Using Nano-Luciferase Binary (NanoBiT) Technology to Assess the Interaction Between Viral Spike Protein and Angiotensin-Converting Enzyme II by Aptamers
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Generation of the HEK293T/LgBiT-hACE2 Cell Line
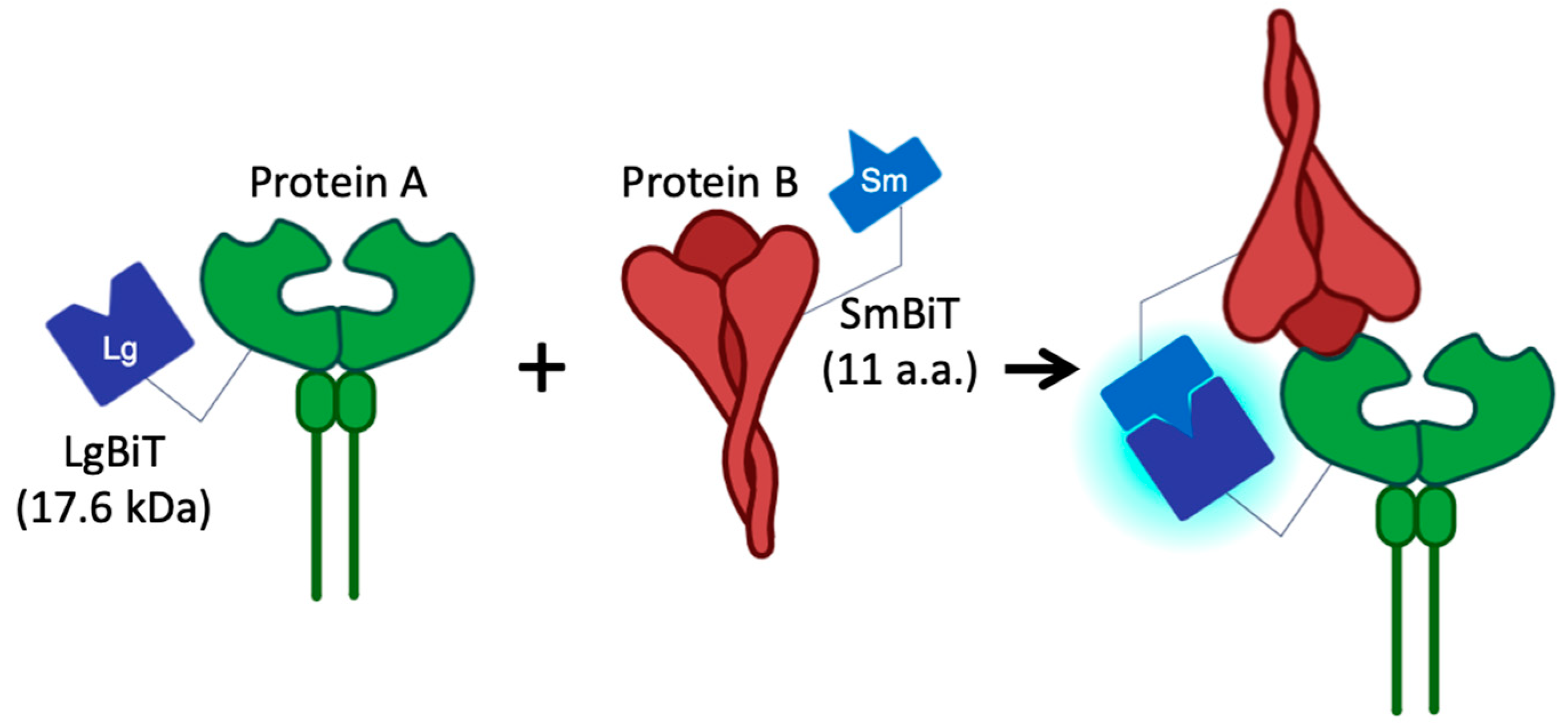
2.3. Production of the NanoBiT-Based Pseudovirus
2.4. NanoBiT Detection
2.5. Production of the GFP- and Luciferase-Based Pseudovirus
2.6. Fluorescence of GFP and Luminescence of Luciferase Detection
2.7. Western Blot Analysis
2.8. Aptamer Sequences and Structure Prediction
2.9. Cy5-Aptamer and Confocal Microscopy
2.10. Statistical Analysis
3. Results
3.1. Validation of HEK293T/LgBiT-hACE2 Cells and the SmBiT-BA.2 Pseudovirus
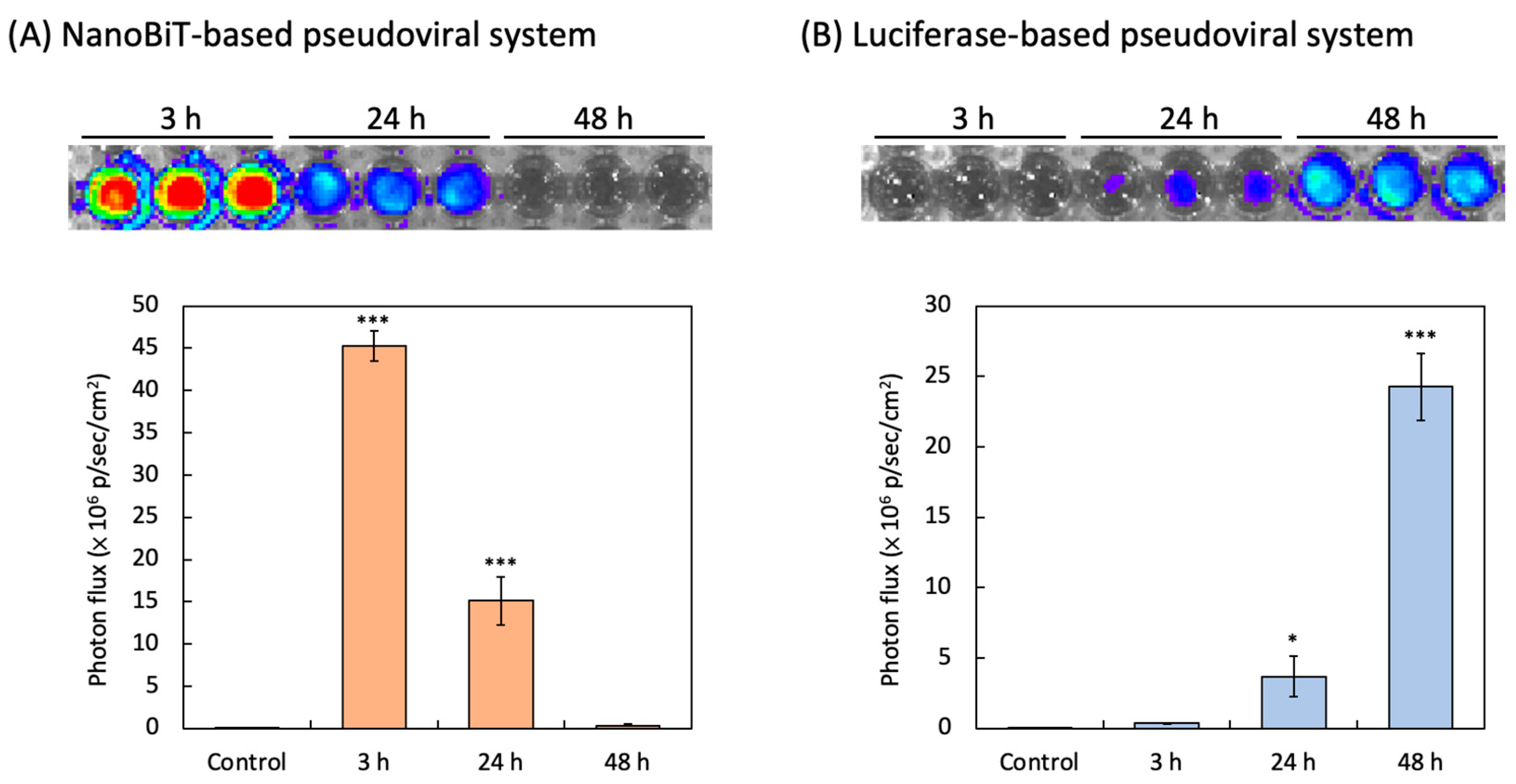
3.2. NanoBiT-Based and Luciferase-Based SARS-CoV-2 Spike Pseudovirus Infection
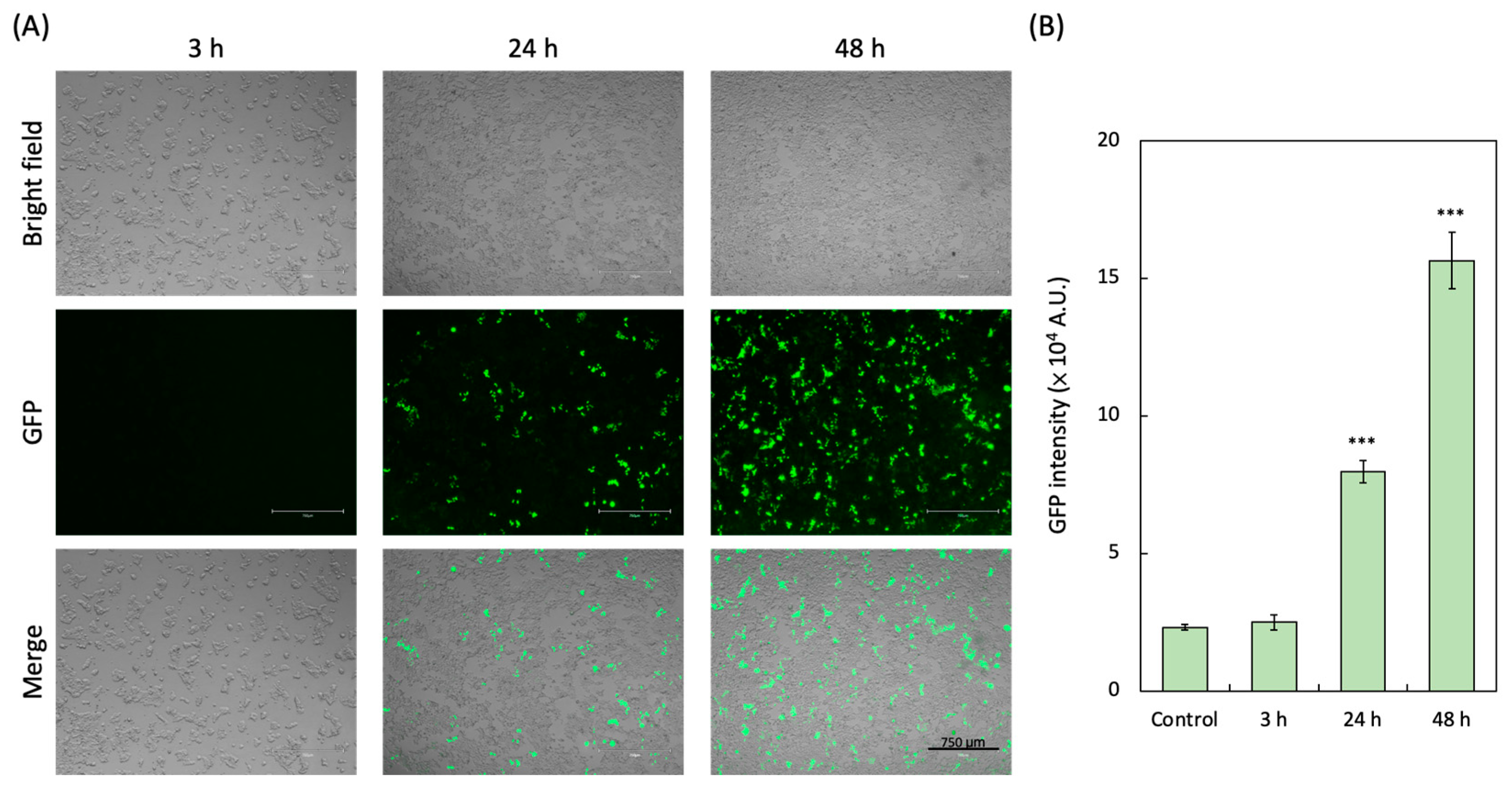
3.3. GFP-Based Pseudoviral SARS-CoV-2 Infection
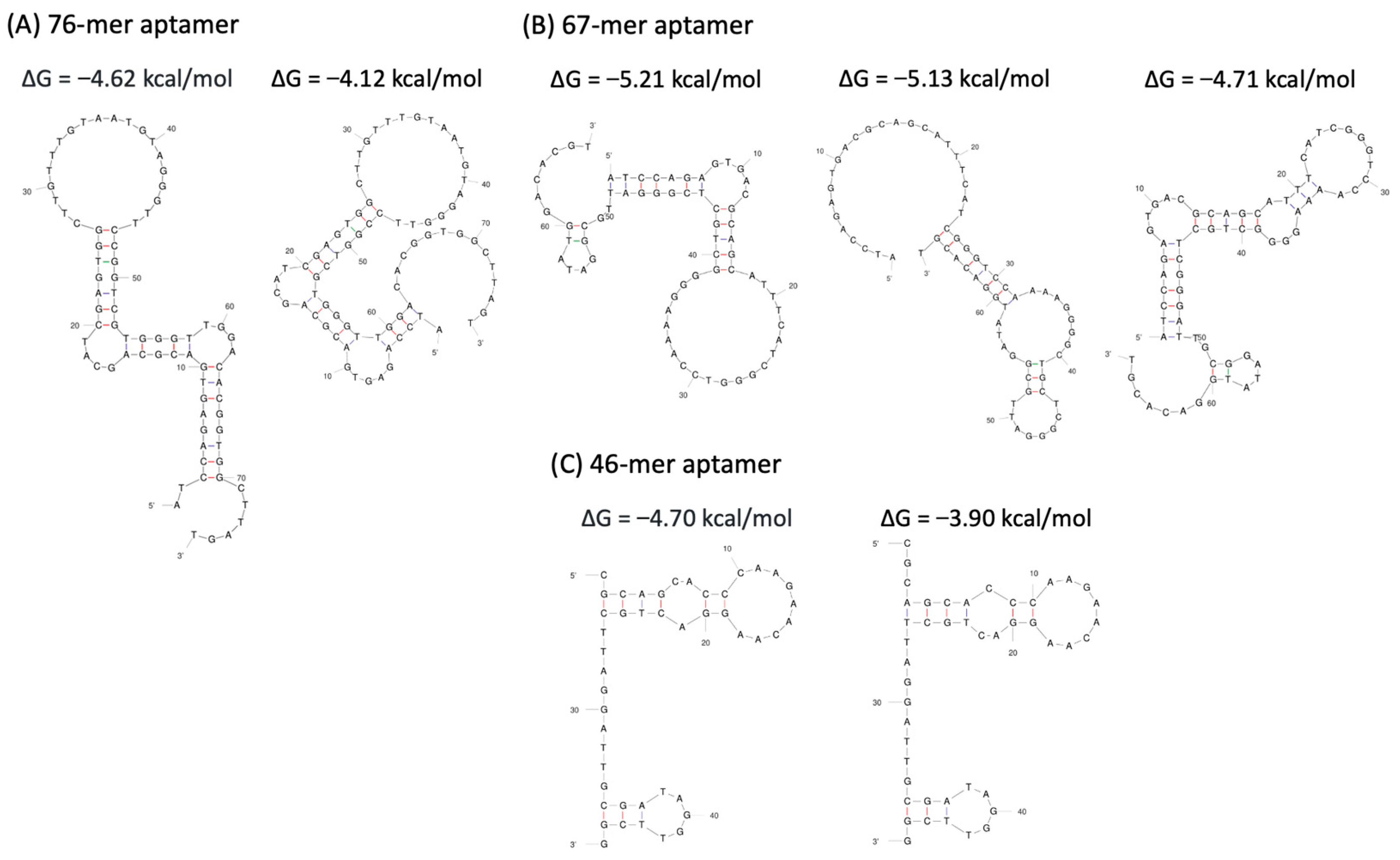
3.4. Prediction of Aptamer Sequences
3.5. Inhibition of the Aptamers on NanoBiT-Based Pseudoviral SARS-CoV-2
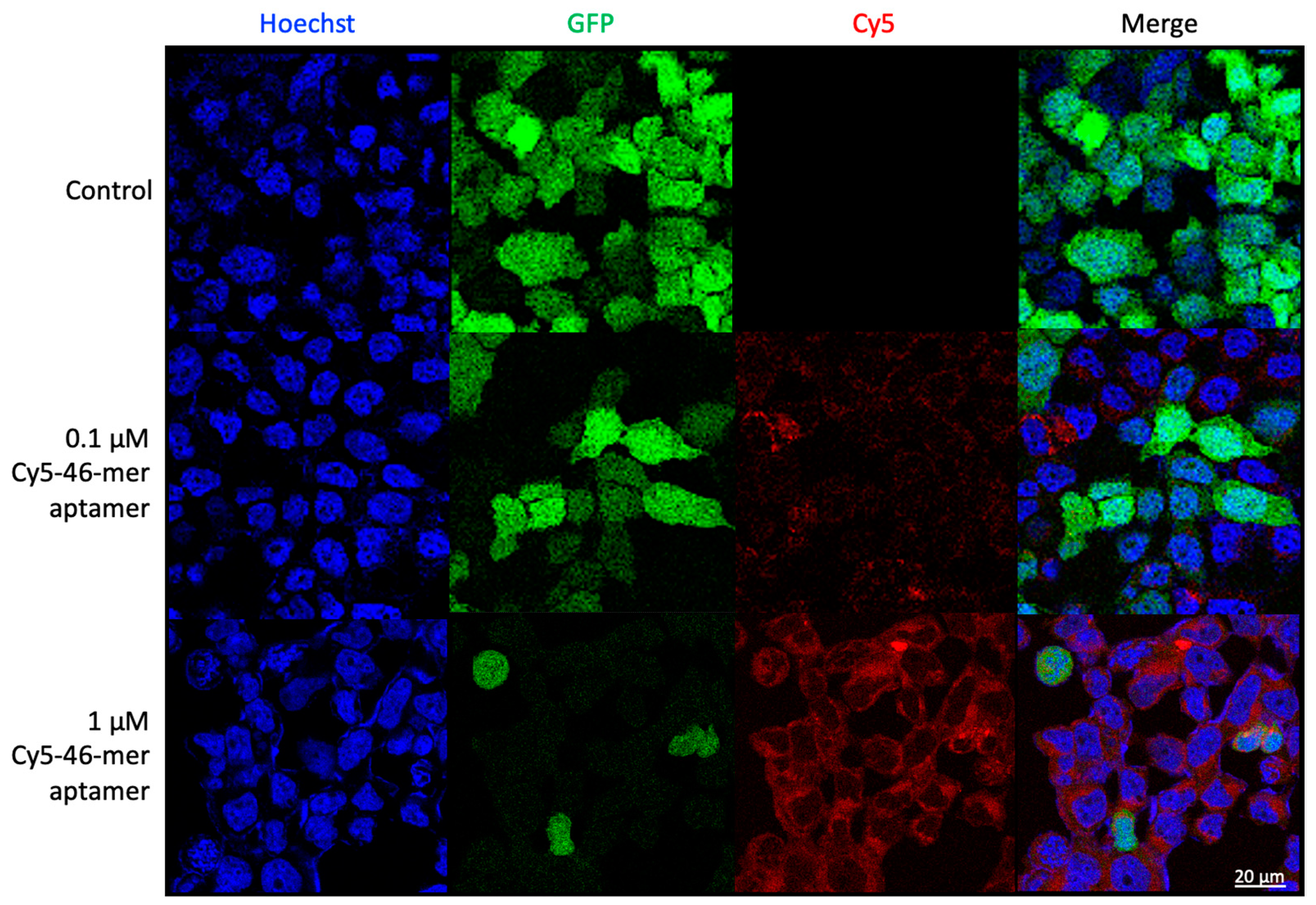
3.6. Aptamers as Detection Reagents for SARS-CoV-2
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Almulla, N.; Soltane, R.; Alasiri, A.; Allayeh, A.K.; Alqadi, T.; Alshehri, F.; Hamad Alrokban, A.; Zaghlool, S.S.; Zayan, A.Z.; Abdalla, K.F.; et al. Advancements in SARS-CoV-2 detection: Navigating the molecular landscape and diagnostic technologies. Heliyon 2024, 10, e29909. [Google Scholar] [CrossRef]
- Brito, A.F.; Pinney, J.W. Protein-protein interactions in virus-host systems. Front. Microbiol. 2017, 8, 1557. [Google Scholar] [CrossRef]
