A Score for Predicting Freedom from Progression of Children and Adolescents with Hodgkin Lymphoma
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Cohorts
2.2. Study Design
2.3. Clinical Trial Registration Information
2.4. HLA-G Genotyping
2.5. Endpoints and Statistical Analysis
2.6. Data-Sharing Statement
3. Results
3.1. Demographics and Clinical Details of the Studied Samples
3.2. Identification of Demographic and Hematological/Biochemical Variables Associated with cHL Progression/Relapse
3.3. FPR Multivariate Modeling for FFP Survival
3.4. Prediction Result of FPR Model for FFP and Comparison with TG and CHIPS Models
3.5. Molecular FPR and TG Performance in the NScHL Set
3.6. Comparative FPR and TG Kaplan–Meier Curves for OS
3.7. Comparison among the Predictive Models for Treatment Response of FPR, TG, and the Interim PET/CT Scanning
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CR | complete response |
| IF | involved fields |
| M/T | mediastinal–thoracic ratio |
| PR | partial response |
| RT | radiotherapy |
| ROC | receiver operating characteristic |
| HLA | human leukocyte antigen |
| SNP | single nucleotide polymorphism |
| FFP | freedom from progression and disease |
| FPR | final prognostic rank model |
| NScHL | nodular sclerosis classical Hodgkin lymphoma histological subtype |
| TG | therapeutic group |
| PET/CT | positron Emission tomography/computed tomography |
References
- Burnelli, R.; Fiumana, G.; Rondelli, R.; Pillon, M.; Sala, A.; Garaventa, A.; D’Amore, E.S.G.; Sabattini, E.; Buffardi, S.; Bianchi, M.; et al. Comparison of Hodgkin’s Lymphoma in Children and Adolescents. A Twenty Year Experience with MH’96 and LH2004 AIEOP (Italian Association of Pediatric Hematology and Oncology) Protocols. Cancers 2020, 12, 1620. [Google Scholar] [CrossRef] [PubMed]
- Lopci, E.; Mascarin, M.; Piccardo, A.; Castello, A.; Elia, C.; Guerra, L.; Borsatti, E.; Sala, A.; Todesco, A.; Zucchetta, P.; et al. FDG PET in Response Evaluation of Bulky Masses in Paediatric Hodgkin’s Lymphoma (HL) Patients Enrolled in the Italian AIEOP-LH2004 Trial. Eur. J. Nucl. Med. Mol. Imaging 2019, 46, 97–106. [Google Scholar] [CrossRef] [Green Version]
- Mascarin, M.; Fiumana, G.; Rondelli, R.; Scarzello, G.; Pillon, M.; Sala, A.; Buffardi, S.; Bianchi, M.; Farruggia, P.; Facchini, E.; et al. Protocollo AIEOP LH-2004 per Bambini e Adolescenti Con Linfoma Di Hodgkin: Si Può Omettere La Radioterapia Nei Pazienti a Basso Rischio, o Ridurne Dosi e Volumi Nei Pazienti a Rischio Standard, Se in Remissione Completa Dopo Chemioterapia Co10. In Proceedings of the XLIV Congresso Nazionale AIEOP, Catania, Italy, 13—15 October 2019. Hematology Reports. [Google Scholar]
