Biogenic Hydrogen Sulfide Production Using Elemental Sulfur and Low-Cost Organic Substrates to Remove Metal Ions from Mining Effluents
Abstract
:1. Introduction
2. Materials and Methods
2.1. Microbial Consortium
2.2. Bioreactors
2.3. Characterization of Microbial Consortia by Fluorescence In Situ Hybridization
2.4. Culture Media
2.5. Batch Cultures
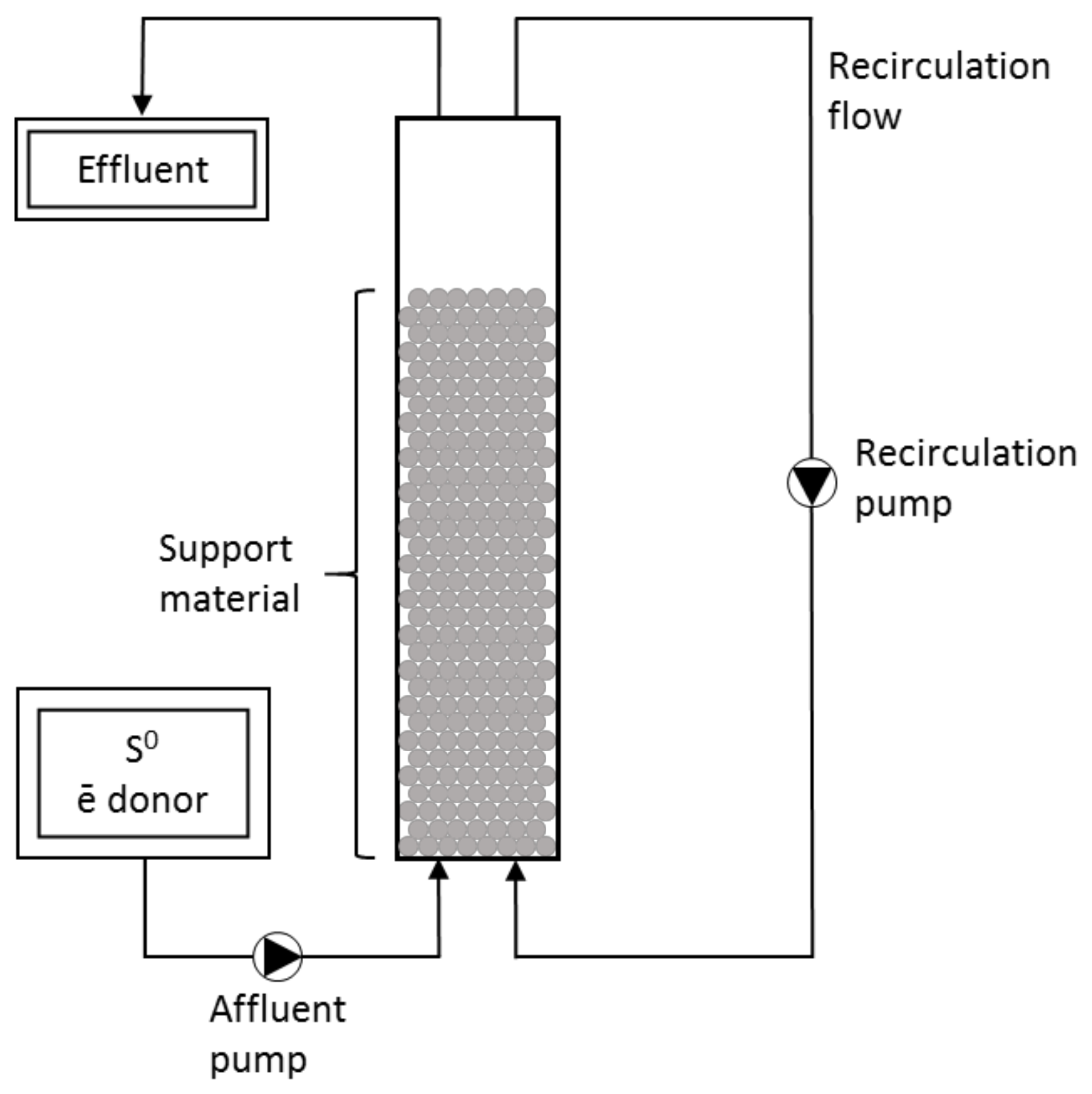
2.6. System Setup and Operation
2.7. Determination of Hydrogen Sulfide in Culture Media
2.8. Quantification of Chemical Oxygen Demand
2.9. Determination of the Grow Capacity of the Microbial Consortium in Bioreactors
2.10. Analysis of Data
2.11. Operation Parameters
3. Results
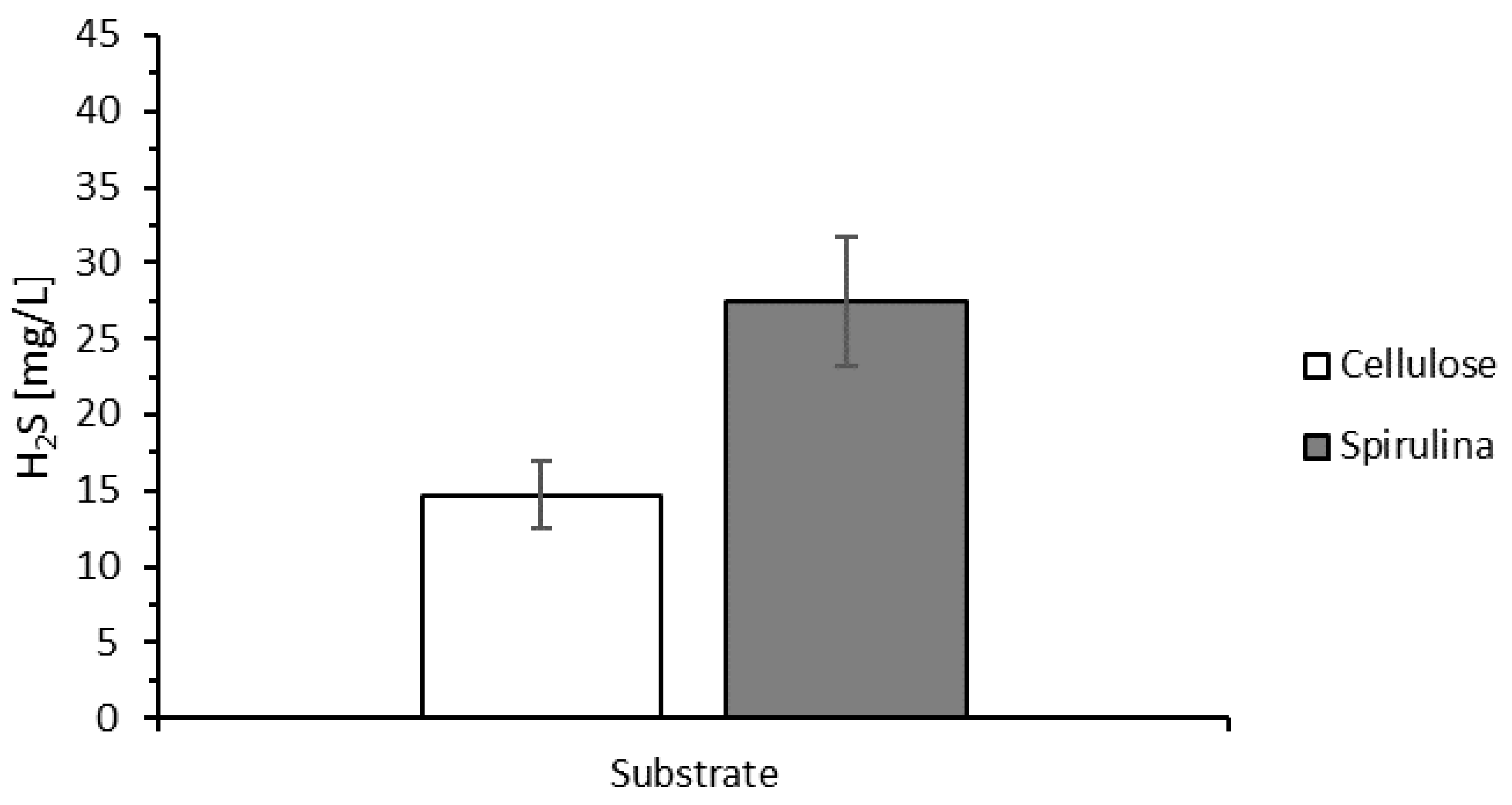
3.1. Growth of the Microbial Consortium and Determination of H2S Generation
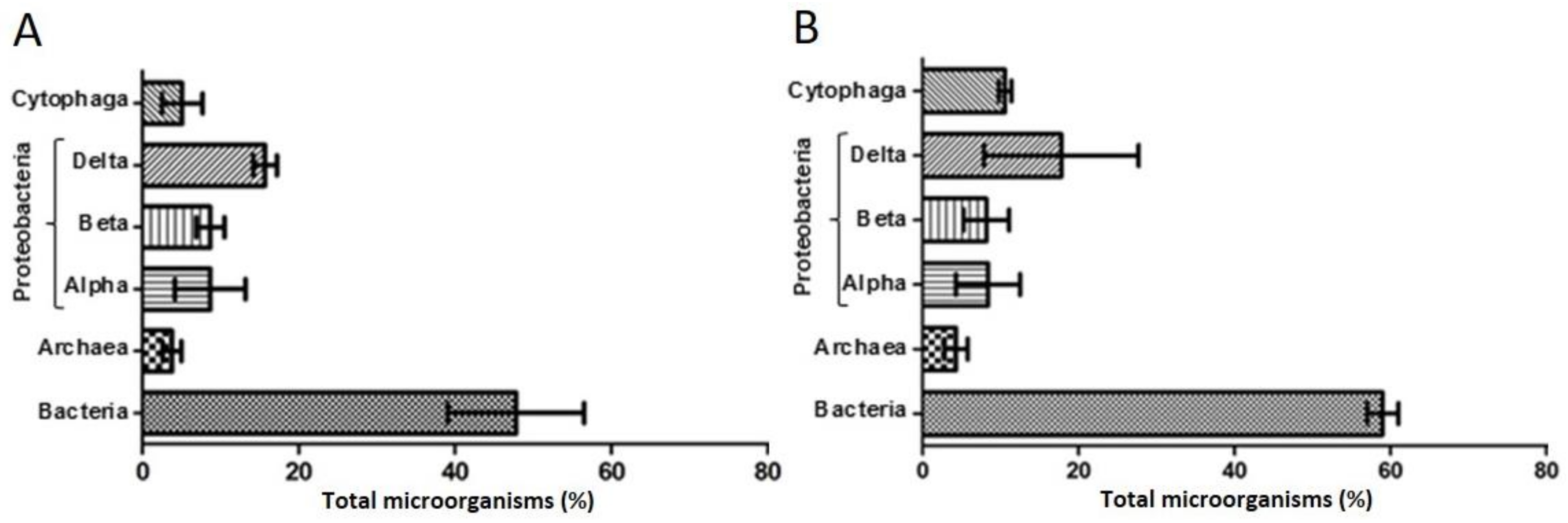
3.2. Microbial Consortium Characterization
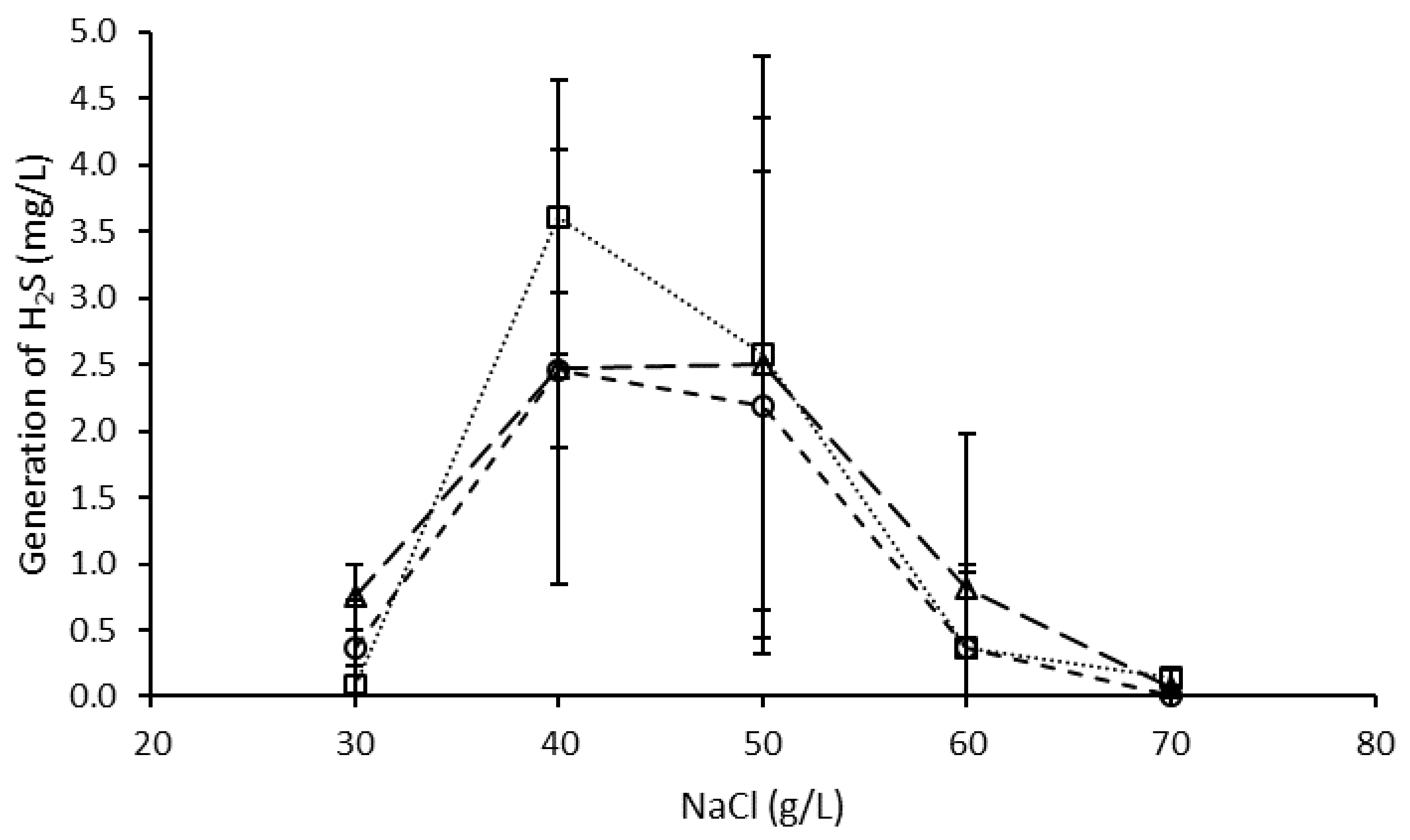
3.3. Effect of NaCl on H2S Generation by the Microbial Consortium
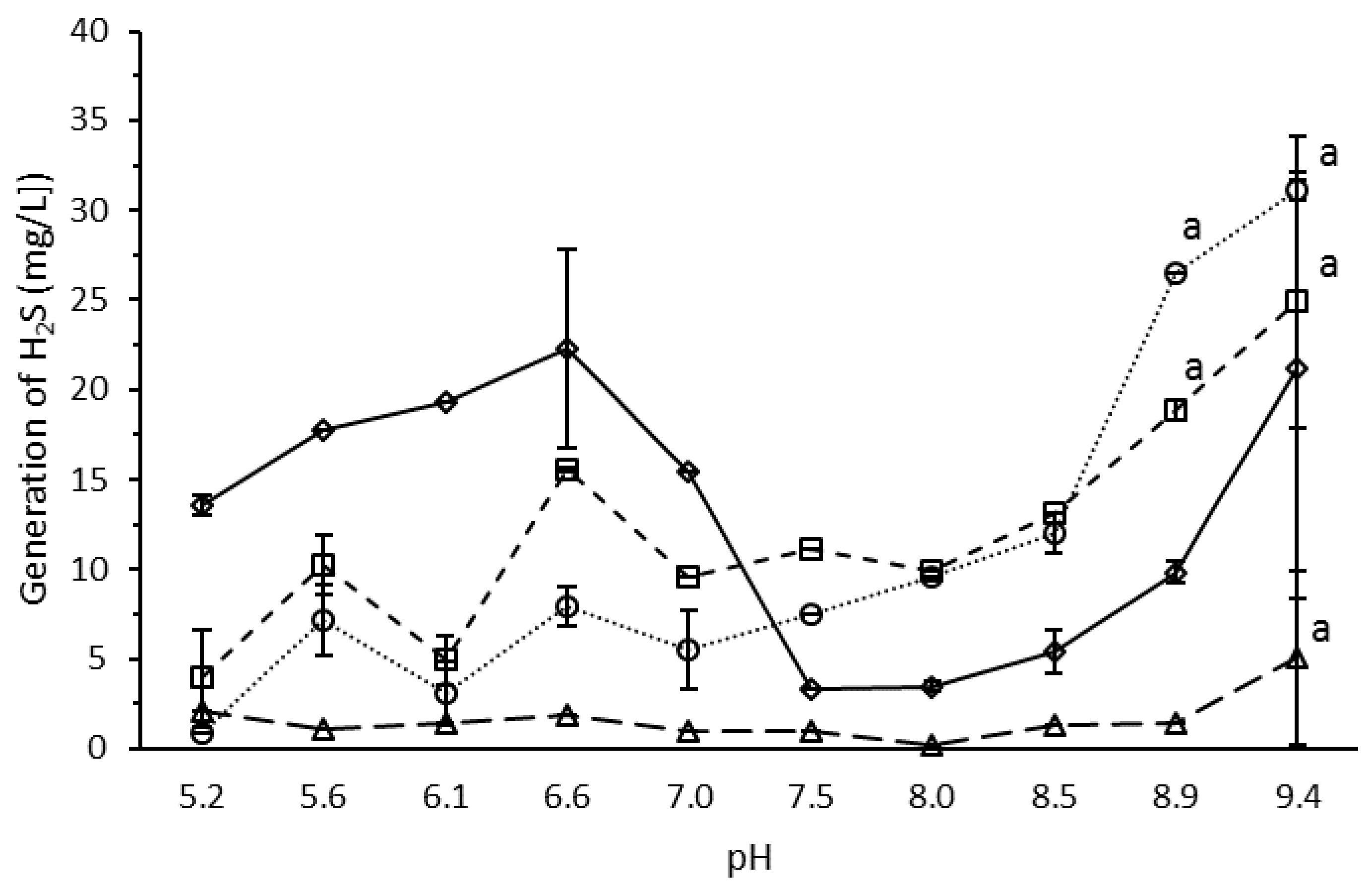
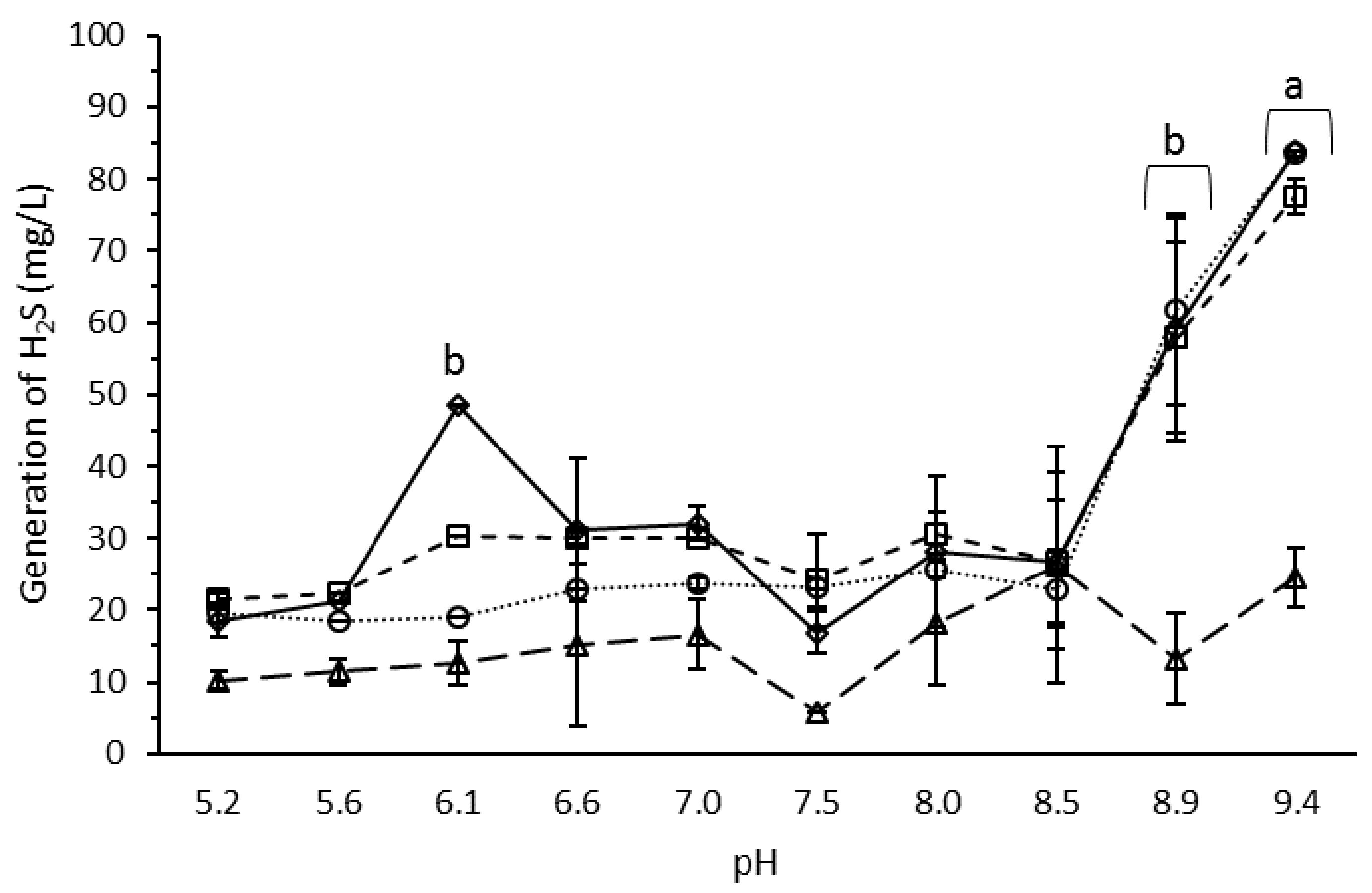
3.4. Effect of pH on H2S Production by the Microbial Consortium
3.5. Determination of the Growth Capacity of the Microbial Consortium in Bioreactors
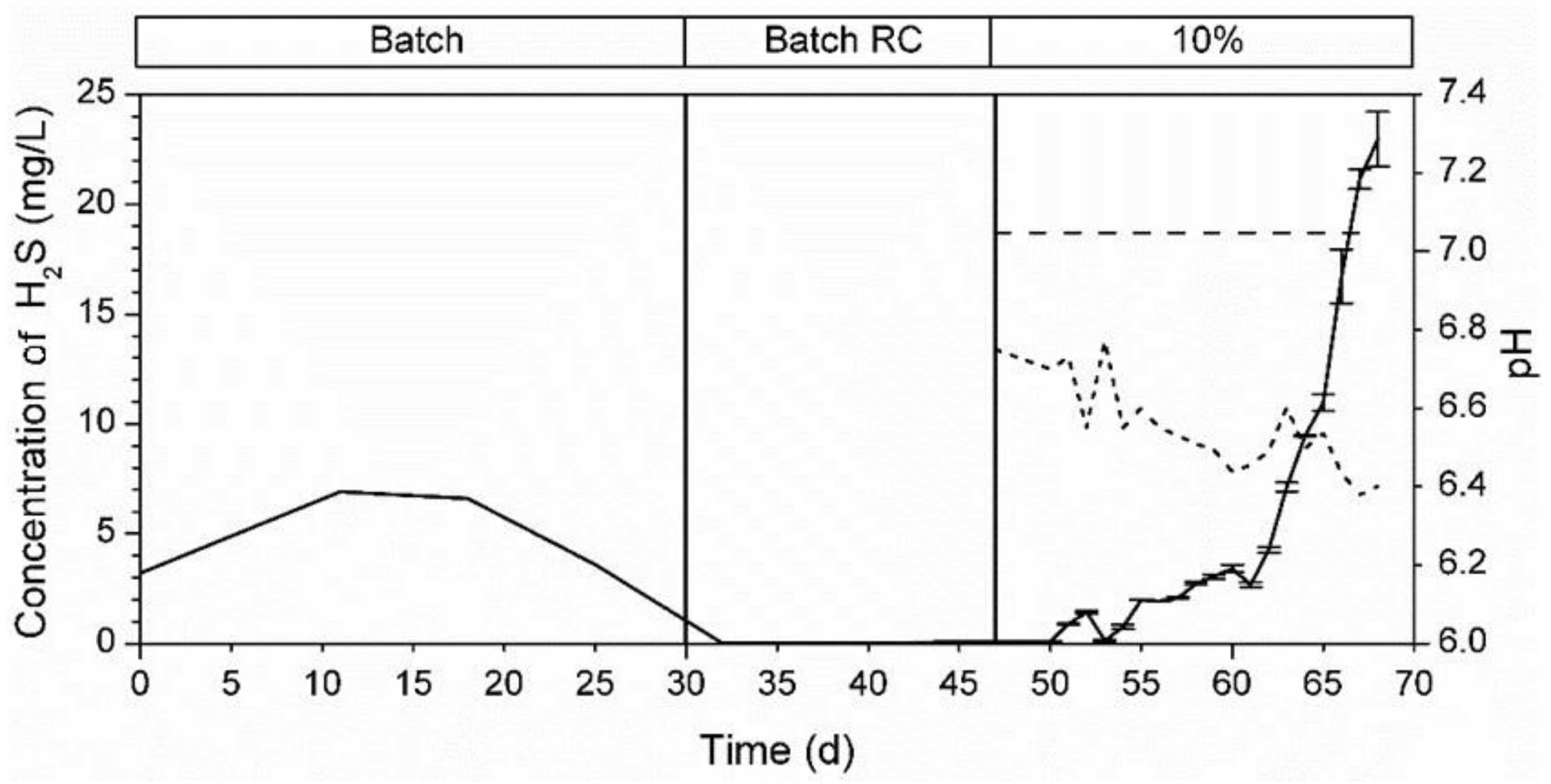
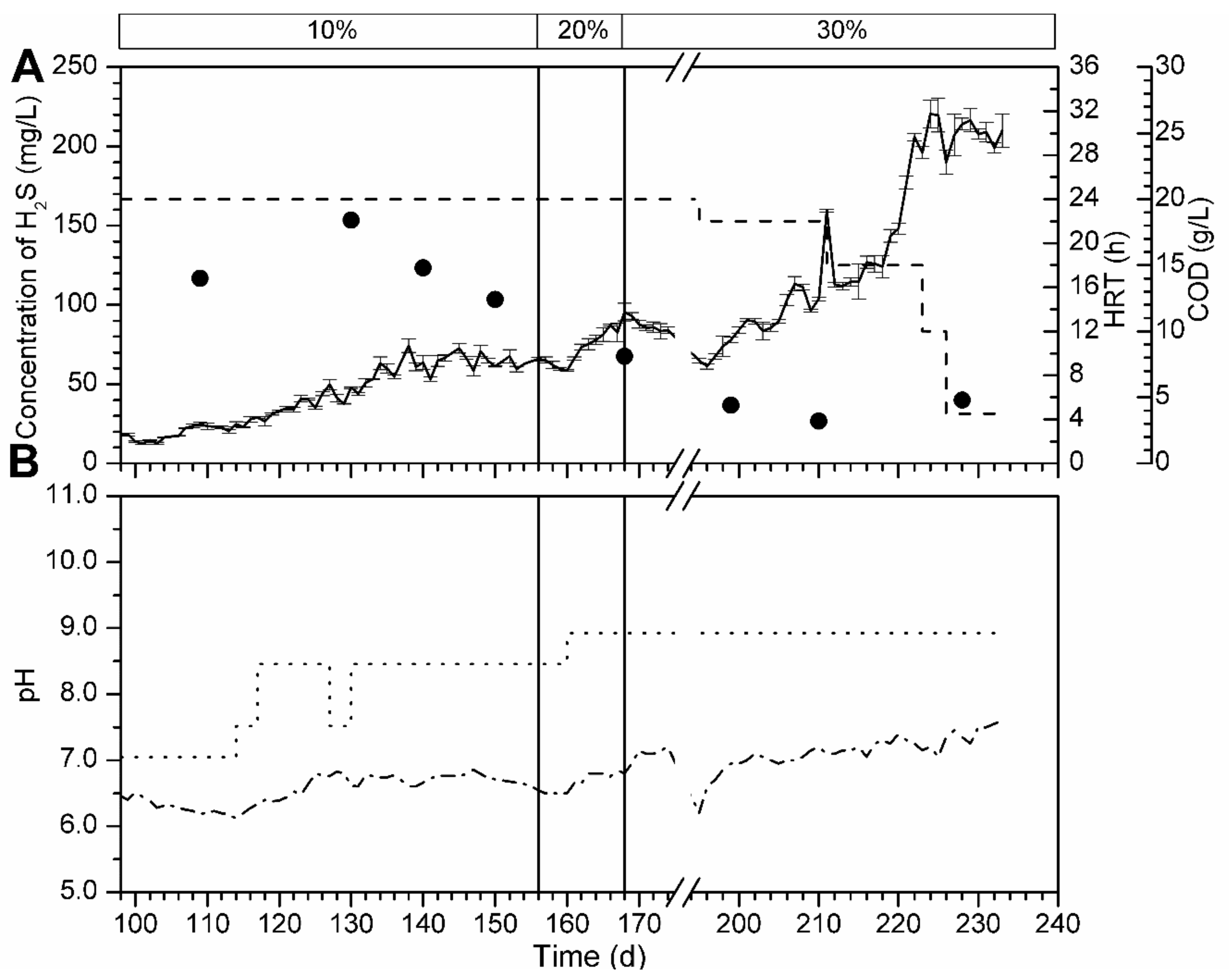
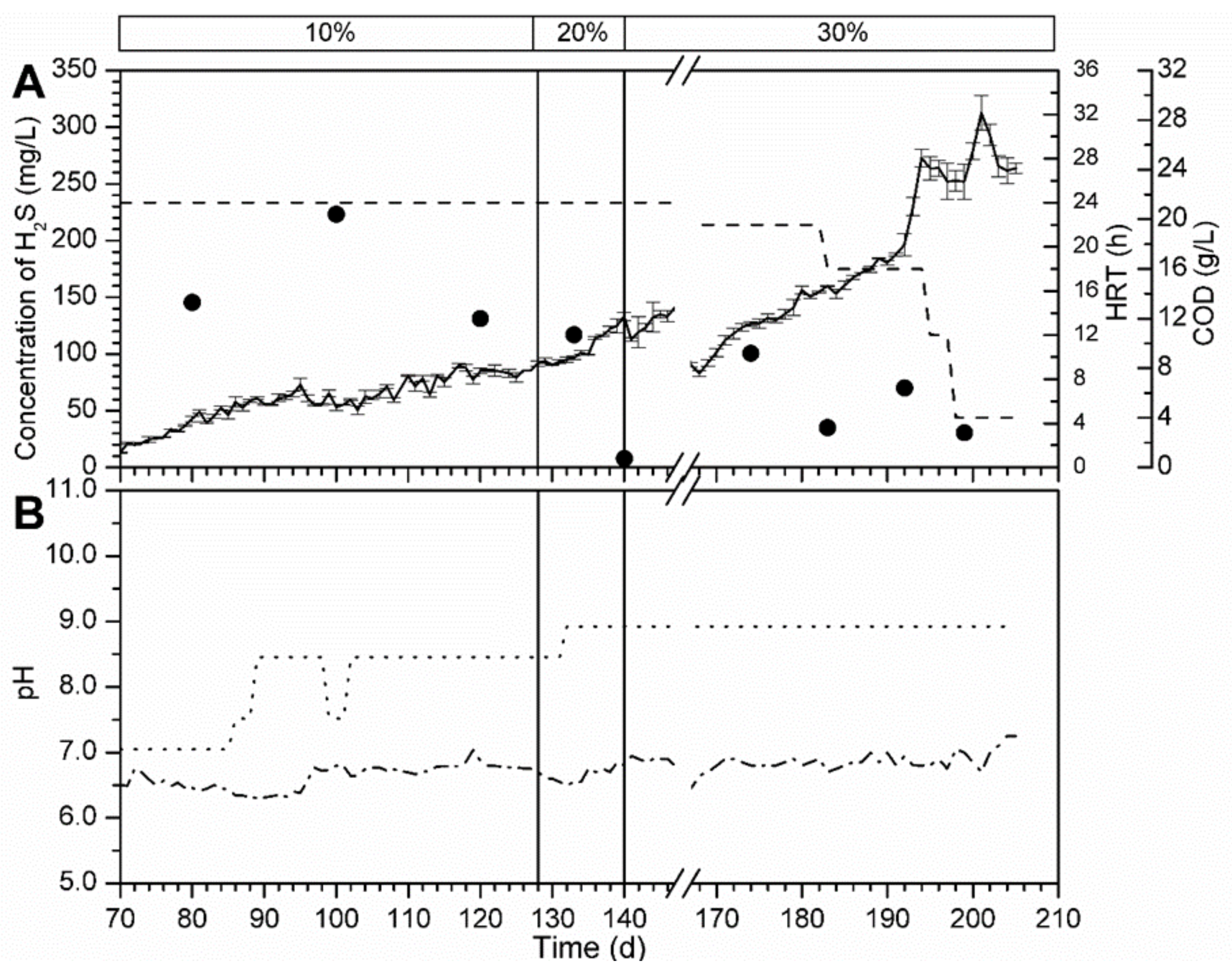
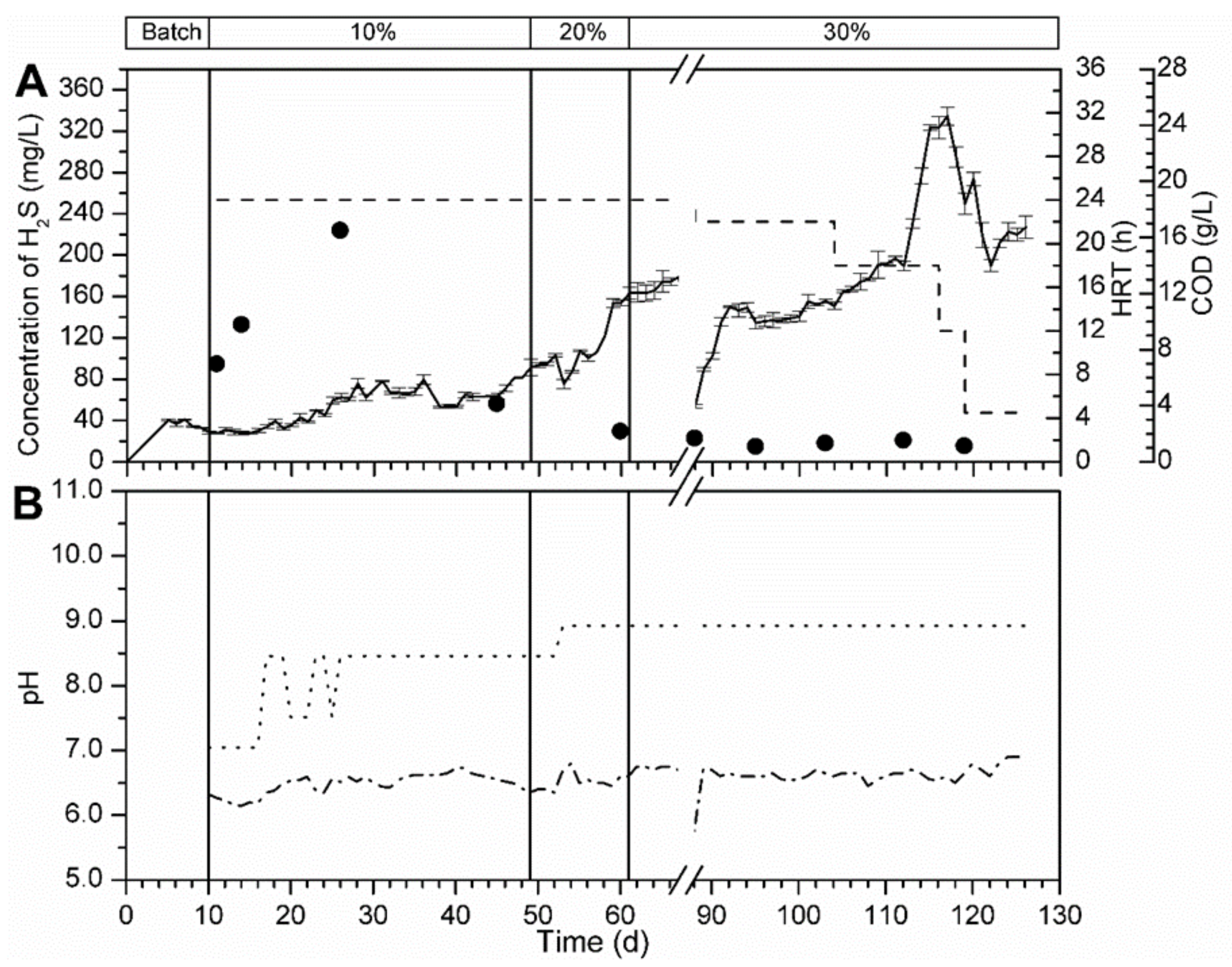
3.5.1. Bioreactor R1 Filled with Celite™ R-635 and Fed with Cellulose
3.5.2. Bioreactor R2 without Support Material and Fed with Spirulina
3.5.3. Bioreactor R3 Filled with Celite™ R-635 and Fed with Spirulina
4. Discussion
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Sheoran, A.S.; Sheoran, V.; Choudhary, R.P. Bioremediation of acid-rock drainage by sulphate-reducing prokaryotes: A review. Miner. Eng. 2010, 23, 1073–1100. [Google Scholar] [CrossRef]
- Barakat, M.A. New trends in removing heavy metals from industrial wastewater. Arab. J. Chem. 2011, 4, 361–377. [Google Scholar] [CrossRef]
- Fu, F.; Wang, Q. Removal of heavy metal ions from wastewaters: A review. J. Environ. Manag. 2011, 92, 407–418. [Google Scholar] [CrossRef] [PubMed]
- Hashim, M.A.; Mukhopadhyay, S.; Sahu, J.N.; Sengupta, B. Remediation technologies for heavy metal contaminated groundwater. J. Environ. Manag. 2011, 92, 2355–2388. [Google Scholar] [CrossRef] [PubMed]
