Biomolecular Liquid–Liquid Phase Separation for Biotechnology
Abstract
:1. Introduction
2. Principles of Biomolecular LLPS
3. Biomolecular Droplets and Their Functions
3.1. Droplets in Cytoplasm
3.2. Droplets in Nucleus
3.3. Droplets in Membranes
3.4. Enzymes and Transcription Factors Undergoing LLPS
3.5. Droplets Discovered in Various Biological Processes
3.6. Artificial Droplet System
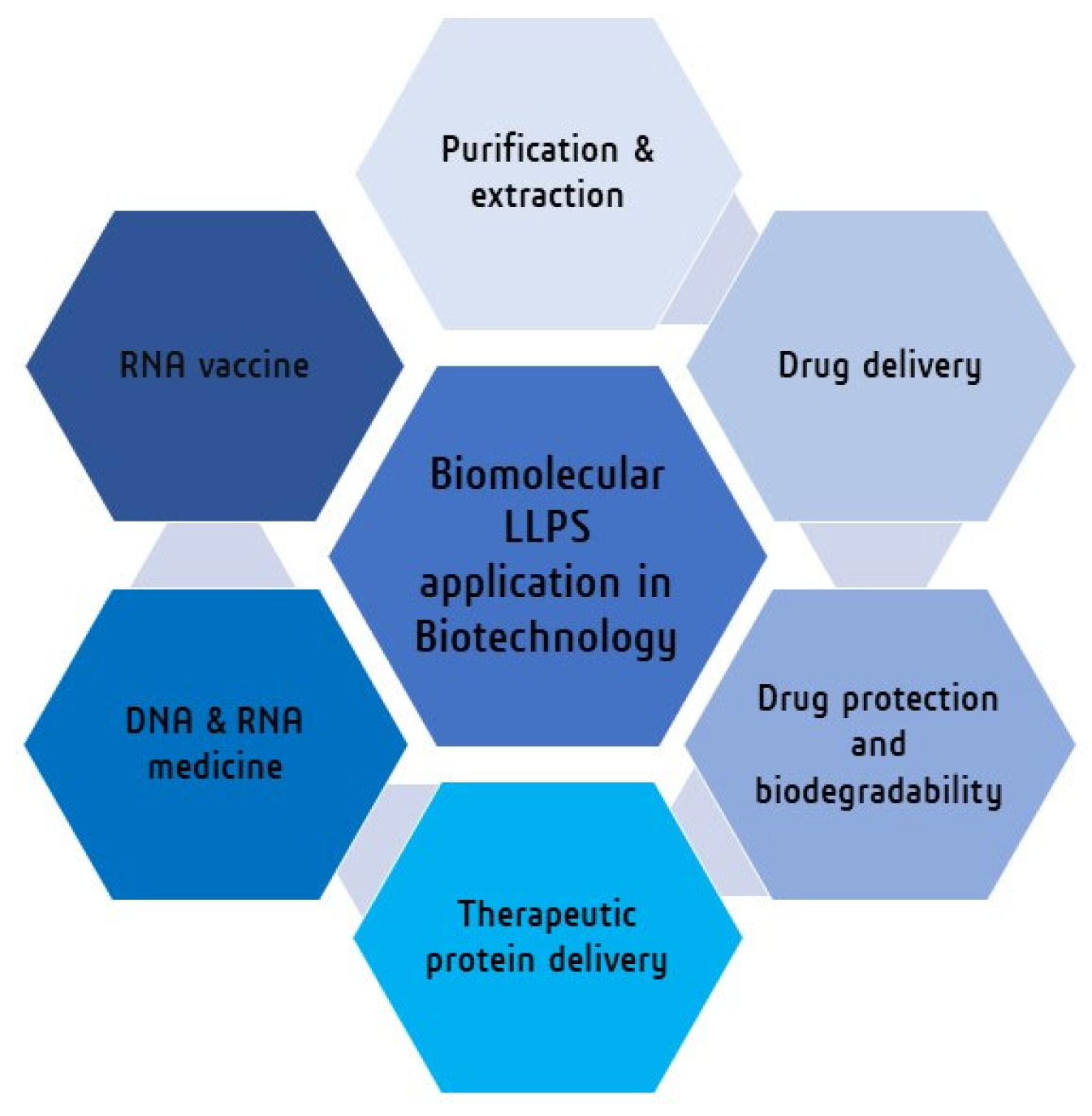
4. Biomolecular LLPS towards Biotechnology
| Biomolecular LLPS System | Application in Biotechnology | Ref. |
|---|---|---|
| ATPS system (PEG-phosphate) | Purification and extraction of biomolecules (DNA, RNA) | [168,169] |
| Sodium-alginate beads combined coacervate system | Drug protection | [162,173,174,175,176] |
| Polyester microspheres and artificial polymer system | Drug delivery | [177,178,179,180,181,182,183,184,185,186] |
| Amylose-based coacervates | Protein delivery | [187,188,189] |
| Lipid nanoparticles | DNA, RNA vaccines | [190,191,192,193,194,195,196,197] |
| Poly-lipoic ester base coactivates | Organic pollutant remover | [199] |
| LLPS system from the gel–sol transition of protein (gelatin solution) in a macromolecular crowding agent (PEG solution). | New material synthesis (protein microgel) | [198] |
| Carbohydrates and proteins-based LLPS system | Encapsulation of active substances (food industry) | [201,202] |
5. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Harmon, T.S.; Holehouse, A.S.; Rosen, M.K.; Pappu, R.V. Intrinsically disordered linkers determine the interplay between phase separation and gelation in multivalent proteins. eLife 2017, 6, e30294. [Google Scholar] [CrossRef]
- Kim, C.G.; Hwang, D.E.; Kumar, R.; Chung, M.; Eom, Y.G.; Kim, H.; Koo, D.H.; Choi, J.M. Recent trends in studies of biomolecular phase separation. BMB Rep. 2022, 55, 363–369. [Google Scholar] [CrossRef] [PubMed]
- Jia, T.Z.; Wang, P.H.; Niwa, T.; Mamajanov, I. Connecting primitive phase separation to biotechnology, synthetic biology, and engineering. J. Biosci. 2021, 46, 79. [Google Scholar] [CrossRef] [PubMed]
- Shin, Y.; Brangwynne, C.P. Liquid phase condensation in cell physiology and disease. Science 2017, 357, eaaf4382. [Google Scholar] [CrossRef] [Green Version]
- Gomes, E.; Shorter, J. The molecular language of membraneless organelles. J. Biol. Chem. 2019, 294, 7115–7127. [Google Scholar] [CrossRef] [Green Version]
- Ryu, J.K.; Hwang, D.E.; Choi, J.M. Current Understanding of Molecular Phase Separation in Chromosomes. Int. J. Mol. Sci. 2021, 22, 10736. [Google Scholar] [CrossRef] [PubMed]
- Banani, S.F.; Rice, A.M.; Peeples, W.B.; Lin, Y.; Jain, S.; Parker, R.; Rosen, M.K. Compositional Control of Phase-Separated Cellular Bodies. Cell 2016, 166, 651–663. [Google Scholar] [CrossRef] [Green Version]
- Feric, M.; Vaidya, N.; Harmon, T.S.; Mitrea, D.M.; Zhu, L.; Richardson, T.M.; Kriwacki, R.W.; Pappu, R.V.; Brangwynne, C.P. Coexisting Liquid Phases Underlie Nucleolar Subcompartments. Cell 2016, 165, 1686–1697. [Google Scholar] [CrossRef] [Green Version]
- Peng, P.H.; Hsu, K.W.; Wu, K.J. Liquid-liquid phase separation (LLPS) in cellular physiology and tumor biology. Am. J. Cancer Res. 2021, 11, 3766–3776. [Google Scholar]
- Cho, W.K.; Spille, J.H.; Hecht, M.; Lee, C.; Li, C.; Grube, V.; Cisse, I.I. Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science 2018, 361, 412–415. [Google Scholar] [CrossRef] [Green Version]
- Su, Q.; Mehta, S.; Zhang, J. Liquid-liquid phase separation: Orchestrating cell signaling through time and space. Mol. Cell 2021, 81, 4137–4146. [Google Scholar] [CrossRef]
- Kroschwald, S.; Alberti, S. Gel or Die: Phase Separation as a Survival Strategy. Cell 2017, 168, 947–948. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Banjade, S.; Rosen, M.K. Phase transitions of multivalent proteins can promote clustering of membrane receptors. eLife 2014, 3, e04123. [Google Scholar] [CrossRef] [PubMed]
- Riback, J.A.; Katanski, C.D.; Kear-Scott, J.L.; Pilipenko, E.V.; Rojek, A.E.; Sosnick, T.R.; Drummond, D.A. Stress-Triggered Phase Separation Is an Adaptive, Evolutionarily Tuned Response. Cell 2017, 168, 1028–1040.e1019. [Google Scholar] [CrossRef] [Green Version]
- Molliex, A.; Temirov, J.; Lee, J.; Coughlin, M.; Kanagaraj, A.P.; Kim, H.J.; Mittag, T.; Taylor, J.P. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 2015, 163, 123–133. [Google Scholar] [CrossRef] [Green Version]
- Nicolas, E.; Parisot, P.; Pinto-Monteiro, C.; de Walque, R.; De Vleeschouwer, C.; Lafontaine, D.L. Involvement of human ribosomal proteins in nucleolar structure and p53-dependent nucleolar stress. Nat. Commun. 2016, 7, 11390. [Google Scholar] [CrossRef] [Green Version]
- Sheu-Gruttadauria, J.; MacRae, I.J. Phase Transitions in the Assembly and Function of Human miRISC. Cell 2018, 173, 946–957.e916. [Google Scholar] [CrossRef]
- Xiao, Q.; McAtee, C.K.; Su, X. Phase separation in immune signalling. Nat. Rev. Immunol. 2022, 22, 188–199. [Google Scholar] [CrossRef] [PubMed]
- Wippich, F.; Bodenmiller, B.; Trajkovska, M.G.; Wanka, S.; Aebersold, R.; Pelkmans, L. Dual specificity kinase DYRK3 couples stress granule condensation/dissolution to mTORC1 signaling. Cell 2013, 152, 791–805. [Google Scholar] [CrossRef] [Green Version]
- Fujioka, Y.; Alam, J.M.; Noshiro, D.; Mouri, K.; Ando, T.; Okada, Y.; May, A.I.; Knorr, R.L.; Suzuki, K.; Ohsumi, Y.; et al. Phase separation organizes the site of autophagosome formation. Nature 2020, 578, 301–305. [Google Scholar] [CrossRef]
- Tong, X.; Tang, R.; Xu, J.; Wang, W.; Zhao, Y.; Yu, X.; Shi, S. Liquid-liquid phase separation in tumor biology. Signal. Transduct. Target. Ther. 2022, 7, 221. [Google Scholar] [CrossRef] [PubMed]
- Sawner, A.S.; Ray, S.; Yadav, P.; Mukherjee, S.; Panigrahi, R.; Poudyal, M.; Patel, K.; Ghosh, D.; Kummerant, E.; Kumar, A.; et al. Modulating α-Synuclein Liquid-Liquid Phase Separation. Biochemistry 2021, 60, 3676–3696. [Google Scholar] [CrossRef] [PubMed]
- Boyko, S.; Surewicz, W.K. Tau liquid-liquid phase separation in neurodegenerative diseases. Trends Cell Biol. 2022, 32, 611–623. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Zhang, L.; Dai, T.; Qin, Z.; Lu, H.; Zhang, L.; Zhou, F. Liquid-liquid phase separation in human health and diseases. Signal. Transduct. Target. Ther. 2021, 6, 290. [Google Scholar] [CrossRef]
- Alberti, S.; Dormann, D. Liquid-Liquid Phase Separation in Disease. Annu. Rev. Genet. 2019, 53, 171–194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abbas, M.; Lipiński, W.P.; Wang, J.; Spruijt, E. Peptide-based coacervates as biomimetic protocells. Chem. Soc. Rev. 2021, 50, 3690–3705. [Google Scholar] [CrossRef]
