermediate | Prostrate |
| 975 | Determinate | Medium | Sparse | Erect branched |
| 978 | Semi-determinate | Large | Dense | Prostrate |
| 979 | Determinate | Med.–large | Intermediate | Prostrate |
| 980A | Determinate | Medium | Intermediate | Erect branched |
| 983 | Determinate | Small–med. | Intermediate | Prostrate branched |
| 984 | Determinate | Med.–large | Intermediate | Prostrate branched |
| 985 | Determinate | Med.–large | Sparse | Erect branched |
| 987 | Determinate | Large | Dense | Prostrate branched |
| 988 | Determinate | Medium | Dense | Less erect |
| 989 | Semi-determinate | Large | Dense | Half erect branched |
| 994A | Determinate | Large | Dense | Prostrate |
| 995 | Determinate | Small–med. | Intermediate | Prostrate branched |
| 996 | Determinate | Large | Dense | Erect branched |
| BAC Accession | Clone Name | SSR Locus | Chr | Primer Sequence (5’–3’) | Repeat Motif | Allele Size (s) (bp) | Tm (°C) |
|---|---|---|---|---|---|---|---|
| AC235792 | C03HBa0029M12 | ju003 | 3 | F-ATGGTGTGTCAGTCCTTTCATC | 8 GA | 254 | 50.4 |
| R-AAAGGTTAAGGGTCCTGCTAGC | 52.9 | ||||||
| AC235795 | C03HBa0036B17 | ju004 | 3 | F-TCGATGTCATTACTCACGTTCC | 5 CA | 238, 242 | 51.7 |
| R-GATACCAAAACGCAGCAAGTTG | 54.1 | ||||||
| AC235804 | C03HBa0224P23 | ju006 | 10 * | F-CATTTCATGAAAGGGGAATTCTAG | 10 TG | 201, 277 | 53.1 |
| R-ACATTTCGTGTTAGCTGGGTTC | 52.6 | ||||||
| AC238438 | C03HBa0031F10 | ju 007 | 3 | F-GAGTTTGATAAAGCAAAAGGC | 6 AG | 163, 182 | 48.2 |
| R-AACAGAACCCGAGTTTGGAC | 50.5 | ||||||
| AC238439 | C03HBa0031P17 | ju008 | 3 | F-CAATTATTAGACAGCCAACCAAG | 5 AAT | 264 | 50.5 |
| R-GGCATTTATTTGGTCAGAAAGC | 52.5 | ||||||
| AC238450 | C03HBa0114P24 | ju010 | 3 | F-TACCCTTTCGTTTACCCAAATTG | 11 AT | 282 | 54.1 |
| R-AATTGACCGATTTTCCCTTCTC | 53.1 | ||||||
| AC238451 | C03HBa0121O11 | ju011 | 3 | F-GTGAAATGATGTTTCCTCTGACAAG | 5 AAC | 246, 253 | 53.7 |
| R-CTTTCGACATCCTTTTGACTCG | 53.2 | ||||||
| AC238457 | C03HBa0166B15 | ju014 | 3 | F-CGGCAATGTAAGAGTTGAGCTC | 6 GA | 243 | 53.4 |
| R-ATCATCCCAAGCGTCAAAATAG | 52.7 | ||||||
| AC238459 | C03HBa0176B05 | ju015 | 3 | F-ACTCTTCATCCGTTGTACAATTC | 6 TTC | 264, 276 | 49.5 |
| R-TTCACTCGGATGATTGTAATCG | 51.9 | ||||||
| AC238462 | C03HBa0203H10 | ju017 | 3 | F-GATTTTATTGGGTGTCTGTTGTC | 5 TGT | 248 | 49.8 |
| R-AGGGAGAAAAGATGAACAGTATC | 48.1 | ||||||
| AC238468b | C12HBa0270F24 | ju022 | 12 ** | F-ATGGATTTACTGTAACAGTGTGAAC | 6 TTC | 293 | 49 |
| R-GTCCAAATTAATAACAGATCCATAG | 48.4 | ||||||
| AC238468c | C12HBa0270F24 | ju023 | 12 ** | F-AATTATTCGTAAGTTTCCGTCTGTC | 25 AT | 308, 320 | 52.2 |
| R-CCTTTATGAATGACCAAAAGCTAC | 51.3 | ||||||
| AC238560 | C03HBa0030A11 | ju026 | 3 | F-AATCAATATCATCGCTTCACTG | 19 TA | 246, 292 | 48.9 |
| R-ATGTTGTGGTATTATTGACTTATGAG | 48.7 | ||||||
| AC238561 | C03HBa0031M05 | ju027 | 3 | F-ATGCTTAAGGTCTCCAAACACC | 5 CAA | 250 | 51.8 |
| R-CTCTCTACTTTTGGGATTACGC | 49.7 | ||||||
| EU124730 | C03HBa0001E24 | ju029 | 3 | F-TGCTGTACATACTGCATAAATGG | 7 TG | 350 | 50.1 |
| R-AACCTGCTGAATTAACTTGTAGTG | 49.5 | ||||||
| EU124737 | C03HBa0054O21 | ju035 | 3 | F-GTTATATAGAAAGACAAGGTAGAAGGTC | 25 AT | 288, 293 | 49.7 |
| R-GGTAGACTTTTTATGTGTTGTTGC | 49.7 | ||||||
| EU124739 | C03HBa0233O20 | ju037 | 3 | F-AAAATTGTTGGTCAACATGGTG | 7 TAT | 241, 246 | 51.6 |