- Perrin-Cocon, L.; Diaz, O.; Jacquemin, C.; Barthel, V.; Ogire, E.; Ramière, C.; André, P.; Lotteau, V.; Vidalain, P.O. The current landscape of coronavirus-host protein–protein interactions. J. Transl. Med. 2020, 18, 319. [Google Scholar] [CrossRef]
- Dixon, A.S.; Schwinn, M.K.; Hall, M.P.; Zimmerman, K.; Otto, P.; Lubben, T.H.; Butler, B.L.; Binkowski, B.F.; Machleidt, T.; Kirkland, T.A.; et al. NanoLuc complementation reporter optimized for accurate measurement of protein interactions in cells. ACS Chem. Biol. 2016, 11, 400–408. [Google Scholar] [CrossRef]
- Kelly, T.; Yang, X. Application of fluorescence- and bioluminescence-based biosensors in cancer drug discovery. Biosensors 2024, 14, 570. [Google Scholar] [CrossRef]
- Rozbeh, R.; Forchhammer, K. Split NanoLuc technology allows quantitation of interactions between PII protein and its receptors with unprecedented sensitivity and reveals transient interactions. Sci. Rep. 2021, 11, 12535. [Google Scholar] [CrossRef]
- Lee, R.K.-L.; Li, T.-N.; Chang, S.-Y.; Chao, T.-L.; Kuo, C.-H.; Pan, M.Y.-C.; Chiou, Y.-T.; Liao, K.-J.; Yang, Y.; Wu, Y.-H.; et al. Identification of entry inhibitors against delta and omicron variants of SARS-CoV-2. Int. J. Mol. Sci. 2022, 23, 4050. [Google Scholar] [CrossRef]
- Lin, C.H.; Yang, X.R.; Lin, M.W.; Chang, H.J.; Lee, C.H.; Lin, C.S. Engineering a NanoBiT biosensor for detecting angiotensin-converting enzyme-2 (hACE2) interaction with SARS-CoV-2 spike protein and screening the inhibitors to block hACE2 and spike interaction. Biosens. Bioelectron. 2024, 263, 116630. [Google Scholar] [CrossRef]