- Verma, A.; Dua, R.; Singh, A.; Bishnoi, N.R. Biogenic sulfides for sequestration of Cr (VI), COD and sulfate from synthetic wastewater. Water Sci. 2015, 29, 19–25. [Google Scholar] [CrossRef]
- Lewis, A.E. Review of metal sulphide precipitation. Hydrometallurgy 2010, 104, 222–234. [Google Scholar] [CrossRef]
- Pohl, A. Removal of heavy metal ions from water and wastewaters by sulfur-containing precipitation agents. Water Air Soil Poll. 2020, 231, 503. [Google Scholar] [CrossRef]
- Huisman, J.L.; Schouten, G.; Schultz, C. Biologically produced sulphide for purification of process streams, effluent treatment and recovery of metals in the metal and mining industry. Hydrometallurgy 2006, 83, 106–113. [Google Scholar] [CrossRef]
- Cohen, R.R.H. Use of microbes for cost reduction of metal removal from metals and mining industry waste streams. J. Clean. Prod. 2006, 14, 1146–1157. [Google Scholar] [CrossRef]
- Cotoras, D.; Valenzuela, F.; Zarzar, M.E.; Viedma, P.L. Plant for the Removal of Metals by Biosorption from Mining or Industrial Effluents. U.S. Patent 7,479,220, 20 January 2009. [Google Scholar]
- Cotoras, D.; Viedma, P.L. Bacterial Strain for a Metal Biosorption Process. U.S. Patent 7,951,578, 31 May 2011. [Google Scholar]
- Geets, J.; Vangronsveld, J.; Diels, L.; Taghavi, S.; van der Lelie, D. Microbial activities, monitoring and application as part of a management strategy for heavy metal-contaminated soil and ground water. Dev. Soil Sci. 2008, 32, 521–559. [Google Scholar] [CrossRef]
- Rabus, R.; Hansen, T.A.; Widdel, F. Dissimilatory sulfate- and sulfur-reducing prokaryotes. In The Prokaryotes: Ecophysiology and Biochemistry; Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K.-H., Stackebrandt, E., Eds.; Springer: New York, NY, USA, 2006; Volume 2, pp. 659–768. [Google Scholar]
- Muyzer, G.; Stams, A.J.M. The ecology and biotechnology of sulphate-reducing bacteria. Nat. Rev. Microbiol. 2008, 6, 441–454. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Wang, J.; Qiu, Y.; Sun, J.; Jiang, F. Realizing a high-rate sulfidogenic reactor driven by sulfur-reducing bacteria with organic substrate dosage minimization and cost-effectiveness maximization. Chemosphere 2019, 236, 124381. [Google Scholar] [CrossRef] [PubMed]