- De Re, V.; Caggiari, L.; Mussolin, L.; d’Amore, E.S.; Famengo, B.; De Zorzi, M.; Martina, L.; Elia, C.; Pillon, M.; Santoro, N.; et al. HLA-G+3027 Polymorphism Is Associated with Tumor Relapse in Pediatric Hodgkin’s Lymphoma. Oncotarget 2017, 8, 105957–105970. [Google Scholar] [CrossRef]
- Castelli, E.C.; Mendes-Junior, C.T.; Deghaide, N.H.S.; de Albuquerque, R.S.; Muniz, Y.C.N.; Simões, R.T.; Carosella, E.D.; Moreau, P.; Donadi, E.A. The Genetic Structure of 3′untranslated Region of the HLA-G Gene: Polymorphisms and Haplotypes. Genes Immun. 2010, 11, 134–141. [Google Scholar] [CrossRef]
- De Zorzi, M.; Caggiari, L.; Crovatto, M.; Talamini, R.; Garziera, M.; De Re, V. A New Human Leukocyte Antigen Class I Allele: HLA-A*02:374. Tissue Antigens 2013, 81, 48–49. [Google Scholar] [CrossRef] [PubMed]
- Schreck, S.; Friebel, D.; Buettner, M.; Distel, L.; Grabenbauer, G.; Young, L.S.; Niedobitek, G. Prognostic Impact of Tumour-Infiltrating Th2 and Regulatory T Cells in Classical Hodgkin Lymphoma. Hematol. Oncol. 2009, 27, 31–39. [Google Scholar] [CrossRef]
- Romano, A.; Parrinello, N.L.; Vetro, C.; Tibullo, D.; Giallongo, C.; La Cava, P.; Chiarenza, A.; Motta, G.; Caruso, A.L.; Villari, L.; et al. The Prognostic Value of the Myeloid-Mediated Immunosuppression Marker Arginase-1 in Classic Hodgkin Lymphoma. Oncotarget 2016, 7, 67333–67346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mirili, C.; Paydas, S.; Kapukaya, T.K.; Yılmaz, A. Systemic Immune-Inflammation Index Predicting Survival Outcome in Patients with Classical Hodgkin Lymphoma. Biomark. Med. 2019, 13, 1565–1575. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, A.; Allavena, P.; Sica, A.; Balkwill, F. Cancer-Related Inflammation. Nature 2008, 454, 436–444. [Google Scholar] [CrossRef]
- Galdiero, M.R.; Varricchi, G.; Loffredo, S.; Mantovani, A.; Marone, G. Roles of Neutrophils in Cancer Growth and Progression. J. Leukoc. Biol. 2018, 103, 457–464. [Google Scholar] [CrossRef]
- Quail, D.F.; Joyce, J.A. Microenvironmental Regulation of Tumor Progression and Metastasis. Nat. Med. 2013, 19, 1423–1437. [Google Scholar] [CrossRef]
- Calabretta, E.; d’Amore, F.; Carlo-Stella, C. Immune and Inflammatory Cells of the Tumor Microenvironment Represent Novel Therapeutic Targets in Classical Hodgkin Lymphoma. Int. J. Mol. Sci. 2019, 20, 5503. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; Shen, Y.; Huang, H.; Pan, S.; Jiang, J.; Chen, W.; Zhang, T.; Zhang, C.; Ni, C. A Rosetta Stone for Breast Cancer: Prognostic Value and Dynamic Regulation of Neutrophil in Tumor Microenvironment. Front. Immunol. 2020, 11, 1779. [Google Scholar] [CrossRef]