- Espinosa, J.R.; Joseph, J.A.; Sanchez-Burgos, I.; Garaizar, A.; Frenkel, D.; Collepardo-Guevara, R. Liquid network connectivity regulates the stability and composition of biomolecular condensates with many components. Proc. Natl. Acad. Sci. USA 2020, 117, 13238–13247. [Google Scholar] [CrossRef]
- Li, P.; Banjade, S.; Cheng, H.C.; Kim, S.; Chen, B.; Guo, L.; Llaguno, M.; Hollingsworth, J.V.; King, D.S.; Banani, S.F.; et al. Phase transitions in the assembly of multivalent signalling proteins. Nature 2012, 483, 336–340. [Google Scholar] [CrossRef] [Green Version]
- Freeman Rosenzweig, E.S.; Xu, B.; Kuhn Cuellar, L.; Martinez-Sanchez, A.; Schaffer, M.; Strauss, M.; Cartwright, H.N.; Ronceray, P.; Plitzko, J.M.; Förster, F.; et al. The Eukaryotic CO(2)-Concentrating Organelle Is Liquid-like and Exhibits Dynamic Reorganization. Cell 2017, 171, 148–162.e119. [Google Scholar] [CrossRef] [Green Version]
- Roden, C.; Gladfelter, A.S. RNA contributions to the form and function of biomolecular condensates. Nat. Rev. Mol. Cell Biol. 2021, 22, 183–195. [Google Scholar] [CrossRef]
- Choi, J.M.; Holehouse, A.S.; Pappu, R.V. Physical Principles Underlying the Complex Biology of Intracellular Phase Transitions. Annu. Rev. Biophys. 2020, 49, 107–133. [Google Scholar] [CrossRef] [Green Version]
- Hong, K.; Song, D.; Jung, Y. Behavior control of membrane-less protein liquid condensates with metal ion-induced phase separation. Nat. Commun. 2020, 11, 5554. [Google Scholar] [CrossRef]
- Onuchic, P.L.; Milin, A.N.; Alshareedah, I.; Deniz, A.A.; Banerjee, P.R. Divalent cations can control a switch-like behavior in heterotypic and homotypic RNA coacervates. Sci. Rep. 2019, 9, 12161. [Google Scholar] [CrossRef] [Green Version]
- Singh, V.; Xu, L.; Boyko, S.; Surewicz, K.; Surewicz, W.K. Zinc promotes liquid-liquid phase separation of tau protein. J. Biol. Chem. 2020, 295, 5850–5856. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vega, I.E.; Umstead, A.; Kanaan, N.M. EFhd2 Affects Tau Liquid-Liquid Phase Separation. Front. Neurosci. 2019, 13, 845. [Google Scholar] [CrossRef] [Green Version]
- Oldfield, C.J.; Dunker, A.K. Intrinsically disordered proteins and intrinsically disordered protein regions. Annu. Rev. Biochem. 2014, 83, 553–584. [Google Scholar] [CrossRef]
- Choi, J.M.; Hyman, A.A.; Pappu, R.V. Generalized models for bond percolation transitions of associative polymers. Phys. Rev. E 2020, 102, 042403. [Google Scholar] [CrossRef]
- Banani, S.F.; Lee, H.O.; Hyman, A.A.; Rosen, M.K. Biomolecular condensates: Organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 2017, 18, 285–298. [Google Scholar] [CrossRef] [PubMed]
- Moser, J.J.; Fritzler, M.J. Cytoplasmic ribonucleoprotein (RNP) bodies and their relationship to GW/P bodies. Int. J. Biochem. Cell Biol. 2010, 42, 828–843. [Google Scholar] [CrossRef]
- Crisp, M.; Liu, Q.; Roux, K.; Rattner, J.B.; Shanahan, C.; Burke, B.; Stahl, P.D.; Hodzic, D. Coupling of the nucleus and cytoplasm: Role of the LINC complex. J. Cell Biol. 2006, 172, 41–53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hofmann, S.; Kedersha, N.; Anderson, P.; Ivanov, P. Molecular mechanisms of stress granule assembly and disassembly. Biochim. Biophys. Acta Mol. Cell Res. 2021, 1868, 118876. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, S.; Cherkasova, V.; Bankhead, P.; Bukau, B.; Stoecklin, G. Translation suppression promotes stress granule formation and cell survival in response to cold shock. Mol. Biol. Cell 2012, 23, 3786–3800. [Google Scholar] [CrossRef] [PubMed]
- Campos-Melo, D.; Hawley, Z.C.E.; Droppelmann, C.A.; Strong, M.J. The Integral Role of RNA in Stress Granule Formation and Function. Front. Cell Dev. Biol. 2021, 9, 621779. [Google Scholar] [CrossRef] [PubMed]
- Wolozin, B.; Ivanov, P. Stress granules and neurodegeneration. Nat. Rev. Neurosci. 2019, 20, 649–666. [Google Scholar] [CrossRef] [PubMed]
- Decker, C.J.; Parker, R. P-bodies and stress granules: Possible roles in the control of translation and mRNA degradation. Cold Spring Harb. Perspect. Biol. 2012, 4, a012286. [Google Scholar] [CrossRef] [Green Version]
- Van Treeck, B.; Parker, R. Principles of Stress Granules Revealed by Imaging Approaches. Cold Spring Harb. Perspect. Biol. 2019, 11, a033068. [Google Scholar] [CrossRef] [Green Version]
- Woodruff, J.B.; Ferreira Gomes, B.; Widlund, P.O.; Mahamid, J.; Honigmann, A.; Hyman, A.A. The Centrosome Is a Selective Condensate that Nucleates Microtubules by Concentrating Tubulin. Cell 2017, 169, 1066–1077.e1010. [Google Scholar] [CrossRef] [Green Version]
- Frikstad, K.M.; Schink, K.O.; Gilani, S.; Pedersen, L.B.; Patzke, S. 3D-Structured Illumination Microscopy of Centrosomes in Human Cell Lines. Bio Protoc. 2022, 12, e4360. [Google Scholar] [CrossRef]
- Gonçalves, A.B.; Hasselbalch, S.K.; Joensen, B.B.; Patzke, S.; Martens, P.; Ohlsen, S.K.; Quinodoz, M.; Nikopoulos, K.; Suleiman, R.; Damsø Jeppesen, M.P.; et al. CEP78 functions downstream of CEP350 to control biogenesis of primary cilia by negatively regulating CP110 levels. eLife 2021, 10, e63731. [Google Scholar] [CrossRef]
- Liu, J.L.; Gall, J.G. U bodies are cytoplasmic structures that contain uridine-rich small nuclear ribonucleoproteins and associate with P bodies. Proc. Natl. Acad. Sci. USA 2007, 104, 11655–11659. [Google Scholar] [CrossRef] [Green Version]
- O’Flynn, B.G.; Mittag, T. The role of liquid-liquid phase separation in regulating enzyme activity. Curr. Opin. Cell Biol. 2021, 69, 70–79. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Xu, Y.; Stoleru, D.; Salic, A. Imaging protein synthesis in cells and tissues with an alkyne analog of puromycin. Proc. Natl. Acad. Sci. USA 2012, 109, 413–418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, M.; Fuller, G.G.; Han, T.; Yao, Y.; Alessi, A.F.; Freeberg, M.A.; Roach, N.P.; Moresco, J.J.; Karnovsky, A.; Baba, M.; et al. Glycolytic Enzymes Coalesce in G Bodies under Hypoxic Stress. Cell Rep. 2017, 20, 895–908. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, Y.; Na, Z.; Slavoff, S.A. P-Bodies: Composition, Properties, and Functions. Biochemistry 2018, 57, 2424–2431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elbaum-Garfinkle, S.; Kim, Y.; Szczepaniak, K.; Chen, C.C.; Eckmann, C.R.; Myong, S.; Brangwynne, C.P. The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proc. Natl. Acad. Sci. USA 2015, 112, 7189–7194. [Google Scholar] [CrossRef] [Green Version]
- Boke, E.; Ruer, M.; Wühr, M.; Coughlin, M.; Lemaitre, R.; Gygi, S.P.; Alberti, S.; Drechsel, D.; Hyman, A.A.; Mitchison, T.J. Amyloid-like Self-Assembly of a Cellular Compartment. Cell 2016, 166, 637–650. [Google Scholar] [CrossRef] [Green Version]
- Jamieson-Lucy, A.; Mullins, M.C. The vertebrate Balbiani body, germ plasm, and oocyte polarity. Curr. Top. Dev. Biol. 2019, 135, 1–34. [Google Scholar] [CrossRef]
