Recent Genome-Editing Approaches toward Post-Implanted Fetuses in Mice
Abstract
1. Introduction
2. In Utero Gene Delivery
2.1. Site-Directed in Utero Gene Delivery and Subsequent in Utero EP
| Type of Method | Genome-Editing Tool (Mode for Gene Modification) | Outcome | Target Gene | Data from |
|---|---|---|---|---|
| In utero gene delivery and subsequent in vivo electroporation (EP) (all-in-one plasmid) | CRISPR/Cas9 (indels) | Performed at E15 of pregnancy; first successful knock-down (KD) in pyramidal neurons in vivo; manipulated cells lacked synaptic current mediated by NMDA-type glutamate receptors | Grin1 | [37] |
| In utero gene delivery and subsequent in vivo EP (all-in-one plasmid) | CRISPR/Cas9 (indels) | Induced abnormalities in axonal projection patterns, which is consistent with the phenotypes previously observed in Satb2 mutant mice | Satb2 | [39] |
| In utero gene delivery and subsequent in vivo EP [all-in-one plasmid or ribonucleoprotein (RNP)] | CRISPR/Cas9 (indels) | Successful disruption of the expression of green fluorescent protein (GFP) or endogenous eomesodermin (Eomes)/T-box brain protein 2 (Tbr2), a gene fundamental for neocortical neurogenesis | GFP Eomes/ Tbr2 | [40] |
| In utero gene delivery and subsequent in vivo EP (all-in-one plasmid + donor plasmid DNA) | CRISPR/Cas9 (KI) | Successful knock-in (KI) of EGFP fragment into the β-actin locus and enhanced GFP (EGFP)-tagged β-actin protein in cortical layer 2/3 pyramidal neurons; provide a useful tool to detect the localization of various endogenous proteins in neurons | β-actin | [41] |
| In utero gene delivery (Cas9-expressing plasmid + gRNA-expressing plasmids + donor plasmid DNA) and subsequent in vivo EP | CRISPR/Cas9 (KI) | Successful de novo targeted KI of EGFP sequence into the target locus through in utero EP into the mammalian brain | βIII-tubulin (Tubb3) | [43] |
| In utero delivery of Ad containing base editor 3 (BE3) and gRNA | CRISPR/BE3-based gene correction | Viral vector–mediated delivery of CRISPR/Cas9 or BE3 through in utero gene delivery through vitelline vein injection of E16 fetus was performed to pursue therapeutic modification of proprotein convertase subtilisin/kexin type 9 (Pcsk9) or hydroxyphenylpyruvate dioxygenase (Hpd) in wild-type mice or a murine model of hereditary tyrosinemia type 1 (HT1), respectively; long-term postnatal persistence of edited cells were observed in both models, with reduction of plasma PCSK9 and cholesterol levels following in utero Pcsk9 targeting and rescue of the lethal phenotype of HT1 following in utero Hpd targeting. | Pcsk9 Hpd | [44] |
| In utero delivery of nanoparticle (NP) carrying peptide nucleic acids (PNAs) + donor DNA | PNA and DNA oligomers-based KI | Intravenous (via the vitelline vein) NP delivery of PNAs and single-stranded DNA (ssDNA) to mouse fetuses (at E15 to E16) harboring mutations in the human β-globin gene, which are recognized as a model for human β-thalassemia, resulted in site-specific genome-editing of fetal liver cells, leading to phenotypic rescue of thalassemia (severe anemia) before birth | β-globin | [45] |
| In utero delivery of adenoviruses (Ad) carrying Cas9, gRNA or donor DNA | CRISPR/Cas9 (KI) | In utero delivery of Ad carrying CRISPR reagents into the amniotic cavity of a fetus (at E16) resulted in transduction of alveolar epithelial cells of fetal lung, extensive pulmonary gene editing, and, finally, rescued a perinatal lethal phenotype in the surfactant protein C (Sftpc)I73T mice, a model for monogenic lung disease | Sftpc | [46] |
| Transplacental gene delivery to acquire genome-edited fetuses (TGPD-GEF) (all-in-one plasmid) | CRISPR/Cas9 (indels) | TPGD-GEF causes indel mutations in embryonic cardiomyocytes of mid-gestational murine fetuses | EGFP | [7] |
| In utero delivery of CRISPR-AAV9 | CRISPR/Cas9 (indels) | Injection of recombinant adeno-associated viruses (rAAVs) carrying short Cas9 variant and gRNA into fetal brain (at E15) of mouse model of Angelman syndrome (AS) resulted in successful unsilencing of paternal ubiquitin–protein ligase E3A (Ube3a) throughout the brain for at least 17 months and rescued abnormal phenotypes associated with AS mice | Snord115 onUBE3A | [47] |
| TPGD-GEF (all-in-one plasmid) | CRISPR/Cas9 (indels) | Hydrodynamics-based gene delivery (HGD) into pregnant female mice at E9.5 resulted in mosaic indel mutations in myosin heavy chain α (MHCα) with an efficiency of 40% | MHCα | [48] |