- Sun, R.; Li, Y.; Lin, N.; Ou, C.; Wang, X.; Zhang, L.; Jiang, F. Removal of heavy metals using a novel sulfidogenic AMD treatment system with sulfur reduction: Configuration, performance, critical parameters and economic analysis. Environ. Int. 2020, 136, 105457. [Google Scholar] [CrossRef]
- Li, G.; Liang, Z.; Sun, J.; Qiu, Y.; Qiu, C.; Liang, X.; Zhu, Y.; Wang, P.; Li, Y.; Jiang, F. A pilot-scale sulfur-based sulfidogenic system for the treatment of Cu-laden electroplating wastewater using real domestic sewage as electron donor. Water Res. 2021, 195, 116999. [Google Scholar] [CrossRef]
- Zhang, L.; Qiu, Y.Y.; Zhou, Y.; Chen, G.H.; van Loosdrecht, M.C.; Jiang, F. Elemental sulfur as electron donor and/or acceptor: Mechanisms, applications and perspectives for biological water and wastewater treatment. Water Res. 2021, 202, 117373. [Google Scholar] [CrossRef] [PubMed]
- Postgate, J.R. The Sulphate-Reducing Bacteria, 2nd ed.; Cambridge University Press: Cambridge, UK; New York, NY, USA, 1984. [Google Scholar]
- Amann, R.I.; Ludwig, W.; Schleifer, K.H.; Amann, R.I.; Ludwig, W. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 1995, 59, 143–169. [Google Scholar] [CrossRef] [PubMed]
- Abràmofff, M.D.; Magalhães, P.J.; Ram, S.J. Image processing with ImageJ Part II. Biophotonics Int. 2005, 11, 36–43. [Google Scholar]
- APHA/AWWA/WEF. Standard Methods for the Examination of Water and Wastewater; American Public Health Association: Washington, DC, USA, 1999; pp. 1–541. [Google Scholar]
- Choudhary, R.P.; Sheoran, A.S. Comparative study of cellulose waste versus organic waste as substrate in a sulfate reducing bioreactor. Bioresour. Technol. 2011, 102, 4319–4324. [Google Scholar] [CrossRef]
- McMahon, M. Development of a Sulfate Reducing Packed Bed Bioreactor for Use in a Sustainable Hydrogen Production Process. Master’s Thesis, Queen’s University, Kingston, ON, Canada, 2007. [Google Scholar]
- Neculita, C.M.; Yim, G.J.; Lee, G.; Ji, S.W.; Jung, J.W.; Park, H.S.; Song, H. Comparative effectiveness of mixed organic substrates to mushroom compost for treatment of mine drainage in passive bioreactors. Chemosphere 2011, 83, 76–82. [Google Scholar] [CrossRef]
- Ayala-Parra, P.; Sierra-Alvarez, R.; Field, J.A. Algae as an electron donor promoting sulfate reduction for the bioremediaton of acid rock drainage. J. Hazard. Mater. 2016, 317, 335–343. [Google Scholar] [CrossRef]