- Wang, J.; Zhou, X.; Liu, Y.; Li, Z.; Li, X. Prognostic Significance of Neutrophil-to-Lymphocyte Ratio in Diffuse Large B-Cell Lymphoma: A Meta-Analysis. PLoS ONE 2017, 12, e0176008. [Google Scholar] [CrossRef] [PubMed]
- Stefaniuk, P.; Szymczyk, A.; Podhorecka, M. The Neutrophil to Lymphocyte and Lymphocyte to Monocyte Ratios as New Prognostic Factors in Hematological Malignancie—A Narrative Review. Cancer Manag. Res. 2020, 12, 2961–2977. [Google Scholar] [CrossRef] [PubMed]
- Mu, S.; Ai, L.; Fan, F.; Sun, C.; Hu, Y. Prognostic Role of Neutrophil-Lymphocyte Ratio in Multiple Myeloma: A Dose-Response Meta-Analysis. Onco Targets Ther. 2018, 11, 499–507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marcheselli, R.; Bari, A.; Tadmor, T.; Marcheselli, L.; Cox, M.C.; Pozzi, S.; Ferrari, A.; Baldini, L.; Gobbi, P.; Aviv, A.; et al. Neutrophil-Lymphocyte Ratio at Diagnosis Is an Independent Prognostic Factor in Patients with Nodular Sclerosis Hodgkin Lymphoma: Results of a Large Multicenter Study Involving 990 Patients. Hematol. Oncol. 2017, 35, 561–566. [Google Scholar] [CrossRef]
- Koh, Y.W.; Kang, H.J.; Park, C.; Yoon, D.H.; Kim, S.; Suh, C.; Kim, J.E.; Kim, C.-W.; Huh, J. Prognostic Significance of the Ratio of Absolute Neutrophil Count to Absolute Lymphocyte Count in Classic Hodgkin Lymphoma. Am. J. Clin. Pathol. 2012, 138, 846–854. [Google Scholar] [CrossRef] [Green Version]
- Dogan, A.; Demircioglu, S. Assessment of the Neutrophil-Lymphocyte Ratio in Classic Hodgkin Lymphoma Patients. Pak. J. Med. Sci. 2019, 35, 1270–1275. [Google Scholar] [CrossRef] [Green Version]
- Romano, A.; Pavoni, C.; Di Raimondo, F.; Tarella, C.; Viviani, S.; Rossi, A.; Patti, C.; Picardi, M.; Cantonetti, M.; La Nasa, G.; et al. The Neutrophil to Lymphocyte Ratio (NLR) and the Presence of Large Nodal Mass Are Independent Predictors of Early Response: A Subanalysis of the Prospective Phase II PET-2-Adapted HD0607 Trial. Cancer Med. 2020, 9, 8735–8746. [Google Scholar] [CrossRef]
- Renner, K.; Singer, K.; Koehl, G.E.; Geissler, E.K.; Peter, K.; Siska, P.J.; Kreutz, M. Metabolic Hallmarks of Tumor and Immune Cells in the Tumor Microenvironment. Front. Immunol. 2017, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bronte, V.; Zanovello, P. Regulation of Immune Responses by L-Arginine Metabolism. Nat. Rev. Immunol. 2005, 5, 641–654. [Google Scholar] [CrossRef] [PubMed]