| In utero gene delivery and subsequent in vivo EP (all-in-one plasmid) | CRISPR/Cas9 (KI) | Development of a novel method, called “Targeted KI with Two”, for precise genome-editing-based tagging in mouse primary cultured neurons with efficiencies up to 42%; when injection of CRISPR reagents into the ateral ventricle of fetal brain (at E15) and subsequent in utero EP was performed, expression of donor DNA was observed along the dendrites of layer 2/3 pyramidal neurons and its expression lasted more than 1 year, indicating the long-term stability of the KI tag | Gria2 | [49] |
| In utero delivery of rAAV9 carrying adenine base editor (ABE) | ABE system | In utero injection of rAAV9 carrying the ABE targeting the Idua G→A (W392X) mutation in the mucopolysaccharidosis type I (MPS-IH) mouse, corresponding to the common IDUA G→A (W402X) mutation in MPS-IH patients, was performed via vitelline vein of a fetus at E15; which resulted in long-term W392X correction in hepatocytes and cardiomyocytes and low-level editing in the brain, highlighting the potential of this approach for MPS-IH and other genetic diseases | Idua | [50] |
| In utero delivery of CRISPR-AAV9-PHP.eB carrying gRNA | CRISPR/Cas9 (indels) | In utero injection of rAAV9-PHP.eB (AAV9-based mutant capsid that is highly efficient in transducing the central nervous system of adult mice) carrying gRNA to the lateral ventricles of fetal mouse brain (at E15) of Cas9 transgenic (Tg) mice resulted in widespread gene KO; suggesting a useful platform for studying brain development and devising genetic intervention for severe developmental diseases | PogZ Depdc5 | [51] |
| In utero delivery of rAAV9 carrying ABE | ABEmax-NG system (ABEmax combined with SpCas9-NG (capable of recognizing NG rather than NGG) | In utero injection of rAAV9 carrying the ABEmax-NG into the vitelline vein of E16 fetus resulted in about 25.3% correction of the pathogenic hypertrophic cardiomyopathy (HCM) mutation (R404Q/+ mutation; Myh6 c.1211C > T) in a mouse model of HCM, and reduced expression of mutant RNA, suggesting that ABEmax-NG has the potential to correct the HCM mutation in vivo | Myh6 | [52] |
2.2. Intraamniotic Injection of Viral Vectors Carrying Genome-Editing Components
2.3. In Utero Injection of Viral Vectors Carrying Genome-Editing Components into the Brain
2.4. Intraamniotic Injection of Non-Viral DNA Encapsulated with Lipids
3. TPGD-GEF Technique
4. Limitations of and Possibilities for In Utero Genome Editing and TPGD-GEF
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AAV | Adeno-associated viruses |
| ABE | Adenine base editor |
| Ad | Adenoviruses |
| AS | Angelman syndrome |
| BE3 | Base editor 3 |
| Cas9 | CRISPR-associated protein 9 |
| CRISPR | Clustered regularly interspaced short palindromic repeats |
| DOGS | 5-Carboxyspermylglycine dioctadecylamide |
| DOPE | 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine |
| dsDNA | Double-stranded DNA |
| DSB | Double-strand break |
| E | Embryonic day |
| EGFP | Enhanced green fluorescent protein |
| Eomes | Eomesodermin |
| EP | Electroporation |
| Fah | Fumarylacetoacetate hydrolase |
| FACS | Fluorescence-activated cell sorting |
| GFP | Green fluorescent protein |
| Gria2 | Glutamate ionotropic receptor AMPA type subunit 2 |
| Grin1 | Glutamate ionotropic receptor NMDA type subunit 1 |
| gRNA | Guide RNA |
| HCM | Hypertrophic cardiomyopathy |
| HDR | Homology directed repair |
| HGD | Hydrodynamics-based gene delivery |
| HIV | Human immunodeficiency virus |
| Hpd | Hydroxyphenylpyruvate dioxygenase |
| HT1 | Hereditary tyrosinemia type 1 |
| Idua | α-L-iduronidase |
| KD | Knock-down |
| KI | Knock-in |
| KO | Knockout |
| MHCα | Myosin heavy chain α |
| miRNA | MicroRNA |
| MPS-IH | Mucopolysaccharidosis type I |
| Myh6 | Myosin heavy chain 6 |
| NHEJ | Non-homologous end joining |
| NMDA | N-methyl-D-aspartate |
| NP | Nanoparticle |
| Pcsk9 | Proprotein convertase subtilisin/kexin type 9 |
| PNAs | Peptide nucleic acids |
| PogZ | Pogo transposable element derived with ZNF domain |
| RNAi | RNA interference |
| RNP | Ribonucleoprotein |
| rAAVs | Recombinant adeno-associated viruses |
| SaCas9 | Staphylococcus aureus Cas9 |
| Satb2 | Special AT-rich sequence-binding protein 2 |
| Sftpc | Surfactant protein C |
| ssDNA | Single-stranded DNA |
| Snord115 | Small nucleolar RNA, C/D box 115 |
| snoRNAs | Small nucleolar RNAs |
| Tbr2 | T-box brain protein 2 |