- Márquez-Reyes, J.M.; López-Chuken, U.J.; Valdez-González, A.; Luna-Olvera, H.A. Removal of chromium and lead by a sulfate-reducing consortium using peat moss as carbon source. Bioresour. Technol. 2013, 144, 128–134. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Wang, H. Biological treatment of acidic coal refuse using sulphate-reducing bacteria with chicken manure as carbon source. Environ. Technol. 2014, 35, 2947–2955. [Google Scholar] [CrossRef] [PubMed]
- Martins, M.; Faleiro, M.L.; Barros, R.J.; Veríssimo, A.R.; Costa, M.C. Biological sulphate reduction using food industry wastes as carbon sources. Biodegradation 2009, 20, 559–567. [Google Scholar] [CrossRef] [PubMed]
- Teclu, D.; Tivchev, G.; Laing, M.; Wallis, M. Determination of the elemental composition of molasses and its suitability as carbon source for growth of sulphate-reducing bacteria. J. Hazard. Mater. 2009, 161, 1157–1165. [Google Scholar] [CrossRef] [PubMed]
- Boshoff, G.; Duncan, J.; Rose, P.D. The use of micro-algal biomass as a carbon source for biological sulphate reducing systems. Water Res. 2004, 38, 2659–2666. [Google Scholar] [CrossRef]
- Hurtado, C.; Viedma, P.; Cotoras, D. Design of a bioprocess for metal and sulfate removal from acid mine drainage. Hydrometallurgy 2018, 180, 72–77. [Google Scholar] [CrossRef]
- Vasquez, Y.; Escobar, M.C.; Saenz, J.S.; Quiceno-Vallejo, M.F.; Neculita, C.M.; Arbeli, Z.; Roldan, F. Effect of hydraulic retention time on microbial community in biochemical passive reactors during treatment of acid mine drainage. Bioresour. Technol. 2018, 247, 624–632. [Google Scholar] [CrossRef]
- Markou, G.; Georgakakis, D. Cultivation of filamentous cyanobacteria (blue-green algae) in agro-industrial wastes and wastewaters: A review. Appl. Energy 2011, 88, 3389–3401. [Google Scholar] [CrossRef]
- Celis, L.; Villa-Gómez, D.; Alpuche-Solís, A.; Ortega-Morales, B.; Razo-Flores, E. Characterization of sulfate-reducing bacteria dominated surface communities during start-up of a down-flow fluidized bed reactor. J. Ind. Microbiol. Biot. 2009, 36, 111–121. [Google Scholar] [CrossRef]
- Brock, T.D.; Madigan, M.T.; Martinko, J.M.; Parker, J. Brock Biology of Microorganisms; Prentice-Hall: Upper Saddle River, NJ, USA, 2003. [Google Scholar]