- Azad, T.; Janse van Rensburg, H.J.; Morgan, J.; Rezaei, R.; Crupi, M.J.F.; Chen, R.; Ghahremani, M.; Jamalkhah, M.; Forbes, N.; Ilkow, C.; et al. Luciferase-based biosensors in the era of the COVID-19 pandemic. ACS Nanosci. Au 2021, 1, 15–37. [Google Scholar] [CrossRef]
- Domsicova, M.; Korcekova, J.; Poturnayova, A.; Breier, A. New insights into aptamers: An alternative to antibodies in the detection of molecular biomarkers. Int. J. Mol. Sci. 2024, 25, 6833. [Google Scholar] [CrossRef]
- Takahashi, M. Nucleic acid aptamers emerging as modulators of G-protein-coupled receptors: Challenge to difficult cell surface proteins. Cells 2022, 11, 1825. [Google Scholar] [CrossRef]
- Kaur, H.; Bruno, J.G.; Kumar, A.; Sharma, T.K. Aptamers in the therapeutics and diagnostics pipelines. Theranostics 2018, 8, 4016–4032. [Google Scholar] [CrossRef]
- Trunzo, N.E.; Hong, K.L. Recent progress in the identification of aptamers against bacterial origins and their diagnostic applications. Int. J. Mol. Sci. 2020, 21, 5074. [Google Scholar] [CrossRef]
- Zhang, Y.; Juhas, M.; Kwok, C.K. Aptamers targeting SARS-CoV-2: A promising tool to fight against COVID-19. Trends Biotechnol. 2023, 41, 528–544. [Google Scholar] [CrossRef]
- Akmal Shukri, A.M.; Wang, S.M.; Feng, C.; Chia, S.L.; Mohd Nawi, S.F.A.; Citartan, M. In silico selection of aptamers against SARS-CoV-2. Analyst 2024, 149, 4770–4788. [Google Scholar] [CrossRef]
- Lin, Y.-C.; Chen, W.-Y.; Hwu, E.-T.; Hu, W.-P. In-Silico Selection of Aptamer Targeting SARS-CoV-2 Spike Protein. Int. J. Mol. Sci. 2022, 23, 5810. [Google Scholar] [CrossRef]
- Liu, X.; Wang, Y.L.; Wu, J.; Qi, J.; Zeng, Z.; Wan, Q.; Chen, Z.; Manandhar, P.; Cavener, V.S.; Boyle, N.R.; et al. Neutralizing Aptamers Block S/RBD-ACE2 Interactions and Prevent Host Cell Infection. Angew. Chem. Int. Ed. 2021, 60, 10273–10278. [Google Scholar] [CrossRef]