| Tg | Transgenic |
| TPGD | Transplacental gene delivery |
| TPGD-GEF | Transplacental gene delivery to acquire genome-edited fetuses |
| UBE3A | Ubiquitin–protein ligase E3A |
| UBE3A-ATS | UBE3A antisense transcript |
| VE | Visceral endoderm |
| YS | Yolk sac |
References
- Harrison, M.M.; Jenkins, B.V.; O’Connor-Giles, K.M.; Wildonger, J. A CRISPR view of development. Genes Dev. 2014, 28, 1859–1872. [Google Scholar] [CrossRef] [PubMed]
- Hsu, P.D.; Lander, E.S.; Zhang, F. Development and applications of CRISPR-Cas9 for genome engineering. Cell 2014, 157, 1262–1278. [Google Scholar] [CrossRef] [PubMed]
- Ran, F.A.; Hsu, P.D.; Wright, J.; Agarwala, V.; Scott, D.A.; Zhang, F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013, 8, 2281–2308. [Google Scholar] [CrossRef] [PubMed]
- National Academies of Sciences, Engineering, and Medicine; National Academy of Medicine; National Academy of Sciences; Committee on Human Gene Editing: Scientific, Medical, and Ethical Considerations. Human Genome Editing: Science, Ethics, and Governance; Copyright 2017 by the National Academy of Sciences; National Academies Press (US): Washington, DC, USA, 2017. [Google Scholar]
- Baliou, S.; Adamaki, M.; Kyriakopoulos, A.M.; Spandidos, D.A.; Panayiotidis, M.; Christodoulou, I.; Zoumpourlis, V. CRISPR therapeutic tools for complex genetic disorders and cancer (Review). Int. J. Oncol. 2018, 53, 443–468. [Google Scholar] [CrossRef]
- Sato, M.; Takabayashi, S.; Akasaka, E.; Nakamura, S. Recent advances and future perspectives of in vivo targeted delivery of genome-editing reagents to germ cells, embryos, and fetuses in mice. Cells 2020, 9, 799. [Google Scholar] [CrossRef]
- Nakamura, S.; Ishihara, M.; Ando, N.; Watanabe, S.; Sakurai, T.; Sato, M. Transplacental delivery of genome editing components causes mutations in embryonic cardiomyocytes of mid-gestational murine fetuses. IUBMB Life 2019, 71, 835–844. [Google Scholar] [CrossRef]
- Nakamura, S.; Watanabe, S.; Ando, N.; Ishihara, M.; Sato, M. Transplacental gene delivery (TPGD) as a noninvasive tool for fetal gene manipulation in mice. Int. J. Mol. Sci. 2019, 20, 5926. [Google Scholar] [CrossRef]
- Liu, C.; Zhang, L.; Liu, H.; Cheng, K. Delivery strategies of the CRISPR-Cas9 gene-editing system for therapeutic applications. J. Control Release 2017, 266, 17–26. [Google Scholar] [CrossRef]
- Fajrial, A.K.; He, Q.Q.; Wirusanti, N.I.; Slansky, J.E.; Ding, X. A review of emerging physical transfection methods for CRISPR/Cas9-mediated gene editing. Theranostics 2020, 10, 5532–5549. [Google Scholar] [CrossRef]
- Zhao, H.; Li, Y.; He, L.; Pu, W.; Yu, W.; Li, Y.; Wu, Y.-T.; Xu, C.; Wei, Y.; Ding, Q.; et al. In vivo AAV-CRISPR/Cas9–mediated gene editing ameliorates atherosclerosis in familial hypercholesterolemia. Circulation 2020, 141, 67–79. [Google Scholar] [CrossRef]
- Ngô-Muller, V.; Muneoka, K. In utero and exo utero surgery on rodent embryos. Methods Enzymol. 2010, 476, 205–226. [Google Scholar] [CrossRef] [PubMed]
- Sheehy, N.T.; Cordes, K.R.; White, M.P.; Ivey, K.N.; Srivastava, D. The neural crest-enriched microRNA miR-452 regulates epithelial-mesenchymal signaling in the first pharyngeal arch. Development 2010, 137, 4307–4316. [Google Scholar] [CrossRef] [PubMed]
- Endo, M.; Henriques-Coelho, T.; Zoltick, P.W.; Stitelman, D.H.; Peranteau, W.H.; Radu, A.; Flake, A.W. The developmental stage determines the distribution and duration of gene expression after early intra-amniotic gene transfer using lentiviral vectors. Gene Ther. 2010, 17, 61–71. [Google Scholar] [CrossRef] [PubMed]
- O’Shea, K.S.; De Boer, L.S.; Slawny, N.A.; Gratsch, T.E. Transplacental RNAi: Deciphering gene function in the postimplantation-staged embryo. J. Biomed. Biotechnol. 2006, 2006, 18657. [Google Scholar] [CrossRef] [PubMed]
- Tabata, H.; Nakajima, K. Efficient in utero gene transfer system to the developing mouse brain using electroporation: Visualization of neuronal migration in the developing cortex. Neuroscience 2001, 103, 865–872. [Google Scholar] [CrossRef] [PubMed]
- Saito, T.; Nakatsuji, N. Efficient gene transfer into the embryonic mouse brain using in vivo electroporation. Dev. Biol. 2001, 240, 237–246. [Google Scholar] [CrossRef]
- Fukuchi-Shimogori, T.; Grove, E.A. Neocortex patterning by the secreted signaling molecule FGF8. Science 2001, 294, 1071–1074. [Google Scholar] [CrossRef]