- Lee, K.L.; Marlow, F.L. Visualizing the Balbiani Body in Zebrafish Oocytes. Methods Mol. Biol. 2019, 1920, 277–293. [Google Scholar] [CrossRef]
- Voronina, E.; Seydoux, G.; Sassone-Corsi, P.; Nagamori, I. RNA granules in germ cells. Cold Spring Harb. Perspect. Biol. 2011, 3, a002774. [Google Scholar] [CrossRef] [Green Version]
- Brangwynne, C.P.; Eckmann, C.R.; Courson, D.S.; Rybarska, A.; Hoege, C.; Gharakhani, J.; Jülicher, F.; Hyman, A.A. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 2009, 324, 1729–1732. [Google Scholar] [CrossRef]
- Saha, S.; Weber, C.A.; Nousch, M.; Adame-Arana, O.; Hoege, C.; Hein, M.Y.; Osborne-Nishimura, E.; Mahamid, J.; Jahnel, M.; Jawerth, L.; et al. Polar Positioning of Phase-Separated Liquid Compartments in Cells Regulated by an mRNA Competition Mechanism. Cell 2016, 166, 1572–1584.e1516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kiebler, M.A.; Bassell, G.J. Neuronal RNA granules: Movers and makers. Neuron 2006, 51, 685–690. [Google Scholar] [CrossRef] [Green Version]
- Bauer, K.E.; Bargenda, N.; Schieweck, R.; Illig, C.; Segura, I.; Harner, M.; Kiebler, M.A. RNA supply drives physiological granule assembly in neurons. Nat. Commun. 2022, 13, 2781. [Google Scholar] [CrossRef] [PubMed]
- Pederson, T. The nucleolus. Cold Spring Harb. Perspect. Biol. 2011, 3, a000638. [Google Scholar] [CrossRef]
- Yang, K.; Yang, J.; Yi, J. Nucleolar Stress: Hallmarks, sensing mechanism and diseases. Cell Stress 2018, 2, 125–140. [Google Scholar] [CrossRef] [PubMed]
- Boisvert, F.M.; van Koningsbruggen, S.; Navascués, J.; Lamond, A.I. The multifunctional nucleolus. Nat. Rev. Mol. Cell Biol. 2007, 8, 574–585. [Google Scholar] [CrossRef]
- Brangwynne, C.P.; Mitchison, T.J.; Hyman, A.A. Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc. Natl. Acad. Sci. USA 2011, 108, 4334–4339. [Google Scholar] [CrossRef] [Green Version]
- Yoneda, M.; Nakagawa, T.; Hattori, N.; Ito, T. The nucleolus from a liquid droplet perspective. J. Biochem. 2021, 170, 153–162. [Google Scholar] [CrossRef]
- Nizami, Z.; Deryusheva, S.; Gall, J.G. The Cajal body and histone locus body. Cold Spring Harb. Perspect. Biol. 2010, 2, a000653. [Google Scholar] [CrossRef] [Green Version]
- Duronio, R.J.; Marzluff, W.F. Coordinating cell cycle-regulated histone gene expression through assembly and function of the Histone Locus Body. RNA Biol. 2017, 14, 726–738. [Google Scholar] [CrossRef] [Green Version]
- Larson, A.G.; Elnatan, D.; Keenen, M.M.; Trnka, M.J.; Johnston, J.B.; Burlingame, A.L.; Agard, D.A.; Redding, S.; Narlikar, G.J. Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature 2017, 547, 236–240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, C.W.; Shen, Y.C.; Yan, S.J. HP1a-mediated heterochromatin formation inhibits high dietary sugar-induced tumor progression. Cell Death Dis. 2021, 12, 1130. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, D. HP1a/KDM4A is involved in the autoregulatory loop of the oncogene gene c-Jun. Epigenetics 2015, 10, 453–459. [Google Scholar] [CrossRef] [Green Version]
- Tatarakis, A.; Behrouzi, R.; Moazed, D. Evolving Models of Heterochromatin: From Foci to Liquid Droplets. Mol. Cell 2017, 67, 725–727. [Google Scholar] [CrossRef] [Green Version]
- Strom, A.R.; Biggs, R.J.; Banigan, E.J.; Wang, X.; Chiu, K.; Herman, C.; Collado, J.; Yue, F.; Ritland Politz, J.C.; Tait, L.J.; et al. HP1α is a chromatin crosslinker that controls nuclear and mitotic chromosome mechanics. eLife 2021, 10, e63972. [Google Scholar] [CrossRef] [PubMed]
- Erdel, F.; Rademacher, A.; Vlijm, R.; Tünnermann, J.; Frank, L.; Weinmann, R.; Schweigert, E.; Yserentant, K.; Hummert, J.; Bauer, C.; et al. Mouse Heterochromatin Adopts Digital Compaction States without Showing Hallmarks of HP1-Driven Liquid-Liquid Phase Separation. Mol. Cell. 2020, 78, 236–249.e237. [Google Scholar] [CrossRef]
- Capitanio, J.S.; Montpetit, B.; Wozniak, R.W. Human Nup98 regulates the localization and activity of DExH/D-box helicase DHX9. eLife 2017, 6, e18825. [Google Scholar] [CrossRef] [PubMed]
- Wong, R.W. New Activities of the Nuclear Pore Complexes. Cells 2021, 10, 2123. [Google Scholar] [CrossRef]
- Beck, M.; Hurt, E. The nuclear pore complex: Understanding its function through structural insight. Nat. Rev. Mol. Cell Biol. 2017, 18, 73–89. [Google Scholar] [CrossRef] [PubMed]
- Kumanski, S.; Viart, B.T.; Kossida, S.; Moriel-Carretero, M. Lipid Droplets Are a Physiological Nucleoporin Reservoir. Cells 2021, 10, 472. [Google Scholar] [CrossRef]
- Spector, D.L.; Lamond, A.I. Nuclear speckles. Cold Spring Harb. Perspect. Biol. 2011, 3, a000646. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.; Han, K.Y.; Khanna, N.; Ha, T.; Belmont, A.S. Nuclear speckle fusion via long-range directional motion regulates speckle morphology after transcriptional inhibition. J. Cell Sci. 2019, 132, jcs226563. [Google Scholar] [CrossRef] [Green Version]
- Rothkamm, K.; Barnard, S.; Moquet, J.; Ellender, M.; Rana, Z.; Burdak-Rothkamm, S. DNA damage foci: Meaning and significance. Environ. Mol. Mutagen. 2015, 56, 491–504. [Google Scholar] [CrossRef]
- Oshidari, R.; Huang, R.; Medghalchi, M.; Tse, E.Y.W.; Ashgriz, N.; Lee, H.O.; Wyatt, H.; Mekhail, K. DNA repair by Rad52 liquid droplets. Nat. Commun. 2020, 11, 695. [Google Scholar] [CrossRef] [Green Version]
- Morimoto, M.; Boerkoel, C.F. The role of nuclear bodies in gene expression and disease. Biology 2013, 2, 976–1033. [Google Scholar] [CrossRef] [Green Version]
- Cauchi, R.J. Gem formation upon constitutive Gemin3 overexpression in Drosophila. Cell Biol. Int. 2011, 35, 1233–1238. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Dreyfuss, G. A novel nuclear structure containing the survival of motor neurons protein. Embo J. 1996, 15, 3555–3565. [Google Scholar] [CrossRef] [PubMed]
- Pirrotta, V.; Li, H.B. A view of nuclear Polycomb bodies. Curr. Opin. Genet. Dev. 2012, 22, 101–109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smigová, J.; Juda, P.; Cmarko, D.; Raška, I. Fine structure of the “PcG body” in human U-2 OS cells established by correlative light-electron microscopy. Nucleus 2011, 2, 219–228. [Google Scholar] [CrossRef] [Green Version]
- Fox, A.H.; Lamond, A.I. Paraspeckles. Cold Spring Harb. Perspect. Biol. 2010, 2, a000687. [Google Scholar] [CrossRef] [Green Version]
- Hennig, S.; Kong, G.; Mannen, T.; Sadowska, A.; Kobelke, S.; Blythe, A.; Knott, G.J.; Iyer, K.S.; Ho, D.; Newcombe, E.A.; et al. Prion-like domains in RNA binding proteins are essential for building subnuclear paraspeckles. J. Cell Biol. 2015, 210, 529–539. [Google Scholar] [CrossRef] [Green Version]