- Oren, A. Thermodynamic limits to microbial life at high salt concentrations. Environ. Microbiol. 2011, 13, 1908–1923. [Google Scholar] [CrossRef]
- Liamleam, W.; Annachhatre, A. Electron donors for biological sulfate reduction. Biotechnol. Adv. 2007, 25, 452–463. [Google Scholar] [CrossRef] [PubMed]
- Kersters, K.; De Vos, P.; Gillis, M.; Swings, J.; Vandamme, P.; Stackebrandt, E. Introduction to the Proteobacteria. In The Prokaryotes; Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K.H., Stackebrandt, E., Eds.; Springer: New York, NY, USA, 2006; pp. 3–37. [Google Scholar]
- Leschine, S.B. Cellulose degradation in anaerobic environments. Annu. Rev. Microbiol. 1995, 49, 399–426. [Google Scholar] [CrossRef] [PubMed]
- Dumova, V.A.; Kruglov, Y.V. A cellulose-decomposing bacterial association. Microbiology 2009, 78, 234–239. [Google Scholar] [CrossRef]
- Singh, G.; Chandoha-Lee, C.; Zhang, W.; Renneckar, S.; Vikesland, P.J.; Pruden, A. Biodegradation of nanocrystalline cellulose by two environmentally-relevant consortia. Water Res. 2016, 104, 137–146. [Google Scholar] [CrossRef] [PubMed]
- Strang, O.; Ács, N.; Wirth, R.; Maróti, G.; Bagi, Z.; Rákhely, G.; Kovács, K.L. Bioaugmentation of the thermophilic anaerobic biodegradation of cellulose and corn stover. Anaerobe 2017, 46, 104–113. [Google Scholar] [CrossRef] [PubMed]
- Oren, A. Bioenergetic aspects of halophilism. Microbiol. Mol. Biol. Rev. 1999, 63, 334–348. [Google Scholar] [CrossRef]
- Tang, K.; Baskaran, V.; Nemati, M. Bacteria of the sulphur cycle: An overview of microbiology, biokinetics and their role in petroleum and mining industries. Biochem. Eng. J. 2009, 44, 73–94. [Google Scholar] [CrossRef]
- Yuan, C.L.; Zhang, L.M.; Wang, J.T.; Hu, H.W.; Shen, J.P.; Cao, P.; He, J.Z. Distributions and environmental drivers of archaea and bacteria in paddy soils. J. Soils Sediments 2019, 19, 23–37. [Google Scholar] [CrossRef]
- Sorokin, D.Y.; Rusanov, I.I.; Pimenov, N.V.; Tourova, T.P.; Abbas, B.; Muyzer, G. Sulfidogenesis under extremely haloalkaline conditions in soda lakes of Kulunda Steppe (Altai, Russia). FEMS Microbiol. Ecol. 2010, 73, 278–290. [Google Scholar] [CrossRef]
- Sorokin, D.Y.; Kuenen, J.G.; Muyzer, G. The microbial sulfur cycle at extremely haloalkaline conditions of soda lakes. Front. Microbiol. 2011, 2, 44. [Google Scholar] [CrossRef]