- Lemos, H.; Huang, L.; Prendergast, G.C.; Mellor, A.L. Immune Control by Amino Acid Catabolism during Tumorigenesis and Therapy. Nat. Rev. Cancer 2019, 19, 162–175. [Google Scholar] [CrossRef]
- Czystowska-Kuzmicz, M.; Sosnowska, A.; Nowis, D.; Ramji, K.; Szajnik, M.; Chlebowska-Tuz, J.; Wolinska, E.; Gaj, P.; Grazul, M.; Pilch, Z.; et al. Small Extracellular Vesicles Containing Arginase-1 Suppress T-Cell Responses and Promote Tumor Growth in Ovarian Carcinoma. Nat. Commun. 2019, 10, 3000. [Google Scholar] [CrossRef] [PubMed]
- Quintero-Fabián, S.; Arreola, R.; Becerril-Villanueva, E.; Torres-Romero, J.C.; Arana-Argáez, V.; Lara-Riegos, J.; Ramírez-Camacho, M.A.; Alvarez-Sánchez, M.E. Role of Matrix Metalloproteinases in Angiogenesis and Cancer. Front. Oncol. 2019, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roy, R.; Yang, J.; Moses, M.A. Matrix Metalloproteinases as Novel Biomarkers and Potential Therapeutic Targets in Human Cancer. J. Clin. Oncol. 2009, 27, 5287–5297. [Google Scholar] [CrossRef] [Green Version]
- Grzywa, T.M.; Sosnowska, A.; Matryba, P.; Rydzynska, Z.; Jasinski, M.; Nowis, D.; Golab, J. Myeloid Cell-Derived Arginase in Cancer Immune Response. Front. Immunol. 2020, 11, 938. [Google Scholar] [CrossRef]
- La Manna, G.; Ghinatti, G.; Tazzari, P.L.; Alviano, F.; Ricci, F.; Capelli, I.; Cuna, V.; Todeschini, P.; Brunocilla, E.; Pagliaro, P.; et al. Neutrophil Gelatinase-Associated Lipocalin Increases HLA-G(+)/FoxP3(+) T-Regulatory Cell Population in an In Vitro Model of PBMC. PLoS ONE 2014, 9, e89497. [Google Scholar] [CrossRef] [Green Version]
- Caocci, G.; Greco, M.; Fanni, D.; Senes, G.; Littera, R.; Lai, S.; Risso, P.; Carcassi, C.; Faa, G.; La Nasa, G. HLA-G Expression and Role in Advanced-Stage Classical Hodgkin Lymphoma. Eur. J. Histochem. 2016, 60, 2606. [Google Scholar] [CrossRef] [Green Version]
- Poras, I.; Yaghi, L.; Martelli-Palomino, G.; Mendes-Junior, C.T.; Muniz, Y.C.N.; Cagnin, N.F.; Sgorla de Almeida, B.; Castelli, E.C.; Carosella, E.D.; Donadi, E.A.; et al. Haplotypes of the HLA-G 3′ Untranslated Region Respond to Endogenous Factors of HLA-G+ and HLA-G-Cell Lines Differentially. PLoS ONE 2017, 12, e0169032. [Google Scholar] [CrossRef] [Green Version]
- Martelli-Palomino, G.; Pancotto, J.A.; Muniz, Y.C.; Mendes-Junior, C.T.; Castelli, E.C.; Massaro, J.D.; Krawice-Radanne, I.; Poras, I.; Rebmann, V.; Carosella, E.D.; et al. Polymorphic Sites at the 3′ Untranslated Region of the HLA-G Gene Are Associated with Differential Hla-g Soluble Levels in the Brazilian and French Population. PLoS ONE 2013, 8, e71742. [Google Scholar] [CrossRef]