- Song, Y.; Song, J.; Wei, X.; Huang, M.; Sun, M.; Zhu, L.; Lin, B.; Shen, H.; Zhu, Z.; Yang, C. Discovery of Aptamers Targeting the Receptor-Binding Domain of the SARS-CoV-2 Spike Glycoprotein. Anal. Chem. 2020, 92, 9895–9900. [Google Scholar] [CrossRef]
- Sun, M.; Liu, S.; Wei, X.; Wan, S.; Huang, M.; Song, T.; Lu, Y.; Weng, X.; Lin, Z.; Chen, H.; et al. Aptamer blocking strategy inhibits SARS-CoV-2 virus infection. Angew. Chem. Int. Ed. 2021, 60, 10266–10272. [Google Scholar] [CrossRef]
- Hung, Y.H.; Hsieh, W.Y.; Hsieh, J.S.; Liu, F.C.; Tsai, C.H.; Lu, L.C.; Huang, C.Y.; Wu, C.L.; Lin, C.S. Alternative roles of STAT3 and MAPK signaling pathways in the MMPs activation and progression of lung injury induced by cigarette smoke exposure in ACE2 knockout mice. Int. J. Biol. Sci. 2016, 12, 454–465. [Google Scholar] [CrossRef]
- Yang, M.; Li, C.; Ye, G.; Shen, C.; Shi, H.; Zhong, L.; Tian, Y.; Zhao, M.; Wu, P.; Hussain, A.; et al. Aptamers targeting SARS-CoV-2 nucleocapsid protein exhibit potential anti pan-coronavirus activity. Signal Transduct. Target. Ther. 2024, 9, 40. [Google Scholar] [CrossRef] [PubMed]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef] [PubMed]
- Bojadzic, D.; Alcazar, O.; Chen, J.; Chuang, S.T.; Condor Capcha, J.M.; Shehadeh, L.A.; Buchwald, P. Small-molecule inhibitors of the Coronavirus spike: ACE2 protein-protein interaction as blockers of viral attachment and entry for SARS-CoV-2. ACS Infect. Dis. 2021, 7, 1519–1534. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yuan, H.; Li, X.; Wang, H. Spike protein mediated membrane fusion during SARS-CoV-2 infection. J. Med. Virol. 2023, 95, e28212. [Google Scholar] [CrossRef]