- Schauder, R.; Müller, E. Polysulfide as a possible substrate for sulfur-reducing bacteria. Arch. Microbiol. 1993, 160, 377–382. [Google Scholar] [CrossRef]
- Florentino, A.P.; Pereira, I.A.; Boeren, S.; van den Born, M.; Stams, A.J.; Sánchez-Andrea, I. Insight into the sulfur metabolism of Desulfurella amilsii by differential proteomics. Environ. Microbiol. 2019, 21, 209–225. [Google Scholar] [CrossRef]
- Wu, C.H.; Schut, G.J.; Poole, F.L.; Haja, D.K.; Adams, M.W.W. Characterization of membrane-bound sulfane reductase: A missing link in the evolution of modern day respiratory complexes. J. Biol. Chem. 2018, 293, 16687–16696. [Google Scholar] [CrossRef]
- Kaksonen, A.H.; Puhakka, J.A. Sulfate reduction based bioprocesses for the treatment of acid mine drainage and the recovery of metals. Eng. Life Sci. 2007, 7, 541–564. [Google Scholar] [CrossRef]
- Durham, D.; Marshall, L.; Miller, J.; Chmurny, A. Characterization of inorganic biocarriers that moderate system upsets during fixed-film biotreatment processes. Appl. Environ. Microb. 1994, 60, 3329–3335. [Google Scholar] [CrossRef] [PubMed]
- McMahon, M.; Daugulis, A. Enhancement of biogenic sulfide production in a packed-bed bioreactor via critical inoculum design and carrier material selection. Biotechnol. Bioeng. 2008, 100, 855–863. [Google Scholar] [CrossRef] [PubMed]
- Kousi, P.; Remoundaki, E.; Hatzikioseyian, A.; Battaglia-Brunet, F.; Joulian, C.; Kousteni, V.; Tsezos, M. Metal precipitation in an ethanol-fed, fixed-bed sulphate-reducing bioreactor. J. Hazard. Mater. 2011, 189, 677–684. [Google Scholar] [CrossRef]
- de Beer, D.; Stoodley, P. Microbial Biofilms. In The Prokaryotes: Symbiotic Associations, Biotechnology, Applied Microbiology, 3rd ed.; Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K.-H., Stackebrandt, E., Eds.; Springer: New York, NY, USA, 2006; Volume 1, pp. 904–937. [Google Scholar]
- Escudié, R.; Cresson, R.; Delgenès, J.P.; Bernet, N. Control of start-up and operation of anaerobic biofilm reactors: An overview of 15 years of research. Water Res. 2011, 45, 1–10. [Google Scholar] [CrossRef]
- Lyberatos, G.; Pullammanappallil, P.C. Anaerobic digestion in suspended growth bioreactors. In Environmental Biotechnology, Handbook of Environmental Engineering; Wang, L.K., Ivanov, V., Tay, J.H., Eds.; Humana Press: Totowa, NJ, USA, 2010; Volume 10, pp. 395–438. [Google Scholar]