- Celik, A.A.; Simper, G.S.; Hiemisch, W.; Blasczyk, R.; Bade-Döding, C. HLA-G Peptide Preferences Change in Transformed Cells: Impact on the Binding Motif. Immunogenetics 2018, 70, 485–494. [Google Scholar] [CrossRef] [Green Version]
- Schwich, E.; Rebmann, V.; Michita, R.T.; Rohn, H.; Voncken, J.W.; Horn, P.A.; Kimmig, R.; Kasimir-Bauer, S.; Buderath, P. HLA-G 3′ Untranslated Region Variants +3187G/G, +3196G/G and +3035T Define Diametrical Clinical Status and Disease Outcome in Epithelial Ovarian Cancer. Sci. Rep. 2019, 9, 5407. [Google Scholar] [CrossRef] [PubMed]
- de Charette, M.; Houot, R. Hide or Defend, the Two Strategies of Lymphoma Immune Evasion: Potential Implications for Immunotherapy. Haematologica 2018, 103, 1256–1268. [Google Scholar] [CrossRef] [PubMed]
- Naji, A.; Menier, C.; Maki, G.; Carosella, E.D.; Rouas-Freiss, N. Neoplastic B-Cell Growth Is Impaired by HLA-G/ILT2 Interaction. Leukemia 2012, 26, 1889–1892. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aldinucci, D.; Borghese, C.; Casagrande, N. Formation of the Immunosuppressive Microenvironment of Classic Hodgkin Lymphoma and Therapeutic Approaches to Counter It. Int. J. Mol. Sci. 2019, 20, 2416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiu, J.; Ernst, D.M.; Keating, A. Acquired Natural Killer Cell Dysfunction in the Tumor Microenvironment of Classic Hodgkin Lymphoma. Front. Immunol. 2018, 9, 267. [Google Scholar] [CrossRef] [PubMed]
- Carosella, E.D.; Rouas-Freiss, N.; Roux, D.T.-L.; Moreau, P.; LeMaoult, J. HLA-G: An Immune Checkpoint Molecule. In Advances in Immunology; Alt, F.W., Ed.; Elsevier Academic Press Inc.: San Diego, CA, USA, 2015; Volume 127, pp. 33–144. ISBN 978-0-12-802245-0. [Google Scholar]
- Hunt, J.S.; Petroff, M.G.; McIntire, R.H.; Ober, C. HLA-G and Immune Tolerance in Pregnancy. FASEB J. 2005, 19, 681–693. [Google Scholar] [CrossRef]
- RouasFreiss, N.; Goncalves, R.M.B.; Menier, C.; Dausset, J.; Carosella, E.D. Direct Evidence to Support the Role of HLA-G in Protecting the Fetus from Maternal Uterine Natural Killer Cytolysis. Proc. Natl. Acad. Sci. USA 1997, 94, 11520–11525. [Google Scholar] [CrossRef] [Green Version]



| cHL (n = 133) | NScHL (n = 108) | |
|---|---|---|
| Age, years | ||
| Median (IQR) | 14 (11.9–15.1) | 14 (12.0–15.6) |
| Gender (%) | ||
| -female | 49 (36.8%) | 42 (38.9%) |
| -male | 84 (63.2) | 66 (61.1%) |