- Takahashi, M.; Sato, K.; Nomura, T.; Osumi, N. Manipulating gene expressions by electroporation in the developing brain of mammalian embryos. Differentiation 2002, 70, 155–162. [Google Scholar] [CrossRef]
- Sato, M.; Tanigawa, M.; Kikuchi, N. Nonviral gene transfer to surface skin of mid-gestational murine embryos by intraamniotic injection and subsequent electroporation. Mol. Reprod. Dev. 2004, 69, 268–277. [Google Scholar] [CrossRef]
- De Vry, J.; Martínez-Martínez, P.; Losen, M.; Temel, Y.; Steckler, T.; Steinbusch, H.W.; De Baets, M.H.; Prickaerts, J. In vivo electroporation of the central nervous system: A non-viral approach for targeted gene delivery. Prog. Neurobiol. 2010, 92, 227–244. [Google Scholar] [CrossRef]
- Matsui, A.; Yoshida, A.C.; Kubota, M.; Ogawa, M.; Shimogori, T. Mouse in utero electroporation: Controlled spatiotemporal gene transfection. J. Vis. Exp. 2011, 54, e3024. [Google Scholar] [CrossRef]
- Nishimura, Y.V.; Shinoda, T.; Inaguma, Y.; Ito, H.; Nagata, K. Application of in utero electroporation and live imaging in the analyses of neuronal migration during mouse brain development. Med. Mol. Morphol. 2012, 45, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Wang, K. Manipulating potassium channel expression and function in hippocampal neurons by in utero electroporation. Methods Mol. Biol. 2018, 1684, 1–5. [Google Scholar] [CrossRef]
- Huang, C.C.; Carcagno, A. Electroporation of postimplantation mouse embryos in utero. Cold Spring Harb. Protoc. 2018, 2018, 99–102. [Google Scholar] [CrossRef] [PubMed]
- Gumus, H.G.; Illa, M.; Pla, L.; Gonzalez, A.; Demicheva, E.; Ley, D.; Crispi, F.; Gratacos, E. Ultrasound-guided intrauterine labeling of rat fetuses. Gynecol. Obstet. Investig. 2018, 83, 116–123. [Google Scholar] [CrossRef] [PubMed]
- Takeda, H.; Miwa, T.; Kim, M.Y.; Choi, B.Y.; Orita, Y.; Minoda, R. Prenatal electroporation-mediated gene transfer restores Slc26a4 knock-out mouse hearing and vestibular function. Sci. Rep. 2019, 9, 17979. [Google Scholar] [CrossRef] [PubMed]
- Rashnonejad, A.; Amini Chermahini, G.; Gündüz, C.; Onay, H.; Aykut, A.; Durmaz, B.; Baka, M.; Su, Q.; Gao, G.; Özkınay, F. Fetal gene therapy using a single injection of recombinant AAV9 rescued SMA phenotype in mice. Mol. Ther. 2019, 27, 2123–2133. [Google Scholar] [CrossRef]
- Shangaris, P.; Loukogeorgakis, S.P.; Subramaniam, S.; Flouri, C.; Jackson, L.H.; Wang, W.; Blundell, M.P.; Liu, S.; Eaton, S.; Bakhamis, N.; et al. In utero gene therapy (IUGT) using GLOBE lentiviral vector phenotypically corrects the heterozygous humanised mouse model and its progress can be monitored using MRI techniques. Sci. Rep. 2019, 9, 11592. [Google Scholar] [CrossRef]
- Papaioannou, V.E. In utero manipulations, in Post Implantation Mammalian Embryos, A Practical Approach. In A Practical Approach; Copp, A.J., Cockroft, D.L., Eds.; Oxford University Press: Oxford, UK, 1990; pp. 61–80. [Google Scholar]
- Woo, Y.J.; Raju, G.P.; Swain, J.L.; Richmond, M.E.; Gardner, T.J.; Balice-Gordon, R.J. In utero cardiac gene transfer via intraplacental delivery of recombinant adenovirus. Circulation 1997, 96, 3561–3569. [Google Scholar] [CrossRef]
- Türkay, A.; Saunders, T.; Kurachi, K. Intrauterine gene transfer: Gestational stage-specific gene delivery in mice. Gene Ther. 1999, 6, 1685–1694. [Google Scholar] [CrossRef]
- Larson, J.E.; Morrow, S.L.; Happel, L.; Sharp, J.F.; Cohen, J.C. Reversal of cystic fibrosis phenotype in mice by gene therapy in utero. Lancet 1997, 349, 619–620. [Google Scholar] [CrossRef] [PubMed]
- Douar, A.M.; Adebakin, S.; Themis, M.; Pavirani, A.; Cook, T.; Coutelle, C. Foetal gene delivery in mice by intra-amniotic administration of retroviral producer cells and adenovirus. Gene Ther. 1997, 4, 883–890. [Google Scholar] [CrossRef]
- Endoh, M.; Koibuchi, N.; Sato, M.; Morishita, R.; Kanzaki, T.; Murata, Y.; Kaneda, Y. Fetal gene transfer by intrauterine injection with microbubble-enhanced ultrasound. Mol. Ther. 2002, 5, 501–508. [Google Scholar] [CrossRef] [PubMed]
- Schachtner, S.; Buck, C.; Bergelson, J.; Baldwin, H. Temporally regulated expression patterns following in utero adenovirus-mediated gene transfer. Gene Ther. 1999, 6, 1249–1257. [Google Scholar] [CrossRef] [PubMed]
- Straub, C.; Granger, A.J.; Saulnier, L.; Sabatini, B.L. CRISPR/Cas9-mediated gene knock-down in post-mitotic neurons. PLoS ONE 2014, 9, e105584. [Google Scholar] [CrossRef] [PubMed]