- Suvarnapathaki, S.; Chauhan, D.; Nguyen, A.; Ramalingam, M.; Camci-Unal, G. Advances in targeting ACE2 for developing COVID-19 therapeutics. Ann. Biomed. Eng. 2022, 50, 1734–1749. [Google Scholar] [CrossRef]
- Sun, S.; Yang, X.; Wang, Y.; Shen, X. In vivo analysis of protein–protein interactions with bioluminescence resonance energy transfer (BRET): Progress and prospects. Int. J. Mol. Sci. 2016, 17, 1704. [Google Scholar] [CrossRef]
- Xiang, Q.; Li, L.; Wu, J.; Tian, M.; Fu, Y. Application of pseudovirus system in the development of vaccine, antiviral-drugs, and neutralizing antibodies. Microbiol. Res. 2022, 258, 126993. [Google Scholar] [CrossRef]
- Csepregi, R.; Temesfői, V.; Poór, M.; Faust, Z.; Kőszegi, T. Green fluorescent protein-based viability assay in a multiparametric configuration. Molecules 2018, 23, 1575. [Google Scholar] [CrossRef]
- Hirano, M.; Ando, R.; Shimozono, S.; Sugiyama, M.; Takeda, N.; Kurokawa, H.; Deguchi, R.; Endo, K.; Haga, K.; Takai-Todaka, R.; et al. A highly photostable and bright green fluorescent protein. Nat. Biotechnol. 2022, 40, 1132–1142. [Google Scholar] [CrossRef]
- Mizuta, K.; Gallagher, S.; Wang, A.; Chang, N.; Morinaga, S.; Sato, M.; Kang, B.M.; Hoffman, R.M. Head-to-head comparison of green fluorescent protein (GFP) imaging with luciferase-luciferin imaging in vivo using single-nanometer laser-excitation tuning and an ultra-low-light-detection camera and optics demonstrates the superiority of GFP. Anticancer Res. 2024, 44, 2823–2826. [Google Scholar] [CrossRef]