- Guyot, J.P.; Calderon, M.; Morlon-Guyot, J. Effect of pH control on lactic acid fermentation of starch by Lactobacillus manihotivorans LMG 18010. J. Appl. Microbiol. 2000, 88, 176–182. [Google Scholar] [CrossRef]


| Probes | Specificity (rRNA, Position) | Sequence |
|---|---|---|
| EUB338 | Bacteria (16S, 338–355) | GCTGCCTCCCGTAGGAGT |
| Archaea | Archaea (16S, 915–934) | GTGCTCCCCCGCCAATTCCT |
| ALF1b | α-Proteobacteria (16S, 19–35) | CGTTCGYTCTGAGCCAG |
| BET42a | β-Proteobacteria (23S, 1027–1043) | GCCTTCCCACTTCGTTT |
| GAM42a | γ-Proteobacteria (23S, 1027–1043) | GCCTTCCCACATCGTTT |
| SRB385 | δ-Proteobacteria (16S, 385–402) | CGGCGTCGCTGCGTCAGG |
| CF319a | Cytophaga-Flavobacterium (16S, 319–336) | TGGTCCGTGTCTCAGTAC |
| Probes | Formamide [%] | NaCl [M] | Tris/HCl (pH 7.2) [mM] | SDS [%] |
|---|---|---|---|---|
| ALF1b/EUB338/Archaea | 20 | 0.9 | 20 | 0.01 |
| BET42a/GAM42a/ CF319a/SRB385 | 35 | 0.9 | 20 | 0.01 |
| Probes | Tris/HCl (pH 7.2) [mM] | SDS [%] | NaCl [M] | EDTA [mM] |
|---|---|---|---|---|
| ALF1b/EUB338/Archaea | 20 | 0.010 | 180 | 5 |
| BET42a/GAM42a/ CF319a/SRB385 | 20 | 0.021 | 40 | 5 |
| Bioreactor | Substrate | Support | Working Volume (mL) | Dimensions (cm) | Operation Time (Days) |
|---|---|---|---|---|---|
| R1 | Cellulose | Celite™ R-635 | 410 | 49 × 3.3 | 233 |
| R2 | Spirulina | – | 410 | 49 × 3.3 | 205 |
| R3 | Spirulina | Celite™ R-635 | 496 | 49 × 3.6 | 126 |
| Experimental Period | |||||||||||||
| Parameter | I | II | III | IV | V | VI | VII | VIII | IX | X | XI | XII | XIII |
| Type of feeding * | Batch | Batch RC | 10% | Batch | 10% | 10% | 20% | 30% | Batch RC | 30% | 30% | 30% | 30% |
| Operation time (days) | 0–30 | 31–47 | 48–68 | 69–97 | 98–113 | 114–155 | 156–167 | 168–174 | 175–194 | 195–210 | 211–222 | 223–225 | 226–233 |
| Operating Conditions | |||||||||||||
| Flow (mL/min) | - | - | 1.08 | - | 1.95 | 1.95 | 1.95 | 1.95 | - | 1.95 | 1.52 | 1.73 | 1.52 |
| HRT recirculated (h) | - | 48 | 24 | - | 24 | 24 | 24 | 24 | 22 | 22 | 18 | 12 | 4.5 |
| pH | 7.1 | 7.1 | 7.1 | 7.1 | 7.1 | 8.5 | 8.9 | 8.9 | 8.9 | 8.9 | 8.9 | 8.9 | 8.9 |
| Experimental Period | |||||||||||||
| Parameter | I | II | III | IV | V | VI | VII | VIII | IX | X | XI | XII | XIII |