- Intson, K.; van Eede, M.C.; Islam, R.; Milenkovic, M.; Yan, Y.; Salahpour, A.; Henkelman, R.M.; Ramsey, A.J. Progressive neuroanatomical changes caused by Grin1 loss-of-function mutation. Neurobiol. Dis. 2019, 132, 104527. [Google Scholar] [CrossRef] [PubMed]
- Shinmyo, Y.; Tanaka, S.; Tsunoda, S.; Hosomichi, K.; Tajima, A.; Kawasaki, H. CRISPR/Cas9-mediated gene knockout in the mouse brain using in utero electroporation. Sci. Rep. 2016, 6, 20611. [Google Scholar] [CrossRef]
- Kalebic, N.; Taverna, E.; Tavano, S.; Wong, F.K.; Suchold, D.; Winkler, S.; Huttner, W.B.; Sarov, M. CRISPR/Cas9-induced disruption of gene expression in mouse embryonic brain and single neural stem cells in vivo. EMBO Rep. 2016, 17, 338–348. [Google Scholar] [CrossRef]
- Uemura, T.; Mori, T.; Kurihara, T.; Kawase, S.; Koike, R.; Satoga, M.; Cao, X.; Li, X.; Yanagawa, T.; Sakurai, T.; et al. Fluorescent protein tagging of endogenous protein in brain neurons using CRISPR/Cas9-mediated knock-in and in utero electroporation techniques. Sci. Rep. 2016, 6, 35861. [Google Scholar] [CrossRef]
- Antoniou, P.; Miccio, A.; Brusson, M. Base and prime editing technologies for blood disorders. Front. Genome Ed. 2021, 3, 618406. [Google Scholar] [CrossRef]
- Tsunekawa, Y.; Terhune, R.K.; Fujita, I.; Shitamukai, A.; Suetsugu, T.; Matsuzaki, F. Developing a de novo targeted knock-in method based on in utero electroporation into the mammalian brain. Development 2016, 143, 3216–3222. [Google Scholar] [CrossRef] [PubMed]
- Rossidis, A.C.; Stratigis, J.D.; Chadwick, A.C.; Hartman, H.A.; Ahn, N.J.; Li, H.; Singh, K.; Coons, B.E.; Li, L.; Lv, W.; et al. In utero CRISPR-mediated therapeutic editing of metabolic genes. Nat. Med. 2018, 24, 1513–1518. [Google Scholar] [CrossRef] [PubMed]
- Ricciardi, A.S.; Bahal, R.; Farrelly, J.S.; Quijano, E.; Bianchi, A.H.; Luks, V.L.; Putman, R.; López-Giráldez, F.; Coşkun, S.; Song, E.; et al. In utero nanoparticle delivery for site-specific genome editing. Nat. Commun. 2018, 9, 2481. [Google Scholar] [CrossRef] [PubMed]
- Alapati, D.; Zacharias, W.J.; Hartman, H.A.; Rossidis, A.C.; Stratigis, J.D.; Ahn, N.J.; Coons, B.; Zhou, S.; Li, H.; Singh, K.; et al. In utero gene editing for monogenic lung disease. Sci. Transl. Med. 2019, 11, eaav8375. [Google Scholar] [CrossRef]
- Wolter, J.M.; Mao, H.; Fragola, G.; Simon, J.M.; Krantz, J.L.; Bazick, H.O.; Oztemiz, B.; Stein, J.L.; Zylka, M.J. Cas9 gene therapy for Angelman syndrome traps Ube3a-ATS long non-coding RNA. Nature 2020, 587, 281–284. [Google Scholar] [CrossRef]
- Nakamura, S.; Ando, N.; Watanabe, S.; Akasaka, E.; Ishihara, M.; Sato, M. Hydrodynamics-based transplacental delivery as a useful noninvasive tool for manipulating fetal genome. Cells 2020, 9, 1744. [Google Scholar] [CrossRef]
- Fang, H.; Bygrave, A.M.; Roth, R.H.; Johnson, R.C.; Huganir, R.L. An optimized CRISPR/Cas9 approach for precise genome editing in neurons. eLife 2021, 10, e65202. [Google Scholar] [CrossRef]
- Bose, S.K.; White, B.M.; Kashyap, M.V.; Dave, A.; De Bie, F.R.; Li, H.; Singh, K.; Menon, P.; Wang, T.; Teerdhala, S.; et al. In utero adenine base editing corrects multi-organ pathology in a lethal lysosomal storage disease. Nat. Commun. 2021, 12, 4291. [Google Scholar] [CrossRef]
- Hu, S.; Yang, T.; Wang, Y. Widespread labeling and genomic editing of the fetal central nervous system by in utero CRISPR AAV9-PHP.eB administration. Development 2021, 148, dev195586. [Google Scholar] [CrossRef]
- Ma, S.; Jiang, W.; Liu, X.; Lu, W.J.; Qi, T.; Wei, J.; Wu, F.; Chang, Y.; Zhang, S.; Song, Y.; et al. Efficient correction of a hypertrophic cardiomyopathy mutation by ABEmax-NG. Circ. Res. 2021, 129, 895–908. [Google Scholar] [CrossRef]
- Rashid, S.; Curtis, D.E.; Garuti, R.; Anderson, N.N.; Bashmakov, Y.; Ho, Y.K.; Hammer, R.E.; Moon, Y.A.; Horton, J.D. Decreased plasma cholesterol and hypersensitivity to statins in mice lacking Pcsk9. Proc. Natl. Acad. Sci. USA 2005, 102, 5374–5379. [Google Scholar] [CrossRef] [PubMed]
- Grompe, M.; al-Dhalimy, M.; Finegold, M.; Ou, C.N.; Burlingame, T.; Kennaway, N.G.; Soriano, P. Loss of fumarylacetoacetate hydrolase is responsible for the neonatal hepatic dysfunction phenotype of lethal albino mice. Genes Dev. 1993, 7, 2298–2307. [Google Scholar] [CrossRef] [PubMed]
- Rossant, J.; Cross, J.C. Placental development: Lessons from mouse mutants. Nat. Rev. Genet. 2001, 2, 538–548. [Google Scholar] [CrossRef] [PubMed]
- Tetro, N.; Moushaev, S.; Rubinchik-Stern, M.; Eyal, S. The placental barrier: The gate and the fate in drug distribution. Pharm. Res. 2018, 35, 71. [Google Scholar] [CrossRef] [PubMed]