- Ohnishi, Y.; Huber, W.; Tsumura, A.; Kang, M.; Xenopoulos, P.; Kurimoto, K.; Oleś, A.K.; Araúzo-Bravo, M.J.; Saitou, M.; Hadjantonakis, A.K.; et al. Cell-to-cell expression variability followed by signal reinforcement progressively segregates early mouse lineages. Nat. Cell Biol. 2014, 16, 27–37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jarc, E.; Petan, T. Lipid Droplets and the Management of Cellular Stress. Yale J. Biol. Med. 2019, 92, 435–452. [Google Scholar] [PubMed]
- Sheng, M.; Kim, E. The postsynaptic organization of synapses. Cold Spring Harb. Perspect. Biol. 2011, 3, a005678. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeng, M.; Shang, Y.; Araki, Y.; Guo, T.; Huganir, R.L.; Zhang, M. Phase Transition in Postsynaptic Densities Underlies Formation of Synaptic Complexes and Synaptic Plasticity. Cell 2016, 166, 1163–1175.e1112. [Google Scholar] [CrossRef] [Green Version]
- Case, L.B.; Waterman, C.M. Integration of actin dynamics and cell adhesion by a three-dimensional, mechanosensitive molecular clutch. Nat. Cell Biol. 2015, 17, 955–963. [Google Scholar] [CrossRef] [PubMed]
- Case, L.B.; De Pasquale, M.; Henry, L.; Rosen, M.K. Synergistic phase separation of two pathways promotes integrin clustering and nascent adhesion formation. eLife 2022, 11, e72588. [Google Scholar] [CrossRef]
- Jones, N.; Blasutig, I.M.; Eremina, V.; Ruston, J.M.; Bladt, F.; Li, H.; Huang, H.; Larose, L.; Li, S.S.; Takano, T.; et al. Nck adaptor proteins link nephrin to the actin cytoskeleton of kidney podocytes. Nature 2006, 440, 818–823. [Google Scholar] [CrossRef]
- Martin, C.E.; New, L.A.; Phippen, N.J.; Keyvani Chahi, A.; Mitro, A.E.; Takano, T.; Pawson, T.; Blasutig, I.M.; Jones, N. Multivalent nephrin-Nck interactions define a threshold for clustering and tyrosine-dependent nephrin endocytosis. J. Cell Sci. 2020, 133, jcs236877. [Google Scholar] [CrossRef]
- Dustin, M.L.; Groves, J.T. Receptor signaling clusters in the immune synapse. Annu. Rev. Biophys. 2012, 41, 543–556. [Google Scholar] [CrossRef] [Green Version]
- Su, X.; Ditlev, J.A.; Hui, E.; Xing, W.; Banjade, S.; Okrut, J.; King, D.S.; Taunton, J.; Rosen, M.K.; Vale, R.D. Phase separation of signaling molecules promotes T cell receptor signal transduction. Science 2016, 352, 595–599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calle, Y.; Burns, S.; Thrasher, A.J.; Jones, G.E. The leukocyte podosome. Eur. J. Cell Biol. 2006, 85, 151–157. [Google Scholar] [CrossRef] [PubMed]
- Vincent, C.; Siddiqui, T.A.; Schlichter, L.C. Podosomes in migrating microglia: Components and matrix degradation. J. Neuroinflamm. 2012, 9, 190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goode, B.L.; Eskin, J.A.; Wendland, B. Actin and endocytosis in budding yeast. Genetics 2015, 199, 315–358. [Google Scholar] [CrossRef] [Green Version]
- Moseley, J.B.; Goode, B.L. The yeast actin cytoskeleton: From cellular function to biochemical mechanism. Microbiol. Mol. Biol. Rev. 2006, 70, 605–645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olzmann, J.A.; Carvalho, P. Dynamics and functions of lipid droplets. Nat. Rev. Mol. Cell Biol. 2019, 20, 137–155. [Google Scholar] [CrossRef] [PubMed]
- Johnson, V.M.; Pakrasi, H.B. Advances in the Understanding of the Lifecycle of Photosystem II. Microorganisms 2022, 10, 836. [Google Scholar] [CrossRef] [PubMed]
- Müh, F.; Zouni, A. Structural basis of light-harvesting in the photosystem II core complex. Protein Sci. 2020, 29, 1090–1119. [Google Scholar] [CrossRef] [Green Version]
- Järvi, S.; Suorsa, M.; Aro, E.M. Photosystem II repair in plant chloroplasts--Regulation, assisting proteins and shared components with photosystem II biogenesis. Biochim. Biophys. Acta 2015, 1847, 900–909. [Google Scholar] [CrossRef] [Green Version]
- Saini, B.; Mukherjee, T.K. Biomolecular Condensates Regulate Enzymatic Activity under a Crowded Milieu: Synchronization of Liquid–Liquid Phase Separation and Enzymatic Transformation. J. Phys. Chem. B 2023, 127, 180–193. [Google Scholar] [CrossRef]
- Tang, S.C.; Vijayakumar, U.; Zhang, Y.; Fullwood, M.J. Super-Enhancers, Phase-Separated Condensates, and 3D Genome Organization in Cancer. Cancers 2022, 14, 2866. [Google Scholar] [CrossRef]
- Richter, W.F.; Nayak, S.; Iwasa, J.; Taatjes, D.J. The Mediator complex as a master regulator of transcription by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 2022, 23, 732–749. [Google Scholar] [CrossRef] [PubMed]
- Sharp, P.A.; Chakraborty, A.K.; Henninger, J.E.; Young, R.A. RNA in formation and regulation of transcriptional condensates. RNA 2022, 28, 52–57. [Google Scholar] [CrossRef]
- Bauer, J.A.; Zámocká, M.; Majtán, J.; Bauerová-Hlinková, V. Glucose Oxidase, an Enzyme “Ferrari”: Its Structure, Function, Production and Properties in the Light of Various Industrial and Biotechnological Applications. Biomolecules 2022, 12, 472. [Google Scholar] [CrossRef]
- Koga, S.; Williams, D.S.; Perriman, A.W.; Mann, S. Peptide-nucleotide microdroplets as a step towards a membrane-free protocell model. Nat. Chem. 2011, 3, 720–724. [Google Scholar] [CrossRef] [PubMed]
- Lindhoud, S.; Norde, W.; Cohen Stuart, M.A. Effects of Polyelectrolyte Complex Micelles and Their Components on the Enzymatic Activity of Lipase. Langmuir 2010, 26, 9802–9808. [Google Scholar] [CrossRef]
- Wang, L.; Lin, Y.; Zhou, Y.; Xie, H.; Song, J.; Li, M.; Huang, Y.; Huang, X.; Mann, S. Autonomic Behaviors in Lipase-Active Oil Droplets. Angew. Chem. Int. Ed. Engl. 2019, 58, 1067–1071. [Google Scholar] [CrossRef] [Green Version]
- Strulson, C.A.; Molden, R.C.; Keating, C.D.; Bevilacqua, P.C. RNA catalysis through compartmentalization. Nat. Chem. 2012, 4, 941–946. [Google Scholar] [CrossRef] [PubMed]
- Drobot, B.; Iglesias-Artola, J.M.; Le Vay, K.; Mayr, V.; Kar, M.; Kreysing, M.; Mutschler, H.; Tang, T.D. Compartmentalised RNA catalysis in membrane-free coacervate protocells. Nat. Commun. 2018, 9, 3643. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fuller, G.G.; Han, T.; Freeberg, M.A.; Moresco, J.J.; Ghanbari Niaki, A.; Roach, N.P.; Yates, J.R., III; Myong, S.; Kim, J.K. RNA promotes phase separation of glycolysis enzymes into yeast G bodies in hypoxia. eLife 2020, 9, e48480. [Google Scholar] [CrossRef]
- Zhu, J.; Zhou, Q.; Xia, Y.; Lin, L.; Li, J.; Peng, M.; Zhang, R.; Zhang, M. GIT/PIX Condensates Are Modular and Ideal for Distinct Compartmentalized Cell Signaling. Mol. Cell 2020, 79, 782–796.e786. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, K.; Ohtsuki, T. Inhibition of HSF1 and SAFB Granule Formation Enhances Apoptosis Induced by Heat Stress. Int. J. Mol. Sci. 2021, 22, 4982. [Google Scholar] [CrossRef] [PubMed]
- Gaglia, G.; Rashid, R.; Yapp, C.; Joshi, G.N.; Li, C.G.; Lindquist, S.L.; Sarosiek, K.A.; Whitesell, L.; Sorger, P.K.; Santagata, S. HSF1 phase transition mediates stress adaptation and cell fate decisions. Nat. Cell Biol. 2020, 22, 151–158. [Google Scholar] [CrossRef]