- Surre, J.; Saint-Ruf, C.; Collin, V.; Orenga, S.; Ramjeet, M.; Matic, I. Strong increase in the autofluorescence of cells signals struggle for survival. Sci. Rep. 2018, 8, 12088. [Google Scholar] [CrossRef]
- van Roessel, P.; Brand, A. Imaging into the future: Visualizing gene expression and protein interactions with fluorescent proteins. Nat. Cell. Biol. 2002, 4, e15–e20. [Google Scholar] [CrossRef] [PubMed]
- Neefjes, M.; Housmans, B.A.C.; van den Akker, G.G.H.; van Rhijn, L.W.; Welting, T.J.M.; van der Kraan, P.M. Reporter gene comparison demonstrates interference of complex body fluids with secreted luciferase activity. Sci. Rep. 2021, 11, 1359. [Google Scholar] [CrossRef] [PubMed]
- Trischitta, P.; Tamburello, M.P.; Venuti, A.; Pennisi, R. Pseudovirus-based systems for screening natural antiviral agents: A comprehensive review. Int. J. Mol. Sci. 2024, 25, 5188. [Google Scholar] [CrossRef]
- Di Mauro, V.; Lauta, F.C.; Modica, J.; Appleton, S.L.; De Franciscis, V.; Catalucci, D. Diagnostic and therapeutic aptamers: A promising pathway to improved cardiovascular disease management. JACC Basic Transl. Sci. 2023, 9, 260–277. [Google Scholar] [CrossRef]
- Moreira, M.J.; Pintado, M.; Almeida, J.M.M.M.D. Are aptamer-based biosensors the future of the detection of the human gut microbiome?—A systematic review and meta-analysis. Biosensors 2024, 14, 423. [Google Scholar] [CrossRef] [PubMed]
- Sujith, S.; Naresh, R.; Srivisanth, B.U.; Sajeevan, A.; Rajaramon, S.; David, H.; Solomon, A.P. Aptamers: Precision tools for diagnosing and treating infectious diseases. Front. Cell. Infect. Microbiol. 2024, 14, 1402932. [Google Scholar] [CrossRef]