| Stage (%) | ||
| 1 | 7 (5.3) | 4 (3.7) |
| 2 | 62 (46.6) | 52 (48.1) |
| 3 | 27 (20.3) | 22 (20.4) |
| 4 | 37 (27.8) | 30 (27.8) |
| Treatment group (%) | ||
| 1 | 14 (10.5) | 8 (7.4%) |
| 2 | 17 (12.8) | 17 (15.7%) |
| 3 | 102 (76.7) | 83 (76.9%) |
| Median follow-up, years (IQR) | 5.55 (4.09–7.93) | 5.89 (4.68–7.95) |
| Histology (%) | ||
| NC | 14 (10.5) | |
| MC | 8 (6.0) | |
| LRCHL | 3 (2.3) | |
| NS | 108 (81.2) | 108 (100.0%) |
| Sedimentation rate (mm/hr) | ||
| median (IQR) | 71.0 (38.7–40.0) | 73 (46.8–101.0) |
| Albumin (g/L) | ||
| median (IQR) | 39.0 (37.0–40.0) | 39.0 (31.5–40.0) |
| Ferritin (ng/mL) | ||
| median (IQR) | 127.5 (61.5–337.5) | 135.0 (63.5–-335.8) |
| Hemoglobin (g/L) | ||
| median (IQR) | 11.5 (11.0–12.6) | 11.7 (10.3–12.6) |
| White blood cell count (109/L) | ||
| median (IQR) | 12.47 (8.115–16.815) | 12.60 (8.352–16.585) |
| Lymphocytes (109/L) | ||
| median (IQR) | 1.81 (1.296–2.312) | 1.81 (1.312–2.297) |
| Neutrophils (109/L) | ||
| median (IQR) | 9.07 (5.623–12.526) | 9.39 (6.263–12.907) |
| Eosinophils (109/L) | ||
| Median (IQR) | 0.17 (0.067–0.339) | 0.21 (0.082–0.338) |
| Basophils (109/L) | ||
| median (IQR) | 0.00 (0.000–0.182) | 0.00 (0.000–0.145) |
| Monocytes (109/L) | ||
| median (IQR) | 0.773 (0.053–1.047) | 0.821 (0.583–1.056) |
| Platelets (109/L) | ||
| median (IQR) | 393.0 (295.7–464.2) | 393.5 (300.5–433.5) |
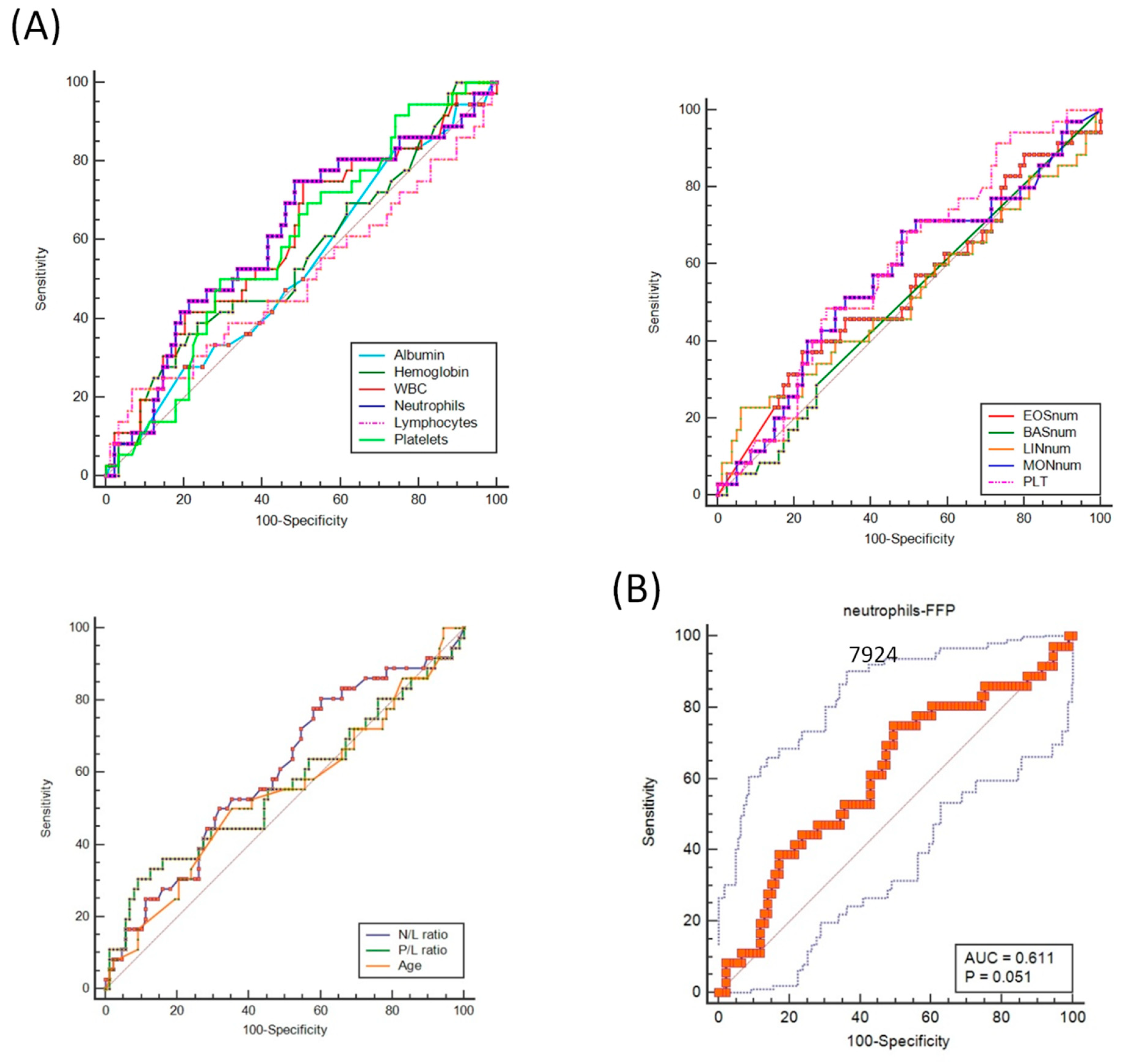
| Variable | AUC. | SE. | 95% CI |
|---|---|---|---|
| Albumin (g/L) | 0.533 | 0.0570 | 0.441 to 0.623 |
| Hemoglobin (g/dl) | 0.563 | 0.0587 | 0.472 to 0.652 |
| White blood-cell count (109/L) | 0.603 | 0.0570 | 0.522 to 0.696 |
| Neutrophils (109/L) | 0.611 | 0.0572 | 0.522 to 0.700 |
| Lymphocytes (109/L) | 0.500 | 0.0626 | 0.409 to 0.591 |
| Platelets (109/L) | 0.598 | 0.0545 | 0.506 to 0.685 |
| ESR (mm/hr) | 0.506 | 0.0761 | 0.384 to 0.628 § |
| Ferritin (ng/mL) | 0.525 | 0.0791 | 0.402 to 0.646 § |
| Eosinophils (109/L) | 0.574 | 0.0782 | 0.450 to 0.692 § |
| Basophils (109/L) | 0.543 | 0.0628 | 0.419 to 0.662 § |
| Monocytes (109/L) | 0.582 | 0.0774 | 0.458 to 0.699 § |
| N/L ratio | 0.601 | 0.0572 | 0.509 to 0.687 |
| P/L ratio | 0.561 | 0.0623 | 0.469 to 0.650 |
| Age | 0.542 | 0.0601 | 0.450 to 0.632 |
| Covariable | Predictive Variable | HR. | 95% CI of Exp(b) | Numerical Point Assigned |
|---|---|---|---|---|
| TG. | TG1 | Reference | ---- | 0 |
| TG2 | --- | ---- | 0 | |
| TG3 | 2.8799 | 0.8647 to 9.5919 | 3 | |
| V1 | HLA-G (C/C) | Reference | 0 | |
| HLA-G (C/A) | 1.1262 | 0.4323 to 2.9335 | 1 | |