- Rawat, P.; Boehning, M.; Hummel, B.; Aprile-Garcia, F.; Pandit, A.S.; Eisenhardt, N.; Khavaran, A.; Niskanen, E.; Vos, S.M.; Palvimo, J.J.; et al. Stress-induced nuclear condensation of NELF drives transcriptional downregulation. Mol. Cell 2021, 81, 1013–1026.e1011. [Google Scholar] [CrossRef]
- Boija, A.; Klein, I.A.; Sabari, B.R.; Dall’Agnese, A.; Coffey, E.L.; Zamudio, A.V.; Li, C.H.; Shrinivas, K.; Manteiga, J.C.; Hannett, N.M.; et al. Transcription Factors Activate Genes through the Phase-Separation Capacity of Their Activation Domains. Cell 2018, 175, 1842–1855.e1816. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Safari, M.S.; Wang, Z.; Tailor, K.; Kolomeisky, A.B.; Conrad, J.C.; Vekilov, P.G. Anomalous Dense Liquid Condensates Host the Nucleation of Tumor Suppressor p53 Fibrils. iScience 2019, 12, 342–355. [Google Scholar] [CrossRef] [Green Version]
- Park, S.K.; Park, S.; Pentek, C.; Liebman, S.W. Tumor suppressor protein p53 expressed in yeast can remain diffuse, form a prion, or form unstable liquid-like droplets. iScience 2021, 24, 102000. [Google Scholar] [CrossRef]
- Kamagata, K.; Kanbayashi, S.; Honda, M.; Itoh, Y.; Takahashi, H.; Kameda, T.; Nagatsugi, F.; Takahashi, S. Liquid-like droplet formation by tumor suppressor p53 induced by multivalent electrostatic interactions between two disordered domains. Sci. Rep. 2020, 10, 580. [Google Scholar] [CrossRef] [Green Version]
- Lemos, C.; Schulze, L.; Weiske, J.; Meyer, H.; Braeuer, N.; Barak, N.; Eberspächer, U.; Werbeck, N.; Stresemann, C.; Lange, M.; et al. Identification of Small Molecules that Modulate Mutant p53 Condensation. iScience 2020, 23, 101517. [Google Scholar] [CrossRef]
- Petronilho, E.C.; Pedrote, M.M.; Marques, M.A.; Passos, Y.M.; Mota, M.F.; Jakobus, B.; Sousa, G.d.S.d.; Pereira da Costa, F.; Felix, A.L.; Ferretti, G.D.S.; et al. Phase separation of p53 precedes aggregation and is affected by oncogenic mutations and ligands. Chem. Sci. 2021, 12, 7334–7349. [Google Scholar] [CrossRef]
- Park, J.E.; Zhang, L.; Bang, J.K.; Andresson, T.; DiMaio, F.; Lee, K.S. Phase separation of Polo-like kinase 4 by autoactivation and clustering drives centriole biogenesis. Nat. Commun. 2019, 10, 4959. [Google Scholar] [CrossRef] [Green Version]
- Vitiello, E.; Moreau, P.; Nunes, V.; Mettouchi, A.; Maiato, H.; Ferreira, J.G.; Wang, I.; Balland, M. Acto-myosin force organization modulates centriole separation and PLK4 recruitment to ensure centriole fidelity. Nat. Commun. 2019, 10, 52. [Google Scholar] [CrossRef]
- Krainer, G.; Welsh, T.J.; Joseph, J.A.; Espinosa, J.R.; Wittmann, S.; de Csilléry, E.; Sridhar, A.; Toprakcioglu, Z.; Gudiškytė, G.; Czekalska, M.A.; et al. Reentrant liquid condensate phase of proteins is stabilized by hydrophobic and non-ionic interactions. Nat. Commun. 2021, 12, 1085. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Gan, W.; Han, X.; Liu, N.; Ma, T.; Li, D. The positive regulation loop between NRF1 and NONO-TFE3 fusion promotes phase separation and aggregation of NONO-TFE3 in NONO-TFE3 tRCC. Int. J. Biol. Macromol. 2021, 176, 437–447. [Google Scholar] [CrossRef]
- Chen, D.; Wang, Z.; Zhao, Y.G.; Zheng, H.; Zhao, H.; Liu, N.; Zhang, H. Inositol Polyphosphate Multikinase Inhibits Liquid-Liquid Phase Separation of TFEB to Negatively Regulate Autophagy Activity. Dev. Cell 2020, 55, 588–602.e587. [Google Scholar] [CrossRef]
- Liu, S.; Wang, T.; Shi, Y.; Bai, L.; Wang, S.; Guo, D.; Zhang, Y.; Qi, Y.; Chen, C.; Zhang, J.; et al. USP42 drives nuclear speckle mRNA splicing via directing dynamic phase separation to promote tumorigenesis. Cell Death Differ. 2021, 28, 2482–2498. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Peng, Z.; Qin, M.; Liu, Y.; Wang, J.; Zhang, C.; Lin, J.; Dong, T.; Wang, L.; Li, S.; et al. Interferon-γ induces tumor resistance to anti-PD-1 immunotherapy by promoting YAP phase separation. Mol. Cell 2021, 81, 1216–1230.e1219. [Google Scholar] [CrossRef]
- Cai, D.; Feliciano, D.; Dong, P.; Flores, E.; Gruebele, M.; Porat-Shliom, N.; Sukenik, S.; Liu, Z.; Lippincott-Schwartz, J. Phase separation of YAP reorganizes genome topology for long-term YAP target gene expression. Nat. Cell Biol. 2019, 21, 1578–1589. [Google Scholar] [CrossRef] [PubMed]
- Barrett, J.; Girr, P.; Mackinder, L.C.M. Pyrenoids: CO(2)-fixing phase separated liquid organelles. Biochim. Biophys. Acta Mol. Cell Res. 2021, 1868, 118949. [Google Scholar] [CrossRef]
- Wunder, T.; Mueller-Cajar, O. Biomolecular condensates in photosynthesis and metabolism. Curr. Opin. Plant Biol. 2020, 58, 1–7. [Google Scholar] [CrossRef]
- Gao, Z.; Zhang, W.; Chang, R.; Zhang, S.; Yang, G.; Zhao, G. Liquid-Liquid Phase Separation: Unraveling the Enigma of Biomolecular Condensates in Microbial Cells. Front. Microbiol. 2021, 12, 751880. [Google Scholar] [CrossRef] [PubMed]
- Lach, R.S.; Qiu, C.; Kajbaf, E.Z.; Baxter, N.; Han, D.; Wang, A.; Lock, H.; Chirikian, O.; Pruitt, B.; Wilson, M.Z. Nucleation of the destruction complex on the centrosome accelerates degradation of β-catenin and regulates Wnt signal transmission. Proc. Natl. Acad. Sci. USA 2022, 119, e2204688119. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Xie, W.; Pickering, B.F.; Chu, K.L.; Savino, A.M.; Yang, X.; Luo, H.; Nguyen, D.T.; Mo, S.; Barin, E.; et al. N(6)-Methyladenosine on mRNA facilitates a phase-separated nuclear body that suppresses myeloid leukemic differentiation. Cancer Cell 2021, 39, 958–972.e958. [Google Scholar] [CrossRef] [PubMed]
- Marschallinger, J.; Iram, T.; Zardeneta, M.; Lee, S.E.; Lehallier, B.; Haney, M.S.; Pluvinage, J.V.; Mathur, V.; Hahn, O.; Morgens, D.W.; et al. Lipid-droplet-accumulating microglia represent a dysfunctional and proinflammatory state in the aging brain. Nat. Neurosci. 2020, 23, 194–208. [Google Scholar] [CrossRef]
- Yue, F.; Oprescu, S.N.; Qiu, J.; Gu, L.; Zhang, L.; Chen, J.; Narayanan, N.; Deng, M.; Kuang, S. Lipid droplet dynamics regulate adult muscle stem cell fate. Cell Rep. 2022, 38, 110267. [Google Scholar] [CrossRef]
- Scholz, P.; Chapman, K.D.; Mullen, R.T.; Ischebeck, T. Finding new friends and revisiting old ones—How plant lipid droplets connect with other subcellular structures. New Phytol. 2022, 236, 833–838. [Google Scholar] [CrossRef]
- Sato, Y.; Takinoue, M. Creation of Artificial Cell-Like Structures Promoted by Microfluidics Technologies. Micromachines 2019, 10, 216. [Google Scholar] [CrossRef] [Green Version]
- Zhao, C.; Li, J.; Wang, S.; Xu, Z.; Wang, X.; Liu, X.; Wang, L.; Huang, X. Membranization of Coacervates into Artificial Phagocytes with Predation toward Bacteria. ACS Nano 2021, 15, 10048–10057. [Google Scholar] [CrossRef]