| Type of feeding (a) | Batch | Batch RC | 10% | Batch | 10% | 10% | 20% | 30% | Batch RC | 30% | 30% | 30% | 30% |
| Operation time (days) | 0–18 | 19–25 | 26–37 | 38–69 | 70–88 | 89–127 | 128–139 | 140–147 | 148–166 | 167–182 | 183–194 | 195–197 | 198–205 |
| Operating Conditions | |||||||||||||
| Flow (mL/min) | - | - | 1.08 | - | 1.95 | 1.95 | 1.95 | 1.95 | - | 1.95 | 1.52 | 1.73 | 1.52 |
| HRT recirculated (h) | - | 48 | 24 | - | 24 | 24 | 24 | 24 | 22 | 22 | 18 | 12 | 4.5 |
| pH | 7.1 | 7.1 | 7.1 | 7.1 | 7.1 | 8.5 | 8.9 | 8.9 | 8.9 | 8.9 | 8.9 | 8.9 | 8.9 |
| Experimental Period | |||||||||||
| Parameter | I | II | III | IV | V | VI | VII | VIII | IX | X | XI |
| Type of feeding (a) | Batch | Batch RC | 10% | 10% | 20% | 30% | Batch RC | 30% | 30% | 30% | 30% |
| Operation time (days) | 0–6 | 7–9 | 10–16 | 17–48 | 49–60 | 61–67 | 68–87 | 88–103 | 104–115 | 116–118 | 119–126 |
| Operating Conditions | |||||||||||
| Flow (mL/min) | ND | ND | 1.08 | 1.08 | 1.95 | 1.95 | ND | 1.95 | 1.52 | 1.73 | 1.52 |
| HRT recirculated (h) | - | 48 | 24 | 24 | 24 | 24 | 22 | 22 | 18 | 12 | 4.5 |
| pH | 7.1 | 7.1 | 7.1 | 8.5 | 8.9 | 8.9 | 8.9 | 8.9 | 8.9 | 8.9 | 8.9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martínez, C.; Viedma, P.; Cárdenas, F.; Cotoras, D. Biogenic Hydrogen Sulfide Production Using Elemental Sulfur and Low-Cost Organic Substrates to Remove Metal Ions from Mining Effluents. Mining 2023, 3, 241-260. https://doi.org/10.3390/mining3020015
Martínez C, Viedma P, Cárdenas F, Cotoras D. Biogenic Hydrogen Sulfide Production Using Elemental Sulfur and Low-Cost Organic Substrates to Remove Metal Ions from Mining Effluents. Mining. 2023; 3(2):241-260. https://doi.org/10.3390/mining3020015
Chicago/Turabian StyleMartínez, Cristian, Pabla Viedma, Franco Cárdenas, and Davor Cotoras. 2023. "Biogenic Hydrogen Sulfide Production Using Elemental Sulfur and Low-Cost Organic Substrates to Remove Metal Ions from Mining Effluents" Mining 3, no. 2: 241-260. https://doi.org/10.3390/mining3020015
APA StyleMartínez, C., Viedma, P., Cárdenas, F., & Cotoras, D. (2023). Biogenic Hydrogen Sulfide Production Using Elemental Sulfur and Low-Cost Organic Substrates to Remove Metal Ions from Mining Effluents. Mining, 3(2), 241-260. https://doi.org/10.3390/mining3020015