| V2 | Neutrophils (≤8 × 109/L) | Reference | 0 | |
| Neutrophils (>8 × 109/L) | 2.3282 | 1.0720 to 5.0564 | 2 |
| (A) cHL n = 133. | |||||||
|---|---|---|---|---|---|---|---|
| Cases Summary | Mean Servival | Hazard Ratio | |||||
| Number of Events | Sample Size | Years | |||||
| FPR model | n | % | Total | Mean | SE. | HR. | 95%CI |
| rank 1 | 2 | 10.00 | 20 | 10.032 | 0.623 | reference | -- |
| rank 2 | 1 | 10.00 | 10 | 9.017 | 0.781 | 0.983 | 0.2556 to 3.7775 |
| rank 3 | 7 | 17.50 | 40 | 9.907 | 0.636 | 1.812 | 0.6913 to 4.7470 |
| rank 4 | 23 | 42.59 | 54 | 9.319 | 1.028 | 5.602 | 2.1584 to 14.5369 |
| rank 5 | 4 | 44.44 | 9 | 7.384 | 1.784 | 5.047 | 1.1628 to 21.9065 |
| TG model | n | % | Total | Mean | SE. | HR. | 95%CI |
| TG 1 | 3 | 21.43 | 14 | 8.995 | 1.003 | reference | -- |
| TG 2 | 0 | 0.00 | 17 | 10.72 | 0.000 | -- | -- |
| TG 3 | 34 | 33.33 | 102 | 10.75 | 0.715 | 1.8277 | 0.6663 to 5.0135 |
| HR, hazard ratio with 95% confidence interval | |||||||
| (B) NScHL n = 108 | |||||||
| Cases Summary | Mean Survival | Hazard Ratio | |||||
| Number of Events | sample Size | Years | |||||
| FPR model | n | % | Total | Mean | SE. | HR. | 95%CI |
| rank 1 | 1 | 6.67 | 15 | 10.211 | 0.712 | reference | -- |
| rank 2 | 0 | 0.00 | 9 | 9.840 | 0.000 | -- | -- |
| rank 3 | 5 | 15.63 | 32 | 10.151 | 0.660 | 2.2661 | 0.6755 to 7.6019 |
| rank 4 | 17 | 38.64 | 44 | 10.010 | 1.111 | 6.9488 | 2.1104 to 22.8794 |
| rank 5 | 3 | 37.50 | 8 | 8.198 | 1.812 | 5.8021 | 1.0392 to 32.3948 |
| FPR model | n | % | Total | Mean | SE. | HR. | 95%CI |
| TG 1 | 1 | 12.50 | 8 | 9.657 | 1.21 | reference | -- |
| TG 2 | 0 | 0.00 | 17 | 10.720 | 0.00 | -- | -- |
| TG 3 | 25 | 30.12 | 83 | 11.314 | 0.76 | 2.6186 | 0.6174 to 11.1053 |
| HR, hazard ratio with 95% confidence interval | |||||||
| (A) | |||||
|---|---|---|---|---|---|
| RISK | Low | Medium | High | ||
| FPR at diagnosis | rank 1 | rank 2 | rank 3 | rank 4 | rank 5 |
| total sample size | 13 | 8 | 27 | 38 | 8 |
| Interim PET/CT scan | |||||
| PET-positive | 0 | 0 | 10 | 21 | 3 |
| PET-negative | 13 | 8 | 17 | 17 | 5 |
| PD/R at follow-up | |||||
| total sample size | 0 | 0 | 4 | 15 | 3 |
| PET-positive | 0 | 0 | 3 | 10 | 2 |
| PET-negative | 0 | 0 | 1 | 5 | 1 |
| PD/R, progressive disease/relapse | |||||
| (B) | |||||
| FPR Model High-Risk | Value | 95% CI | |||
| Sensitivity | 81.82% | 59.72% to 94.81% | |||
| Specificity | 61.11% | 48.89% to 72.38% | |||
| Positive predictive value | 39.12% | 31.17% to 47.71% | |||