- Sato, Y.; Takinoue, M. Capsule-like DNA Hydrogels with Patterns Formed by Lateral Phase Separation of DNA Nanostructures. JACS Au 2022, 2, 159–168. [Google Scholar] [CrossRef]
- Abbas, M.; Lipiński, W.P.; Nakashima, K.K.; Huck, W.T.S.; Spruijt, E. A short peptide synthon for liquid–liquid phase separation. Nat. Chem. 2021, 13, 1046–1054. [Google Scholar] [CrossRef]
- Wang, Z.; Huang, P.; Jacobson, O.; Wang, Z.; Liu, Y.; Lin, L.; Lin, J.; Lu, N.; Zhang, H.; Tian, R.; et al. Biomineralization-Inspired Synthesis of Copper Sulfide-Ferritin Nanocages as Cancer Theranostics. ACS Nano 2016, 10, 3453–3460. [Google Scholar] [CrossRef] [Green Version]
- Nakatani, N.; Sakuta, H.; Hayashi, M.; Tanaka, S.; Takiguchi, K.; Tsumoto, K.; Yoshikawa, K. Specific Spatial Localization of Actin and DNA in a Water/Water Microdroplet: Self-Emergence of a Cell-Like Structure. Chembiochem 2018, 19, 1370–1374. [Google Scholar] [CrossRef] [Green Version]
- Bachler, S.; Haidas, D.; Ort, M.; Duncombe, T.A.; Dittrich, P.S. Microfluidic platform enables tailored translocation and reaction cascades in nanoliter droplet networks. Commun. Biol. 2020, 3, 769. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Huang, Y.; Lin, Y.; Chen, H.; Li, J.; Zhao, C.; Liu, X.; Yang, L.; Huang, X. Life-Inspired Endogenous Dynamic Behavior of Lipid Droplet-like Microcompartments in Artificial Adipocyte-like Structures. CCS Chem. 2020, 3, 2782–2794. [Google Scholar] [CrossRef]
- Tsuruta, M.; Torii, T.; Kohata, K.; Kawauchi, K.; Tateishi-Karimata, H.; Sugimoto, N.; Miyoshi, D. Controlling liquid-liquid phase separation of G-quadruplex-forming RNAs in a sequence-specific manner. Chem. Commun. 2022, 58, 12931–12934. [Google Scholar] [CrossRef]
- Shin, H.; Park, Y.H.; Kim, Y.G.; Lee, J.Y.; Park, J. Aqueous two-phase system to isolate extracellular vesicles from urine for prostate cancer diagnosis. PLoS ONE 2018, 13, e0194818. [Google Scholar] [CrossRef] [PubMed]
- Mastiani, M.; Firoozi, N.; Petrozzi, N.; Seo, S.; Kim, M. Polymer-Salt Aqueous Two-Phase System (ATPS) Micro-Droplets for Cell Encapsulation. Sci. Rep. 2019, 9, 15561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chao, Y.; Shum, H.C. Emerging aqueous two-phase systems: From fundamentals of interfaces to biomedical applications. Chem. Soc. Rev. 2020, 49, 114–142. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Song, Y.; Sun, W.; Cao, J.; Yuan, H.; Wang, X.; Sun, Y.; Shum, H.C. Cell-Inspired All-Aqueous Microfluidics: From Intracellular Liquid-Liquid Phase Separation toward Advanced Biomaterials. Adv. Sci. 2020, 7, 1903359. [Google Scholar] [CrossRef]
- Iqbal, M.; Tao, Y.; Xie, S.; Zhu, Y.; Chen, D.; Wang, X.; Huang, L.; Peng, D.; Sattar, A.; Shabbir, M.A.; et al. Aqueous two-phase system (ATPS): An overview and advances in its applications. Biol. Proced. Online 2016, 18, 18. [Google Scholar] [CrossRef] [Green Version]
- Diamond, A.D.; Hsu, J.T. Aqueous two-phase systems for biomolecule separation. Adv. Biochem. Eng. Biotechnol. 1992, 47, 89–135. [Google Scholar] [CrossRef]
- Albertsson, P.A. Partition of cell particles and macromolecules in polymer two-phase systems. Adv. Protein Chem. 1970, 24, 309–341. [Google Scholar] [CrossRef] [PubMed]
- Jia, T.Z.; Fraccia, T.P. Liquid Crystal Peptide/DNA Coacervates in the Context of Prebiotic Molecular Evolution. Crystals 2020, 10, 964. [Google Scholar] [CrossRef]
- Frankel, E.A.; Bevilacqua, P.C.; Keating, C.D. Polyamine/Nucleotide Coacervates Provide Strong Compartmentalization of Mg2+, Nucleotides, and RNA. Langmuir 2016, 32, 2041–2049. [Google Scholar] [CrossRef] [PubMed]
- Jia, T.Z.; Hentrich, C.; Szostak, J.W. Rapid RNA exchange in aqueous two-phase system and coacervate droplets. Orig. Life Evol. Biosph. 2014, 44, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Keating, C.D. Aqueous phase separation as a possible route to compartmentalization of biological molecules. Acc. Chem. Res. 2012, 45, 2114–2124. [Google Scholar] [CrossRef] [PubMed]
- McQueen, L.; Lai, D. Ionic Liquid Aqueous Two-Phase Systems From a Pharmaceutical Perspective. Front. Chem. 2019, 7, 135. [Google Scholar] [CrossRef] [Green Version]
- Yau, Y.K.; Ooi, C.W.; Ng, E.-P.; Lan, J.C.-W.; Ling, T.C.; Show, P.L. Current applications of different type of aqueous two-phase systems. Bioresour. Bioprocess. 2015, 2, 49. [Google Scholar] [CrossRef] [Green Version]
- Frerix, A.; Müller, M.; Kula, M.R.; Hubbuch, J. Scalable recovery of plasmid DNA based on aqueous two-phase separation. Biotechnol. Appl. Biochem. 2005, 42, 57–66. [Google Scholar] [CrossRef]
- Frerix, A.; Schönewald, M.; Geilenkirchen, P.; Müller, M.; Kula, M.R.; Hubbuch, J. Exploitation of the coil-globule plasmid DNA transition induced by small changes in temperature, pH salt, and poly(ethylene glycol) compositions for directed partitioning in aqueous two-phase systems. Langmuir 2006, 22, 4282–4290. [Google Scholar] [CrossRef] [Green Version]
- Indulkar, A.S.; Gao, Y.; Raina, S.A.; Zhang, G.G.; Taylor, L.S. Exploiting the Phenomenon of Liquid-Liquid Phase Separation for Enhanced and Sustained Membrane Transport of a Poorly Water-Soluble Drug. Mol. Pharm. 2016, 13, 2059–2069. [Google Scholar] [CrossRef]
- Feng, C.; Song, R.; Sun, G.; Kong, M.; Bao, Z.; Li, Y.; Cheng, X.; Cha, D.; Park, H.; Chen, X. Immobilization of Coacervate Microcapsules in Multilayer Sodium Alginate Beads for Efficient Oral Anticancer Drug Delivery. Biomacromolecules 2014, 15, 985–996. [Google Scholar] [CrossRef]
- Zhao, M.; Zacharia, N.S. Protein encapsulation via polyelectrolyte complex coacervation: Protection against protein denaturation. J. Chem. Phys. 2018, 149, 163326. [Google Scholar] [CrossRef]
- Nojima, T.; Niwa, T.; Taguchi, H. Proteome Analysis of Phase-Separated Condensed Proteins with Ionic Surfactants Revealed Versatile Formation of Artificial Biomolecular Condensates. Biomacromolecules 2019, 20, 539–545. [Google Scholar] [CrossRef] [PubMed]
- Nojima, T.; Iyoda, T. Water-Rich Fluid Material Containing Orderly Condensed Proteins. Angew. Chem. Int. Ed. 2017, 56, 1308–1312. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, F.; van der Walle, C.F. Engineering biodegradable polyester particles with specific drug targeting and drug release properties. J. Pharm. Sci. 2008, 97, 71–87. [Google Scholar] [CrossRef] [PubMed]
- Husmann, M.; Schenderlein, S.; Lück, M.; Lindner, H.; Kleinebudde, P. Polymer erosion in PLGA microparticles produced by phase separation method. Int. J. Pharm. 2002, 242, 277–280. [Google Scholar] [CrossRef]
- Karthick, V.; Panda, S.; Kumar, V.G.; Kumar, D.; Shrestha, L.K.; Ariga, K.; Vasanth, K.; Chinnathambi, S.; Dhas, T.S.; Suganya, K.S.U. Quercetin loaded PLGA microspheres induce apoptosis in breast cancer cells. Appl. Surf. Sci. 2019, 487, 211–217. [Google Scholar] [CrossRef]