- Setlem, K.; Mondal, B.; Ramlal, S.; Kingston, J. Immuno affinity SELEX for simple, rapid, and cost-effective aptamer enrichment and identification against aflatoxin B1. Front. Microbiol. 2016, 7, 1909. [Google Scholar] [CrossRef]
- Tamaian, R. Aptamer-based biosensor design for simultaneous detection of cervical cancer-related microRNAs. Eng. Proc. 2023, 58, 88. [Google Scholar] [CrossRef]
- Pantsar, T.; Poso, A. Binding affinity via docking: Fact and fiction. Molecules 2018, 23, 1899. [Google Scholar] [CrossRef]
- Thafar, M.A.; Alshahrani, M.; Albaradei, S.; Gojobori, T.; Essack, M.; Gao, X. Affinity2Vec: Drug-target binding affinity prediction through representation learning, graph mining, and machine learning. Sci. Rep. 2022, 12, 4751. [Google Scholar] [CrossRef]
- Wilson, D.R.; Routkevitch, D.; Rui, Y.; Mosenia, A.; Wahlin, K.J.; Quinones-Hinojosa, A.; Zack, D.J.; Green, J.J. A triple-fluorophore-labeled nucleic acid ph nanosensor to investigate non-viral gene delivery. Mol. Ther. 2017, 25, 1697–1709. [Google Scholar] [CrossRef] [PubMed]
- Altman, R.B.; Terry, D.S.; Zhou, Z.; Zheng, Q.; Geggier, P.; Kolster, R.A.; Zhao, Y.; Javitch, J.A.; Warren, J.D.; Blanchard, S.C. Cyanine fluorophore derivatives with enhanced photostability. Nat. Methods 2011, 9, 68–71. [Google Scholar] [CrossRef] [PubMed]
- Alkhamis, O.; Canoura, J.; Bukhryakov, K.V.; Tarifa, A.; DeCaprio, A.P.; Xiao, Y. DNA Aptamer-Cyanine Complexes as Generic Colorimetric Small-Molecule Sensors. Angew. Chem. Int. Ed. 2022, 61, e202112305. [Google Scholar] [CrossRef] [PubMed]


| Pseudoviral System | Reporter | Activation | Substrate | Optimal Detection Time | Strongest Intensity |
|---|---|---|---|---|---|
| NanoBiT | NanoBiT luciferase fragments, including pseudovirus-SmBiT and live cell-LgBiT | PPI between pseudovirus and cells | Furimazine | 3 h | 45.26 × 106 p/sec/cm2 |