- Figueroa-Espada, C.G.; Hofbauer, S.; Mitchell, M.J.; Riley, R.S. Exploiting the placenta for nanoparticle-mediated drug delivery during pregnancy. Adv. Drug Deliv. Rev. 2020, 160, 244–261. [Google Scholar] [CrossRef]
- Tüzel-Kox, S.N.; Patel, H.M.; Kox, W.J. Uptake of drug-carrier liposomes by placenta: Transplacental delivery of drugs and nutrients. J. Pharmacol. Exp. Ther. 1995, 274, 104–109. [Google Scholar]
- Tsukamoto, M.; Ochiya, T.; Yoshida, S.; Sugimura, T.; Terada, M. Gene transfer and expression in progeny after intravenous DNA injection into pregnant mice. Nat. Genet. 1995, 9, 243–248. [Google Scholar] [CrossRef]
- Wu, N.; Yu, A.B.; Zhu, H.B.; Lin, X.K. Effective silencing of Sry gene with RNA interference in developing mouse embryos resulted in feminization of XY gonad. J. Biomed. Biotechnol. 2012, 2012, 343891. [Google Scholar] [CrossRef]
- Li, J.; Zhang, Y.; Li, D.; Liu, Y.; Chu, D.; Jiang, X.; Hou, D.; Zen, K.; Zhang, C.Y. Small non-coding RNAs transfer through mammalian placenta and directly regulate fetal gene expression. Protein Cell 2015, 6, 391–396. [Google Scholar] [CrossRef]
- Okuda, K.; Xin, K.Q.; Haruki, A.; Kawamoto, S.; Kojima, Y.; Hirahara, F.; Okada, H.; Klinman, D.; Hamajima, K. Transplacental genetic immunization after intravenous delivery of plasmid DNA to pregnant mice. J. Immunol. 2001, 167, 5478–5484. [Google Scholar] [CrossRef]
- Ochiya, T.; Takahama, Y.; Baba-Toriyama, H.; Tsukamoto, M.; Yasuda, Y.; Kikuchi, H.; Terada, M. Evaluation of cationic liposome suitable for gene transfer into pregnant animals. Biochem. Biophys. Res. Commun. 1999, 258, 358–365. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, N.; Nakamura, S.; Ohtsuka, M.; Kimura, M.; Sato, M. Possible mechanism of gene transfer into early to mid-gestational mouse fetuses by tail vein injection. Gene Ther. 2002, 9, 1529–1541. [Google Scholar] [CrossRef] [PubMed]
- Maeda-Mamiya, R.; Noiri, E.; Isobe, H.; Nakanishi, W.; Okamoto, K.; Doi, K.; Sugaya, T.; Izumi, T.; Homma, T.; Nakamura, E. In vivo gene delivery by cationic tetraamino fullerene. Proc. Natl. Acad. Sci. USA 2010, 107, 5339–5344. [Google Scholar] [CrossRef]
- Cornford, E.M.; Hyman, S.; Cornford, M.E.; Chytrova, G.; Rhee, J.; Suzuki, T.; Yamagata, T.; Yamakawa, K.; Penichet, M.L.; Pardridge, W.M. Non-invasive gene targeting to the fetal brain after intravenous administration and transplacental transfer of plasmid DNA using PEGylated immunoliposomes. J. Drug Target. 2016, 24, 58–67. [Google Scholar] [CrossRef] [PubMed]
- Picconi, J.L.; Muff-Luett, M.A.; Wu, D.; Bunchman, E.; Schaefer, F.; Brophy, P.D. Kidney-specific expression of GFP by in-utero delivery of pseudotyped adeno-associated virus 9. Mol. Ther. Methods Clin. Dev. 2014, 1, 14014. [Google Scholar] [CrossRef] [PubMed]
- Durand, J.B.; Abchee, A.B.; Roberts, R. Molecular and clinical aspects of inherited cardiomyopathies. Ann. Med. 1995, 27, 311–317. [Google Scholar] [CrossRef]
- Lino, C.A.; Harper, J.C.; Carney, J.P.; Timlin, J.A. Delivering CRISPR: A review of the challenges and approaches. Drug Deliv. 2018, 25, 1234–1257. [Google Scholar] [CrossRef]
- Yin, H.; Kanasty, R.L.; Eltoukhy, A.A.; Vegas, A.J.; Dorkin, J.R.; Anderson, D.G. Non-viral vectors for gene-based therapy. Nat. Rev. Genet. 2014, 15, 541–555. [Google Scholar] [CrossRef]
- Wang, M.; Glass, Z.A.; Xu, Q. Non-viral delivery of genome-editing nucleases for gene therapy. Gene Ther. 2017, 24, 144–150. [Google Scholar] [CrossRef]
- Dammes, N.; Peer, D. Paving the road for RNA therapeutics. Trends Pharmacol. Sci. 2020, 41, 755–775. [Google Scholar] [CrossRef]
- Duan, L.; Ouyang, K.; Xu, X.; Xu, L.; Wen, C.; Zhou, X.; Qin, Z.; Xu, Z.; Sun, W.; Liang, Y. Nanoparticle Delivery of CRISPR/Cas9 for genome editing. Front. Genet. 2021, 12, 673286. [Google Scholar] [CrossRef] [PubMed]
- Ashok, B.; Peppas, N.A.; Wechsler, M.E. Lipid- and polymer-based nanoparticle systems for the delivery of CRISPR/Cas9. J. Drug Deliv. Sci. Technol. 2021, 65, 102728. [Google Scholar] [CrossRef] [PubMed]
- Rouatbi, N.; McGlynn, T.; Al-Jamal, K.T. Pre-clinical non-viral vectors exploited for in vivo CRISPR/Cas9 gene editing: An overview. Biomater. Sci. 2022, 10, 3410–3432. [Google Scholar] [CrossRef]