| R-TTATCTCCTTTCCCTTTCATTC | 49 | ||||||
| EU124741 | C04HBa0318C22 | ju039 | 4 | F-GATGGTGTCATAGATCTAGCCTTAG | 6 TTAA | 355, 421 | 50.4 |
| R-TGGGGAATTATGTAGTGTTGAG | 48.7 | ||||||
| EU124742 | C03HBa0323D22 | ju040 | 3 | F-GCGATCCTGTTTGAGAAGAAGG | 5 CA | 340, 345 | 54.6 |
| R-ATGAACAAATGCTTAAGAGGGG | 52 | ||||||
| EU124743 | C03HBa0007J09 | ju041 | 3 | F-TTCCAAAAACACTTACGAAAGTTAG | 26 AT | 292, 316, 330 | 51.4 |
| R-CATGTAAGTCAAAAGAATGGAGG | 50.2 |
| Locus | No. of Alleles | Ho | He | PIC | PD |
|---|---|---|---|---|---|
| ju003 | 1 | 0 | 0 | 0 | 0.32 |
| ju004 | 2 | 0.24 | 0.41 | 0.32 | 0.72 |
| ju006 | 2 | 0.59 | 0.49 | 0.36 | 0.64 |
| ju007 | 2 | 0.81 | 0.5 | 0.37 | 0.34 |
| ju008 | 1 | 0 | 0 | 0 | 0.61 |
| ju010 | 1 | 0 | 0 | 0 | 0.73 |
| ju011 | 2 | 0.31 | 0.27 | 0.23 | 0.68 |
| ju014 | 1 | 0 | 0 | 0 | 0.52 |
| ju015 | 2 | 0.32 | 0.27 | 0.23 | 0.75 |
| ju017 | 1 | 0 | 0 | 0 | 0.37 |
| ju022 | 1 | 0 | 0 | 0 | 0.48 |
| ju023 | 2 | 0 | 0.42 | 0.32 | 0.96 |
| ju026 | 2 | 0.71 | 0.54 | 0.38 | 0.96 |
| ju027 | 1 | 0 | 0 | 0 | 0.73 |
| ju029 | 1 | 0 | 0 | 0 | 0.52 |
| ju035 | 2 | 0 | 0.42 | 0.32 | 0.94 |
| ju037 | 2 | 0 | 0.5 | 0.37 | 0.92 |
| ju039 | 2 | 0.48 | 0.37 | 0.3 | 0.72 |
| ju040 | 2 | 0.09 | 0.09 | 0.08 | 0.83 |
| ju041 | 3 | 0.26 | 0.51 | 0.38 | 0.91 |
| Total | 33 | - | - | - | |
| Mean | 1.65 | 0.19 | 0.24 | 0.18 | 0.68 |
| Max | 3 | 0.81 | 0.54 | 0.38 | 0.96 |
| Min | 1 | 0 | 0 | 0 | 0.32 |
| Locus | Nearby Gene | Functions | Reference |
|---|---|---|---|
| ju014, ju015, ju035 | Serine/threonine-protein kinase | Central processor unit (cpu): accepting input information from receptors that sense environmental conditions, phytohormones, and other external factors, and converting it into appropriate outputs, such as changes in metabolism, gene expression, and cell growth and division | [40] |
| ju017 | Serine carboxypeptidase | Stress response, growth, development, and pathogen defense | [41] |
| ju023 | Pirin | Role in seed germination and transcription of a light- and ABA-regulated gene under specific conditions in Arabidopsis thaliana | [42] |
| ju010. ju029 | F-box family protein | Plant hormonal signal transduction, floral development, secondary metabolism, senescence, circadian rhythms, and responses to both biotic and abiotic stresses | [43] |
| ju014, ju040 | Polygalacturonase | Major role in cell wall degradation and fruit softening | [44] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brake, M.; Al-Qadumii, L.; Hamasha, H.; Migdadi, H.; Awad, A.; Haddad, N.; Sadder, M.T. Development of SSR Markers Linked to Stress Responsive Genes along Tomato Chromosome 3 (Solanum lycopersicum L.). BioTech 2022, 11, 34. https://doi.org/10.3390/biotech11030034
Brake M, Al-Qadumii L, Hamasha H, Migdadi H, Awad A, Haddad N, Sadder MT. Development of SSR Markers Linked to Stress Responsive Genes along Tomato Chromosome 3 (Solanum lycopersicum L.). BioTech. 2022; 11(3):34. https://doi.org/10.3390/biotech11030034
Chicago/Turabian StyleBrake, Mohammad, Lana Al-Qadumii, Hassan Hamasha, Hussein Migdadi, Abi Awad, Nizar Haddad, and Monther T. Sadder. 2022. "Development of SSR Markers Linked to Stress Responsive Genes along Tomato Chromosome 3 (Solanum lycopersicum L.)" BioTech 11, no. 3: 34. https://doi.org/10.3390/biotech11030034