| GFP | GFP pseudovirual plasmid and expressed GFP in host cells | Reporter expression in the infected cell | Not applicable | 48 h | 15.6 × 104 A.U. |
| Luciferase | Luciferase pseudoviral plasmid and expressed liciferase in host cells | Reporter expression in the infected cell | D-luciferin | 48 h | 24.26 × 106 p/sec/cm2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, M.-W.; Lin, C.-H.; Chiang, H.-H.; Quintela, I.A.; Wu, V.C.H.; Lin, C.-S. Using Nano-Luciferase Binary (NanoBiT) Technology to Assess the Interaction Between Viral Spike Protein and Angiotensin-Converting Enzyme II by Aptamers. BioTech 2025, 14, 20. https://doi.org/10.3390/biotech14010020
Lin M-W, Lin C-H, Chiang H-H, Quintela IA, Wu VCH, Lin C-S. Using Nano-Luciferase Binary (NanoBiT) Technology to Assess the Interaction Between Viral Spike Protein and Angiotensin-Converting Enzyme II by Aptamers. BioTech. 2025; 14(1):20. https://doi.org/10.3390/biotech14010020
Chicago/Turabian StyleLin, Meng-Wei, Cheng-Han Lin, Hua-Hsin Chiang, Irwin A. Quintela, Vivian C. H. Wu, and Chih-Sheng Lin. 2025. "Using Nano-Luciferase Binary (NanoBiT) Technology to Assess the Interaction Between Viral Spike Protein and Angiotensin-Converting Enzyme II by Aptamers" BioTech 14, no. 1: 20. https://doi.org/10.3390/biotech14010020
APA StyleLin, M.-W., Lin, C.-H., Chiang, H.-H., Quintela, I. A., Wu, V. C. H., & Lin, C.-S. (2025). Using Nano-Luciferase Binary (NanoBiT) Technology to Assess the Interaction Between Viral Spike Protein and Angiotensin-Converting Enzyme II by Aptamers. BioTech, 14(1), 20. https://doi.org/10.3390/biotech14010020