- Wei, T.; Cheng, Q.; Min, Y.L.; Olson, E.N.; Siegwart, D.J. Systemic nanoparticle delivery of CRISPR-Cas9 ribonucleoproteins for effective tissue specific genome editing. Nat. Commun. 2020, 11, 3232. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Li, B.; Luo, X.; Zhao, W.; Jiang, J.; Zhang, C.; Gao, M.; Chen, X.; Dong, Y. Biodegradable amino-ester nanomaterials for Cas9 mRNA delivery in vitro and in vivo. ACS Appl. Mater. Interfaces 2017, 9, 25481–25487. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.B.; Zhang, S.; Kos, P.; Xiong, H.; Zhou, K.; Perelman, S.S.; Zhu, H.; Siegwart, D.J. Non-viral CRISPR/Cas gene editing in vitro and in vivo enabled by synthetic nanoparticle co-delivery of Cas9 mRNA and sgRNA. Angew. Chem. Int. Ed. Engl. 2017, 56, 1059–1063. [Google Scholar] [CrossRef]
- Finn, J.D.; Smith, A.R.; Patel, M.C.; Shaw, L.; Youniss, M.R.; van Heteren, J.; Dirstine, T.; Ciullo, C.; Lescarbeau, R.; Seitzer, J.; et al. A Single Administration of CRISPR/Cas9 lipid nanoparticles achieves robust and persistent in vivo genome editing. Cell Rep. 2018, 22, 2227–2235. [Google Scholar] [CrossRef]
- Shi, J.; Yang, X.; Li, Y.; Wang, D.; Liu, W.; Zhang, Z.; Liu, J.; Zhang, K. MicroRNA-responsive release of Cas9/sgRNA from DNA nanoflower for cytosolic protein delivery and enhanced genome editing. Biomaterials 2020, 256, 120221. [Google Scholar] [CrossRef]
- Yin, L.; Song, Z.; Kim, K.H.; Zheng, N.; Tang, H.; Lu, H.; Gabrielson, N.; Cheng, J. Reconfiguring the architectures of cationic helical polypeptides to control non-viral gene delivery. Biomaterials 2013, 34, 2340–2349. [Google Scholar] [CrossRef]
- Luo, Y.L.; Xu, C.F.; Li, H.J.; Cao, Z.T.; Liu, J.; Wang, J.L.; Du, X.J.; Yang, X.Z.; Gu, Z.; Wang, J. Macrophage-specific in vivo gene editing using cationic lipid-assisted polymeric nanoparticles. ACS Nano 2018, 12, 994–1005. [Google Scholar] [CrossRef]
- Liu, B.Y.; He, X.Y.; Xu, C.; Xu, L.; Ai, S.L.; Cheng, S.X.; Zhuo, R.X. A dual-targeting delivery system for effective genome editing and in situ detecting related protein expression in edited cells. Biomacromolecules 2018, 19, 2957–2968. [Google Scholar] [CrossRef] [PubMed]
- Qiao, J.; Sun, W.; Lin, S.; Jin, R.; Ma, L.; Liu, Y. Cytosolic delivery of CRISPR/Cas9 ribonucleoproteins for genome editing using chitosan-coated red fluorescent protein. Chem. Commun. 2019, 55, 4707–4710. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Zhao, K.; Wang, C.; Zhang, Z.; Zheng, C.; Zhao, Y.; Zheng, Y.; Liu, C.; An, Y.; Shi, L.; et al. Multistage delivery nanoparticle facilitates efficient CRISPR/dCas9 activation and tumor growth suppression in vivo. Adv. Sci. 2019, 6, 1801423. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Wan, T.; Chen, Y.; Chen, Y.; Sun, H.; Cao, T.; Songyang, Z.; Tang, G.; Wu, C.; Ping, Y.; et al. Cationic polymer-mediated CRISPR/Cas9 plasmid delivery for genome editing. Macromol. Rapid. Commun. 2019, 40, e1800068. [Google Scholar] [CrossRef]
- Zhang, B.-C.; Wu, P.-Y.; Zou, J.-J.; Jiang, J.-L.; Zhao, R.-R.; Luo, B.-Y.; Liao, Y.-Q.; Shao, J.-W. Efficient CRISPR/Cas9 gene-chemo synergistic cancer therapy via a stimuli-responsive chitosan-based nanocomplex elicits anti-tumorigenic pathway effect. Chem. Eng. J. 2020, 393, 124688. [Google Scholar] [CrossRef]
- Lee, K.; Conboy, M.; Park, H.M.; Jiang, F.; Kim, H.J.; Dewitt, M.A.; Mackley, V.A.; Chang, K.; Rao, A.; Skinner, C.; et al. Nanoparticle delivery of Cas9 ribonucleoprotein and donor DNA in vivo induces homology-directed DNA repair. Nat. Biomed. Eng. 2017, 1, 889–901. [Google Scholar] [CrossRef]
- Mout, R.; Ray, M.; Yesilbag Tonga, G.; Lee, Y.W.; Tay, T.; Sasaki, K.; Rotello, V.M. Direct cytosolic delivery of CRISPR/Cas9-ribonucleoprotein for efficient gene editing. ACS Nano 2017, 11, 2452–2458. [Google Scholar] [CrossRef]
- Mout, R.; Rotello, V.M. Cytosolic and nuclear delivery of CRISPR/Cas9-ribonucleoprotein for gene editing using arginine functionalized gold nanoparticles. Bio Protoc. 2017, 7, e2586. [Google Scholar] [CrossRef]
- Lee, B.; Lee, K.; Panda, S.; Gonzales-Rojas, R.; Chong, A.; Bugay, V.; Park, H.M.; Brenner, R.; Murthy, N.; Lee, H.Y. Nanoparticle delivery of CRISPR into the brain rescues a mouse model of fragile X syndrome from exaggerated repetitive behaviours. Nat. Biomed. Eng. 2018, 2, 497–507. [Google Scholar] [CrossRef]
- Wang, P.; Zhang, L.; Zheng, W.; Cong, L.; Guo, Z.; Xie, Y.; Wang, L.; Tang, R.; Feng, Q.; Hamada, Y.; et al. Thermo-triggered Release of CRISPR-Cas9 system by lipid-encapsulated gold nanoparticles for tumor therapy. Angew. Chem. Int. Ed. Engl. 2018, 57, 1491–1496. [Google Scholar] [CrossRef]