| Negative predictive value | 91.67% | 81.65% to 96.45% | |||
| Accuracy | 65.96% | 55.46% to 75.42% | |||
| PET Evaluation from FPR High-Risk Group | |||||
| Sensitivity | 66.67% | 40.99% to 86.66% | |||
| Specificity | 57.14% | 37.18% to 75.54% | |||
| Positive predictive value | 32.21% | 21.72% to 44.87% | |||
| Negative predictive value | 84.88% | 73.05% to 92.08% | |||
| Accuracy | 59.37% | 43.89% to 73.60% | |||
| Disease prevalence 23.40% | |||||
| (C) | |||||
| FPR Model Medium-Risk + High-Risk | Value | 95% CI | |||
| Sensitivity | 100.00% | 84.56% to 100.00% | |||
| Specificity | 29.17% | 19.05% to 41.07% | |||
| Positive predictive value | 30.13% | 27.11% to 33.34% | |||
| Negative predictive value | 100.00% | ||||
| Accuracy | 45.74% | 35.42% to 56.34% | |||
| PET Evaluation from Medium-Risk + High-Risk | Value | 95% CI | |||
| Sensitivity | 68.18% | 45.13% to 86.14% | |||
| Specificity | 73.61% | 61.90% to 83.30% | |||
| Positive predictive value | 44.11% | 32.82% to 56.05% | |||
| Negative predictive value | 88.34% | 80.18% to 93.41% | |||
| Accuracy | 72.34% | 62.15% to 81.07% | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
De Re, V.; Caggiari, L.; Mascarin, M.; De Zorzi, M.; Elia, C.; Repetto, O.; Mussolin, L.; Pillon, M.; Muggeo, P.; Buffardi, S.; et al. A Score for Predicting Freedom from Progression of Children and Adolescents with Hodgkin Lymphoma. Hemato 2021, 2, 264-280. https://doi.org/10.3390/hemato2020016
De Re V, Caggiari L, Mascarin M, De Zorzi M, Elia C, Repetto O, Mussolin L, Pillon M, Muggeo P, Buffardi S, et al. A Score for Predicting Freedom from Progression of Children and Adolescents with Hodgkin Lymphoma. Hemato. 2021; 2(2):264-280. https://doi.org/10.3390/hemato2020016
Chicago/Turabian StyleDe Re, Valli, Laura Caggiari, Maurizio Mascarin, Mariangela De Zorzi, Caterina Elia, Ombretta Repetto, Lara Mussolin, Marta Pillon, Paola Muggeo, Salvatore Buffardi, and et al. 2021. "A Score for Predicting Freedom from Progression of Children and Adolescents with Hodgkin Lymphoma" Hemato 2, no. 2: 264-280. https://doi.org/10.3390/hemato2020016
APA StyleDe Re, V., Caggiari, L., Mascarin, M., De Zorzi, M., Elia, C., Repetto, O., Mussolin, L., Pillon, M., Muggeo, P., Buffardi, S., Bianchi, M., Sala, A., Vinti, L., Farruggia, P., Facchini, E., Lopci, E., d’Amore, E. S. G., Burnelli, R., & with the A.I.E.O.P. Consortium. (2021). A Score for Predicting Freedom from Progression of Children and Adolescents with Hodgkin Lymphoma. Hemato, 2(2), 264-280. https://doi.org/10.3390/hemato2020016