- Washington, K.E.; Kularatne, R.N.; Karmegam, V.; Biewer, M.C.; Stefan, M.C. Recent advances in aliphatic polyesters for drug delivery applications. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2017, 9, e1446. [Google Scholar] [CrossRef]
- Abu Ghalia, M. Biodegradable poly(lactic acid)-based scaffolds: Synthesis and biomedical applications. J. Polym. Res. 2017, 24, 74. [Google Scholar] [CrossRef]
- Shive, M.S.; Anderson, J.M. Biodegradation and biocompatibility of PLA and PLGA microspheres. Adv. Drug Deliv. Rev. 1997, 28, 5–24. [Google Scholar] [CrossRef]
- Kucharczyk, P.; Pavelková, A.; Stloukal, P.; Sedlarík, V. Degradation behaviour of PLA-based polyesterurethanes under abiotic and biotic environments. Polym. Degrad. Stab. 2016, 129, 222–230. [Google Scholar] [CrossRef] [Green Version]
- Carvalho, J.R.G.; Conde, G.; Antonioli, M.L.; Dias, P.P.; Vasconcelos, R.O.; Taboga, S.R.; Canola, P.A.; Chinelatto, M.A.; Pereira, G.T.; Ferraz, G.C. Biocompatibility and biodegradation of poly(lactic acid) (PLA) and an immiscible PLA/poly(ε-caprolactone) (PCL) blend compatibilized by poly(ε-caprolactone-b-tetrahydrofuran) implanted in horses. Polym. J. 2020, 52, 629–643. [Google Scholar] [CrossRef]
- Mikos, A.G.; Thorsen, A.J.; Czerwonka, L.A.; Bao, Y.; Langer, R.; Winslow, D.N.; Vacanti, J.P. Preparation and characterization of poly(l-lactic acid) foams. Polymer 1994, 35, 1068–1077. [Google Scholar] [CrossRef]
- Urbánek, T.; Jäger, E.; Jäger, A.; Hrubý, M. Selectively Biodegradable Polyesters: Nature-Inspired Construction Materials for Future Biomedical Applications. Polymers 2019, 11, 1061. [Google Scholar] [CrossRef] [Green Version]
- Xiao, W.; Jakimowicz, M.D.; Zampetakis, I.; Neely, S.; Scarpa, F.; Davis, S.A.; Williams, D.S.; Perriman, A.W. Biopolymeric Coacervate Microvectors for the Delivery of Functional Proteins to Cells. Adv. Biosyst. 2020, 4, 2000101. [Google Scholar] [CrossRef]
- Lagassé, H.A.; Alexaki, A.; Simhadri, V.L.; Katagiri, N.H.; Jankowski, W.; Sauna, Z.E.; Kimchi-Sarfaty, C. Recent advances in (therapeutic protein) drug development. F1000Res 2017, 6, 113. [Google Scholar] [CrossRef] [Green Version]
- Iwata, T.; Hirose, H.; Sakamoto, K.; Hirai, Y.; Arafiles, J.V.V.; Akishiba, M.; Imanishi, M.; Futaki, S. Cover Picture: Liquid Droplet Formation and Facile Cytosolic Translocation of IgG in the Presence of Attenuated Cationic Amphiphilic Lytic Peptides (Angew. Chem. Int. Ed. 36/2021). Angew. Chem. Int. Ed. 2021, 60, 19493. [Google Scholar] [CrossRef]
- Manolio, T.A.; Bult, C.J.; Chisholm, R.L.; Deverka, P.A.; Ginsburg, G.S.; Goldrich, M.; Jarvik, G.P.; Mensah, G.A.; Ramos, E.M.; Relling, M.V.; et al. Genomic medicine year in review: 2021. Am. J. Hum. Genet. 2021, 108, 2210–2214. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Zhong, L.; Weng, Y.; Peng, L.; Huang, Y.; Zhao, Y.; Liang, X.J. Therapeutic siRNA: State of the art. Signal Transduct. Target. Ther. 2020, 5, 101. [Google Scholar] [CrossRef]
- Horton, R.H.; Lucassen, A.M. Recent developments in genetic/genomic medicine. Clin. Sci. 2019, 133, 697–708. [Google Scholar] [CrossRef] [Green Version]
- Nair, S.R. Personalized medicine: Striding from genes to medicines. Perspect. Clin. Res. 2010, 1, 146–150. [Google Scholar] [CrossRef]
- Silveira, M.M.; Moreira, G.; Mendonça, M. DNA vaccines against COVID-19: Perspectives and challenges. Life Sci. 2021, 267, 118919. [Google Scholar] [CrossRef] [PubMed]
- Ura, T.; Yamashita, A.; Mizuki, N.; Okuda, K.; Shimada, M. New vaccine production platforms used in developing SARS-CoV-2 vaccine candidates. Vaccine 2021, 39, 197–201. [Google Scholar] [CrossRef]
- Walsh, E.E.; Frenck, R.W., Jr.; Falsey, A.R.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Neuzil, K.; Mulligan, M.J.; Bailey, R.; et al. Safety and Immunogenicity of Two RNA-Based Covid-19 Vaccine Candidates. N. Engl. J. Med. 2020, 383, 2439–2450. [Google Scholar] [CrossRef]
- Mi, X.; Blocher McTigue, W.C.; Joshi, P.U.; Bunker, M.K.; Heldt, C.L.; Perry, S.L. Thermostabilization of viruses via complex coacervation. Biomater. Sci. 2020, 8, 7082–7092. [Google Scholar] [CrossRef]
- Xu, Y.; Qi, R.; Zhu, H.; Li, B.; Shen, Y.; Krainer, G.; Klenerman, D.; Knowles, T.P.J. Liquid-Liquid Phase-Separated Systems from Reversible Gel-Sol Transition of Protein Microgels. Adv. Mater. 2021, 33, e2008670. [Google Scholar] [CrossRef]
- Zhang, Z.; Liu, Q.; Sun, Z.; Phillips, B.K.; Wang, Z.; Al-Hashimi, M.; Fang, L.; Olson, M.A. Poly-Lipoic Ester-Based Coacervates for the Efficient Removal of Organic Pollutants from Water and Increased Point-of-Use Versatility. Chem. Mater. 2019, 31, 4405–4417. [Google Scholar] [CrossRef]
- Bunce, J.T.; Ndam, E.; Ofiteru, I.D.; Moore, A.; Graham, D.W. A Review of Phosphorus Removal Technologies and Their Applicability to Small-Scale Domestic Wastewater Treatment Systems. Front. Environ. Sci. 2018, 6, 8. [Google Scholar] [CrossRef] [Green Version]
- Tolstoguzov, V. Texturising by phase separation. Biotechnol. Adv. 2006, 24, 626–628. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.C.; Degner, B.; McClements, D.J. Soft matter strategies for controlling food texture: Formation of hydrogel particles by biopolymer complex coacervation. J. Phys. Condens. Matter 2014, 26, 464104. [Google Scholar] [CrossRef] [PubMed]
- Eghbal, N.; Choudhary, R. Complex coacervation: Encapsulation and controlled release of active agents in food systems. LWT 2018, 90, 254–264. [Google Scholar] [CrossRef]


| Droplet | Main Component | Role | Observation | Ref. |
|---|---|---|---|---|
| Stress granule | Proteins and RNAs | Translational regulation mRNA storage | CM (1), smFISH (2) | [42,43] |
| Centrosome | Pericentriolar material | Formation of mitotic spindles during mitosis | CM (1) | [44,45] |
| U body | Uridine-rich small nuclear ribonucleoproteins | Storage and assembly of snRNPs | CM (1) | [46,47] |
| G body | Lipid and protein | Controlling the rate of glycolysis | CM (1) | [48,49] |
| P body | Translationally repressed mRNAs and proteins related to mRNA decay | mRNA decay and silencing | CM (1) | [50,51] |
| Balbiani body (germ cells) | Endoplasmic reticulum/Golgi-like vesicles, mitochondria, and specific RNAs transporter. | Storage and facilitating the organization of the oocyte into a polarized cell | CM (1), EM (3) | [52,53,54,55] |
| Germ granules (germ cells) | Proteins and RNAs | Storage of proteins and RNAs that are required for germ cell development | EM (3) | [56,57,58] |
| RNA transport granule (neuronal cell) | mRNAs and proteins | Storage and transport of mRNAs | CM (1) | [59,60] |
| Droplet | Main Component | Role | Observation | Ref. |
|---|---|---|---|---|
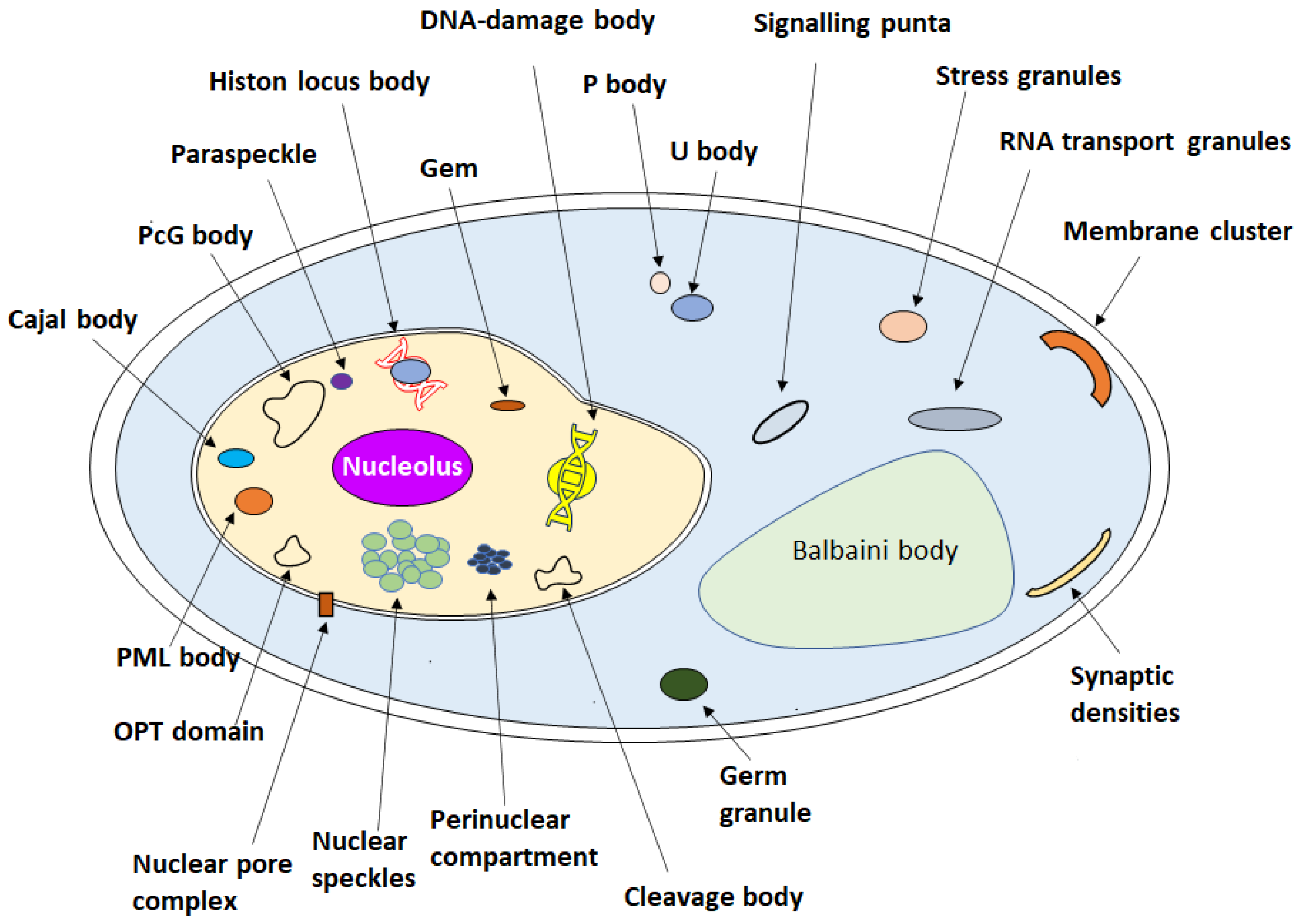
| Nucleolus | Proteins and canonical nucleic acids, non-coding RNA | Ribosome biogenesis | FM 1 | [8,63,64] |