- Tao, Y.; Yi, K.; Hu, H.; Shao, D.; Li, M. Coassembly of nucleus-targeting gold nanoclusters with CRISPR/Cas9 for simultaneous bioimaging and therapeutic genome editing. J. Mater. Chem. B 2021, 9, 94–100. [Google Scholar] [CrossRef] [PubMed]
- Usman, W.M.; Pham, T.C.; Kwok, Y.Y.; Vu, L.T.; Ma, V.; Peng, B.; Chan, Y.S.; Wei, L.; Chin, S.M.; Azad, A.; et al. Efficient RNA drug delivery using red blood cell extracellular vesicles. Nat. Commun. 2018, 9, 2359. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zhou, X.; Wei, M.; Gao, X.; Zhao, L.; Shi, R.; Sun, W.; Duan, Y.; Yang, G.; Yuan, L. In vitro and in vivo RNA inhibition by CD9-HuR functionalized exosomes encapsulated with miRNA or CRISPR/dCas9. Nano Lett. 2019, 19, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Xu, Q.; Zi, Z.; Liu, Z.; Wan, C.; Crisman, L.; Shen, J.; Liu, X. Programmable extracellular vesicles for macromolecule delivery and genome modifications. Dev. Cell 2020, 55, 784–801.e789. [Google Scholar] [CrossRef] [PubMed]
- Gee, P.; Lung, M.S.Y.; Okuzaki, Y.; Sasakawa, N.; Iguchi, T.; Makita, Y.; Hozumi, H.; Miura, Y.; Yang, L.F.; Iwasaki, M.; et al. Extracellular nanovesicles for packaging of CRISPR-Cas9 protein and sgRNA to induce therapeutic exon skipping. Nat. Commun. 2020, 11, 1334. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Liang, X.; Xie, H.; Kumar, S.; Ravinder, N.; Potter, J.; de Mollerat du Jeu, X.; Chesnut, J.D. Improved delivery of Cas9 protein/gRNA complexes using lipofectamine CRISPRMAX. Biotechnol. Lett. 2016, 38, 919–929. [Google Scholar] [CrossRef]
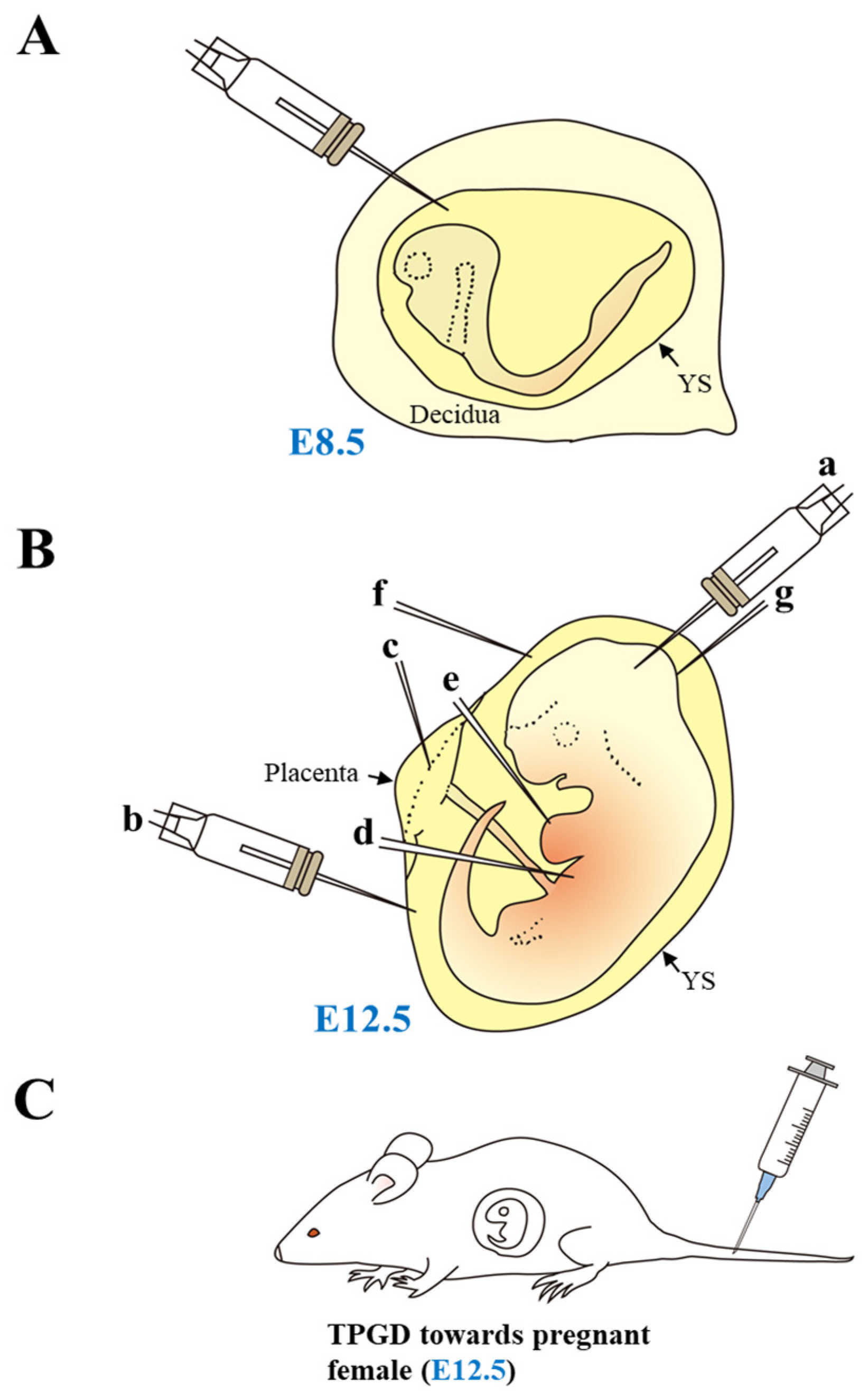
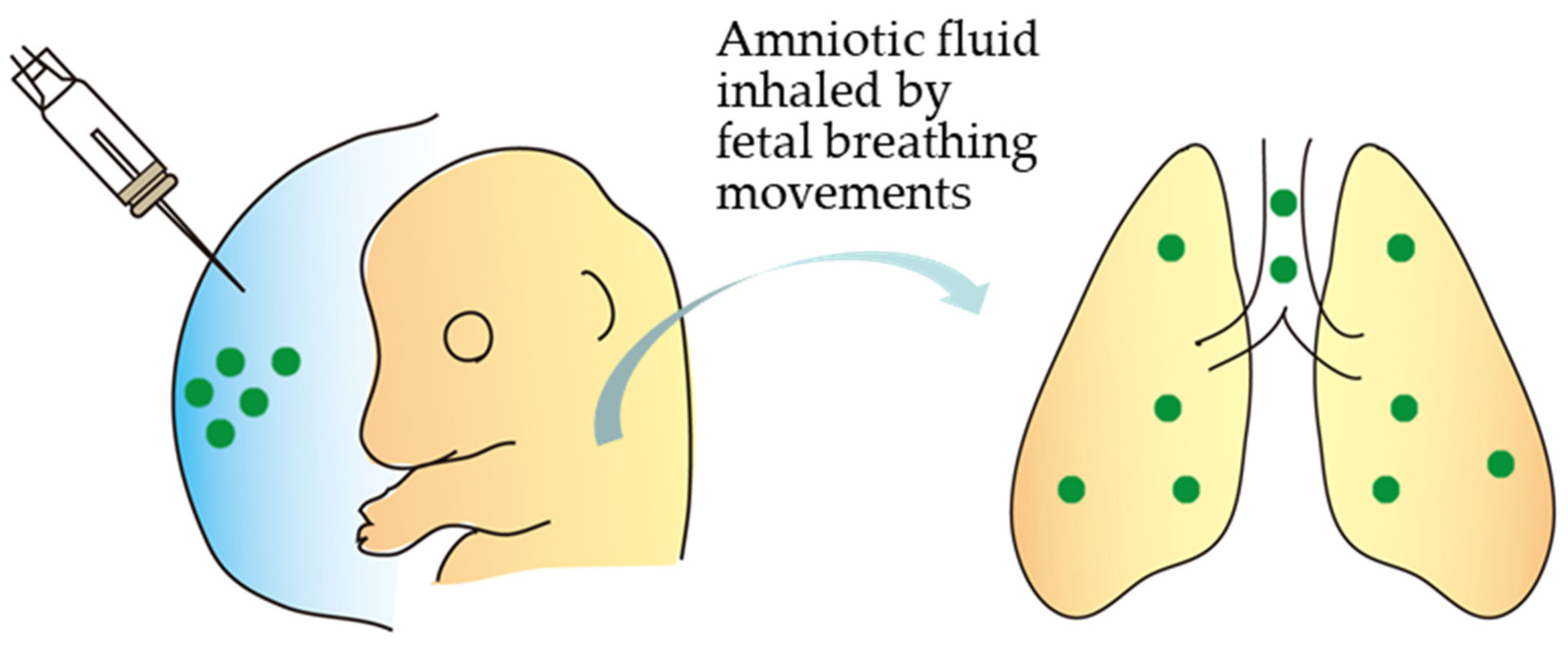
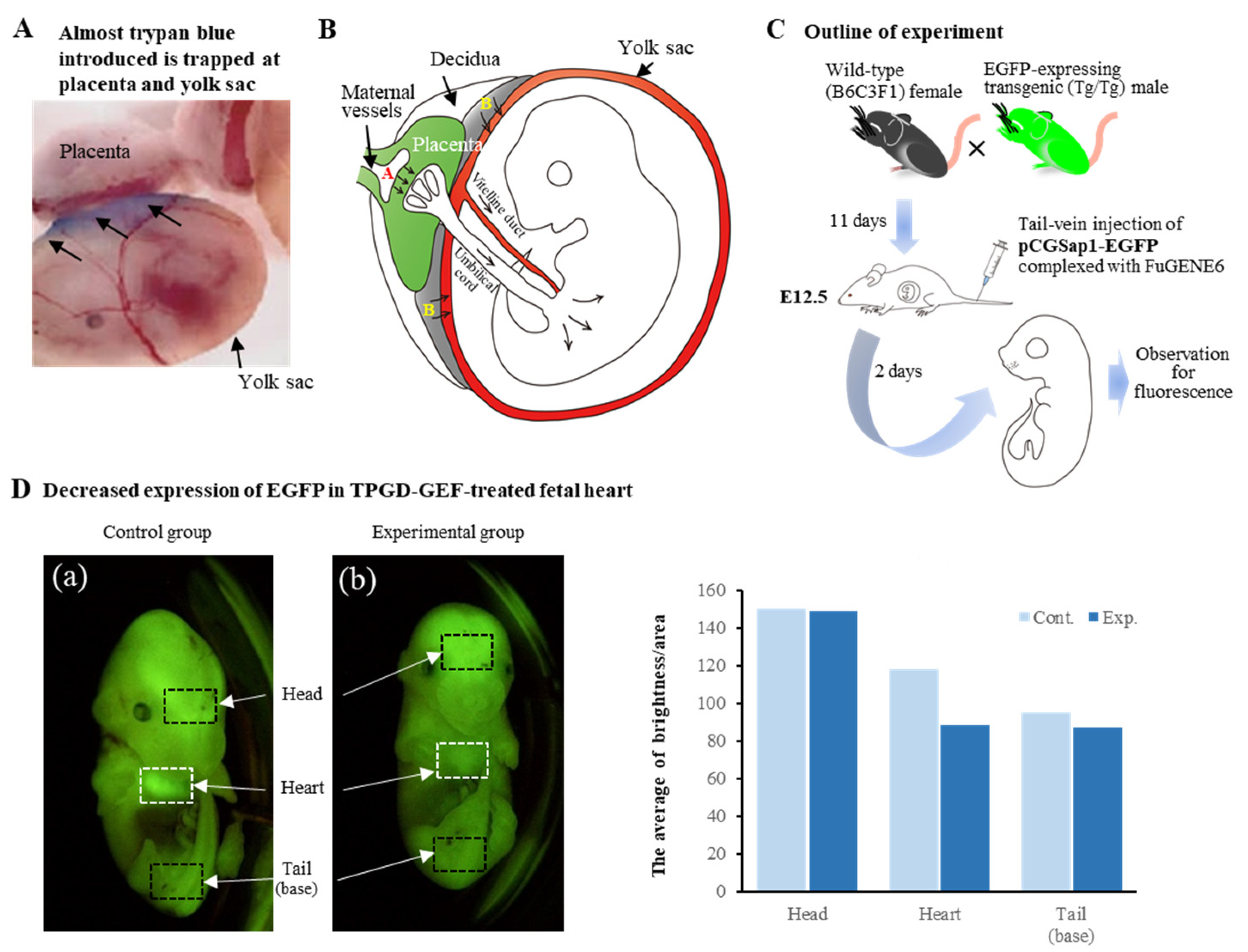
| Procedure | Ease of Procedure | Tissue Type | Efficacy | Safety | Cost | Data from |
|---|---|---|---|---|---|---|
| In utero injection of plasmid DNA and subsequent in vivo EP | Relatively easy but accompanied with surgical procedure | Brain | Depending on the technique of the researchers | Risk of embryonic loss and tissue damage during surgery | Requires electroporator and microinjector device (in some cases) | [37,39,40,41,43,49] |
| In utero injection of Ad containing BE3-based components | Relatively easy but accompanied with surgical procedure | Lung, intestine | High | Risk of embryonic loss and tissue damage during surgery and also for immunologic response | Requires microinjector device (in some cases) | [44,46] |
| In utero injection of NP containing PNAs and donor DNA | Relatively easy but accompanied with surgical procedure | Whole fetus (especially accumulated abundantly in fetal liver) | Relatively high | Risk of embryonic loss and tissue damage during surgery | Requires microinjector device (in some cases) | [45] |
| TPGD-GEF using liposome-encapsulated plasmid DNA | Easy | Whole fetus (especially accumulated abundantly in fetal heart) | Low | Very low risk for both mother and fetuses (however, HGD, poses risk of damage in both mother and fetus) | Requires only tail-injection technique | [7,48] |
| In utero injection of rAAVs containing CRISPR/Cas9 components | Relatively easy but accompanied with surgical procedure | Brain | High | Risk for embryonic loss and tissue damage during surgery; risk of immune response | Requires microinjector device (in some cases) | [47,49,51] |
| In utero injection of rAAVs containing ABE | Relatively easy but accompanied with surgical procedure | Whole fetus (especially abundantly accumulated in fetal liver) | High | Risk for embryonic loss and tissue damage during surgery; risk of immune response | Requires microinjector device (in some cases) | [50,52] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nakamura, S.; Inada, E.; Saitoh, I.; Sato, M. Recent Genome-Editing Approaches toward Post-Implanted Fetuses in Mice. BioTech 2023, 12, 37. https://doi.org/10.3390/biotech12020037
Nakamura S, Inada E, Saitoh I, Sato M. Recent Genome-Editing Approaches toward Post-Implanted Fetuses in Mice. BioTech. 2023; 12(2):37. https://doi.org/10.3390/biotech12020037
Chicago/Turabian StyleNakamura, Shingo, Emi Inada, Issei Saitoh, and Masahiro Sato. 2023. "Recent Genome-Editing Approaches toward Post-Implanted Fetuses in Mice" BioTech 12, no. 2: 37. https://doi.org/10.3390/biotech12020037
APA StyleNakamura, S., Inada, E., Saitoh, I., & Sato, M. (2023). Recent Genome-Editing Approaches toward Post-Implanted Fetuses in Mice. BioTech, 12(2), 37. https://doi.org/10.3390/biotech12020037