| Histone locus body | NPAT 10, SLBP 11, the U7 spliceosomal snRNP-specific components, such as Sm proteins, LSm10 and LSm11, and the U7 spliceosomal snRNA, FLASH 12 | Histone mRNA biogenesis | BFM 2 | [65,66] |
| Heterochromatin | HP1 13, nucleosomal DNA | Promote the formation of heterochromatin | CM 3 | [67,68,69,70,71,72] |
| Nuclear pore central transport channel and nuclear pore complex | Nups 14, FG 15 | Chromosomal translocations, change in protein expression levels. Fuse with oncoproteins, nuclear import/export | HS-AFM 4, FM 1 | [73,74,75,76] |
| Nuclear speckles | RNAs and proteins | mRNA splicing | CM 3 | [77,78] |
| DNA damage foci | Rad52 DNA repair proteins | DNA damage repair | Live-cell CM 5 | [79,80] |
| Gem | SMN complex, ZPR1, GEMIN2–8 16. | Storage aid histone, mRNA processing | CM 3 | [81,82,83] |
| PcG body | Transcriptional repression | IM 6, EM 7 | [84,85] | |
| Paraspeckle | NONO 17, PSP1 17, PSP2 17, SFPQ 18, CFIm68 19, CFIm, hnRNPs 20 NEAT1 21 | RNA processing | CM 3 | [86,87,88] |
| OPT domain | The RNA polymerases and the general transcription factors | Transcriptional regulation | FM 1, EM 7 | [89,90,91] |
| Cajal body | Coilin, CB-specific RNAs | Assembly and/or modification of splicing machinery | BFM 2 | [66,92,93] |
| Perinuclear compartment | RNA-binding proteins and pol III RNA | Associated with malignancy | EM 7, IM 6 | [92] |
| Cleavage body | snRNPs 22, p80-coilin protein, RNA polymerases, transcriptional factors, nucleolar constituents | mRNA processing | IL 8 | [94] |
| Nuclear bodies (NBs) | Protein and non-protein components, heat shock transcription factors, HSF1 23 and HSF2 24, SAF-B 25, Sam68 26, SRSF1 27, SRSF7 27 and SRSF9 27. RNA Pol II. | Regulation of genome function | IM 6, SRM 9 | [66,95] |
| PML body | DAXX, SUMO 28 | Transcriptional regulation; apoptosis signaling; antiviral defense | EM 7 | [82,96] |
| Droplet | Main Component | Role | Observation | Ref. |
|---|---|---|---|---|
| Membrane cluster | Triacylglycerols, phospholipid, protein | Lipid uptake, distribution, storage, and use in the cell. | - | [98] |
| Synaptic densities | Actin’s cytoskeleton, kinases, phosphatases, and regulators, GTPases, subunits of AMPA and NMDA receptors, Catenin, N-Cadherin | Neurotransmission | AEM 1 | [99,100] |
| Focal adhesions | p130Cas (‘Cas’) and FAK 7 | Cell adhesion/migration | SDCM 2 | [101,102] |
| Nephrin clusters | Cytoplasmic adaptor protein Nck, the nephrin–Nck–N-WASp complex | Glomerular filtration barrier | SRSIM 5 | [13,28,103] |
| TCR clusters | LAT 8 | Immune synapse | TIRF 3 | [104,105,106] |
| Podosomes | F-actin and its regulatory molecules, structural proteins | Cell adhesion/migration | PCM 4 | [107,108] |
| Actin patches | Actin-associated proteins, upstream signaling molecules | Endocytosis | EM 6 | [109,110] |
| Enzyme | Role | Observation | Ref. |
|---|---|---|---|
| Horseradish peroxidase (HRP) | Catalyst (horseradish peroxidase (HRP) | CM 1 | [115] |
| Glucose oxidase (GOx) | Catalyst (oxidation of β-d-glucose to d-glucono-δ-lactone) | CM 1 | [119] |
| Hexokinase | Catalyst, catalyzing the phosphorylation of keto- and aldohexoses | OM 2 | [120] |
| Lipase | Fat breakdown | CM 1 | [121,122] |
| Hammerhead ribozyme | Cleavage and ligation of RNA molecule | FRET 6, CD 4, CM 1 | [123,124] |
| Pfk2, Eno1, Eno2, Fba1 | Glycolysis | FM 3 | [125] |
| GIT1 | GTPase activator | FRAP 5, CM 1 | [126] |
| HSF1 | Transcription factor | FM 3 | [127,128] |
| NELFE | Transcriptional regulation | FM 3 | [129] |
| p53 | Transcription factor | FM 3 | [130,131,132,133,134,135] |
| PLK4 | Serine/threonine-protein kinase | CM 1 | [136,137] |
| SOX-2 | Transcription factor | FM 3 | [138] |
| TFE3 | Transcription factor | FM 3 | [139] |
| TFEB | Transcription factor | FRAP 5 | [140] |
| USP42 | Deubiquitinating enzyme | FM 3 | [141] |
| YAP | Transcription factor | FM 3 | [142,143] |
| Droplet | Location | Role | Main Component | Observation | Ref. |
|---|---|---|---|---|---|
| Pyrenoids (Rubisco), carboxysomes | Chloroplast | Photosynthesis, metabolism (Carbon fixation) | Carboxysomal linker proteins CsoS2 and CcmM, Rubisco large subunit | Microscopy and sedimentation assay | [144,145,146] |
| Wnt droplet | Cell cytoplasm | Stem cell differentiation, controlling Wnt pathway | Scaffold proteins and kinases that regulate β-catenin stability | CRISPR-engineered fluorescent tags, optogenetic tools | [147] |
| YTHDC droplet (nuclear bodies) | Nucleus | AML cell survival, differentiation state, leukemogenesis | YTHDC1 protein, m6 A-containing RNA | IF 1, SEM 2. | [148] |
| LDAM 3 | Hippocampus | Promotion of pathogenesis, neuroinflammation | Lipid | CARS 4 | [149] |
| Lipid droplets | Cell cytoplasm (Stem cell) | Skeletal muscle satellite cell fate determination | Lipid | TEM 5 | [150] |
| Plant lipid droplets: LD-Erm LD-Peroxisomes | Plant cell | Unknown | Triacylglycerols (TAGs), sterol esters (SEs) | FM 6, CM 7 | [151] |
| Droplet | Component | Observation | Application | Ref. |
|---|---|---|---|---|
| Adiposomes (artificial lipid droplets (ALDs)) | Phospholipids and neutral lipids such as TAG | LM 1, EM 2 | Potential usage in drug delivery. | [156] |
| Cell-sized aqueous/aqueous microdroplets (CAMDs) | PEG 3 and DEX 4, actin | FM 5 | Provide cell-like crowded microenvironments | [157] |
| Microfluidic platform in a defined pattern | Hexadecane/squalene with dissolved lipids | Broad range of applications in the field of artificial cells, bioreactors, and pharmacological studies. | [158] | |
| Lipase-stabilized tributyrin microcompartment and amylose-polymer-stabilized 2-ethyl-1-hexanol microcompartment | Amy-PNIPAAm 6, BSA-PNIPAAm 7, Lipase | OM 8 | Synthetic biology, bottom-up reaction | [159] |
| G-quadruplex-forming oligonucleotides and R-rich oligopeptides | FMR1 RNA, C9orf72 RNA, peptide derived from FMRP | CM 9 | Droplet redissolution in a sequence-specific manner | [160] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shil, S.; Tsuruta, M.; Kawauchi, K.; Miyoshi, D. Biomolecular Liquid–Liquid Phase Separation for Biotechnology. BioTech 2023, 12, 26. https://doi.org/10.3390/biotech12020026
Shil S, Tsuruta M, Kawauchi K, Miyoshi D. Biomolecular Liquid–Liquid Phase Separation for Biotechnology. BioTech. 2023; 12(2):26. https://doi.org/10.3390/biotech12020026
Chicago/Turabian StyleShil, Sumit, Mitsuki Tsuruta, Keiko Kawauchi, and Daisuke Miyoshi. 2023. "Biomolecular Liquid–Liquid Phase Separation for Biotechnology" BioTech 12, no. 2: 26. https://doi.org/10.3390/biotech12020026
APA StyleShil, S., Tsuruta, M., Kawauchi, K., & Miyoshi, D. (2023). Biomolecular Liquid–Liquid Phase Separation for Biotechnology. BioTech, 12(2), 26. https://doi.org/10.3390/biotech12020026








