Perils of Underestimating Species Diversity: Revisiting Systematics of Psammocambeva Catfishes (Siluriformes: Trichomycteridae) from the Rio Paraíba do Sul Basin, South-Eastern Brazil †
Abstract
:1. Introduction
Historical Review of PAC Species in RPSA
2. Materials and Methods
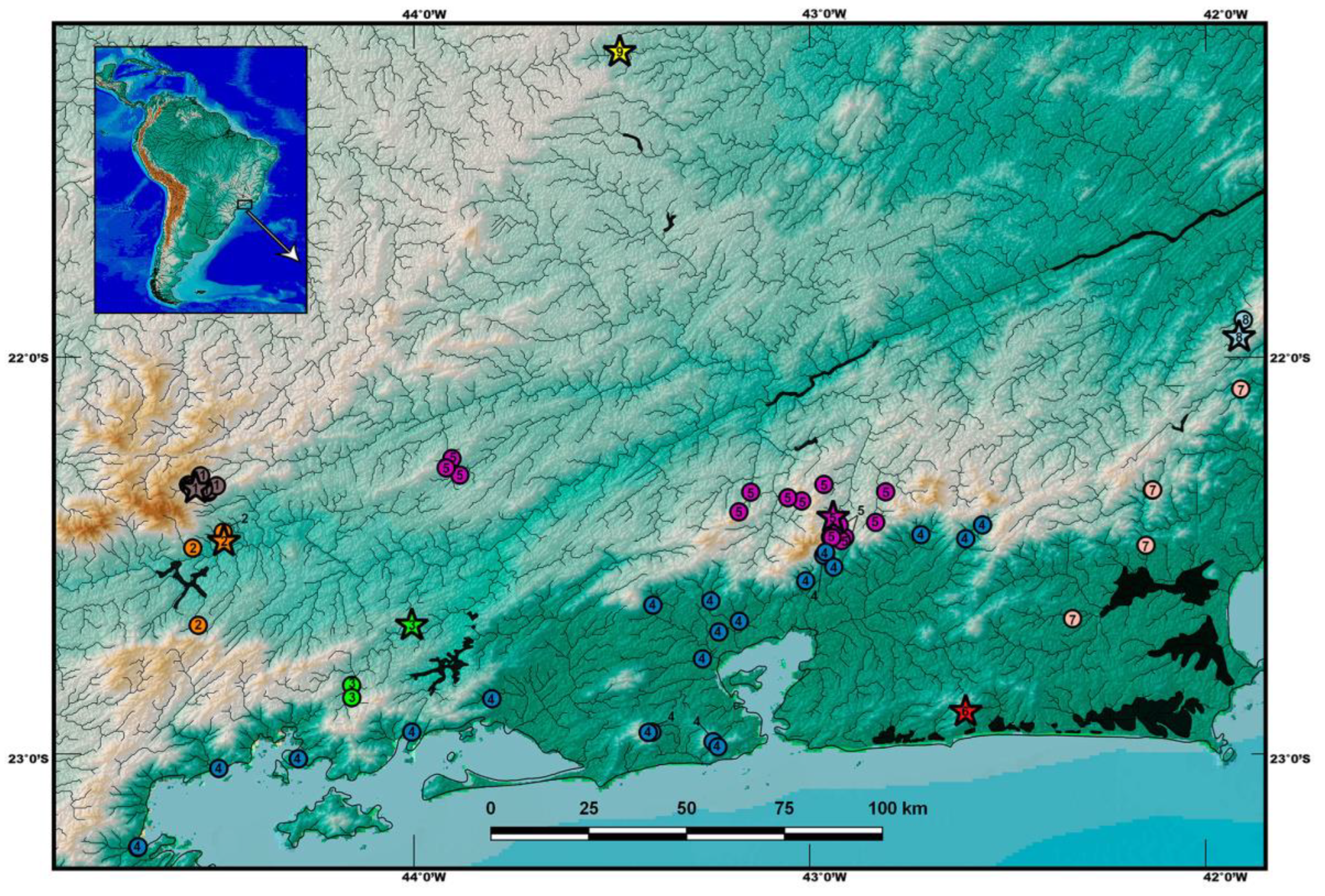
2.1. Specimens
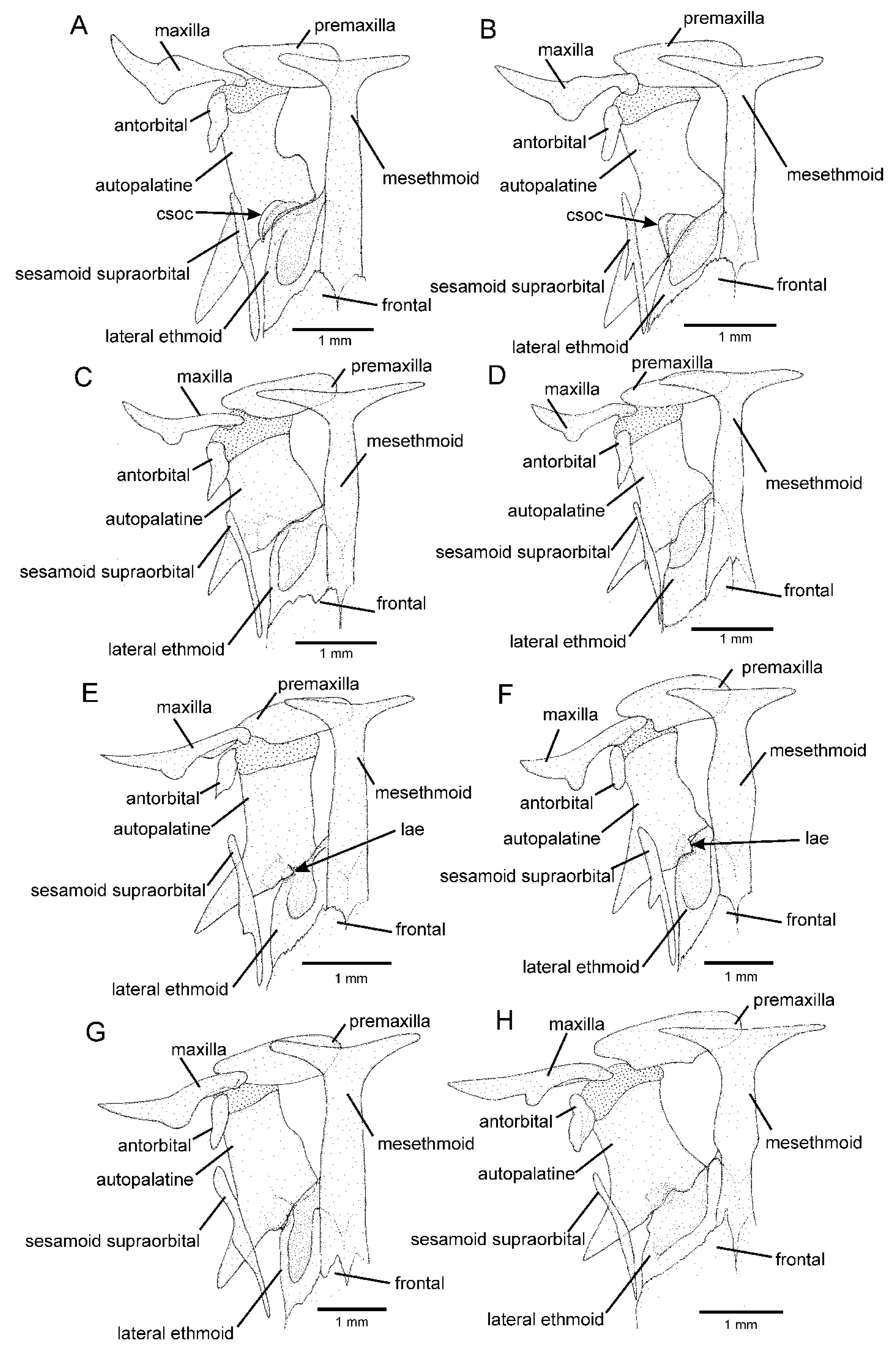
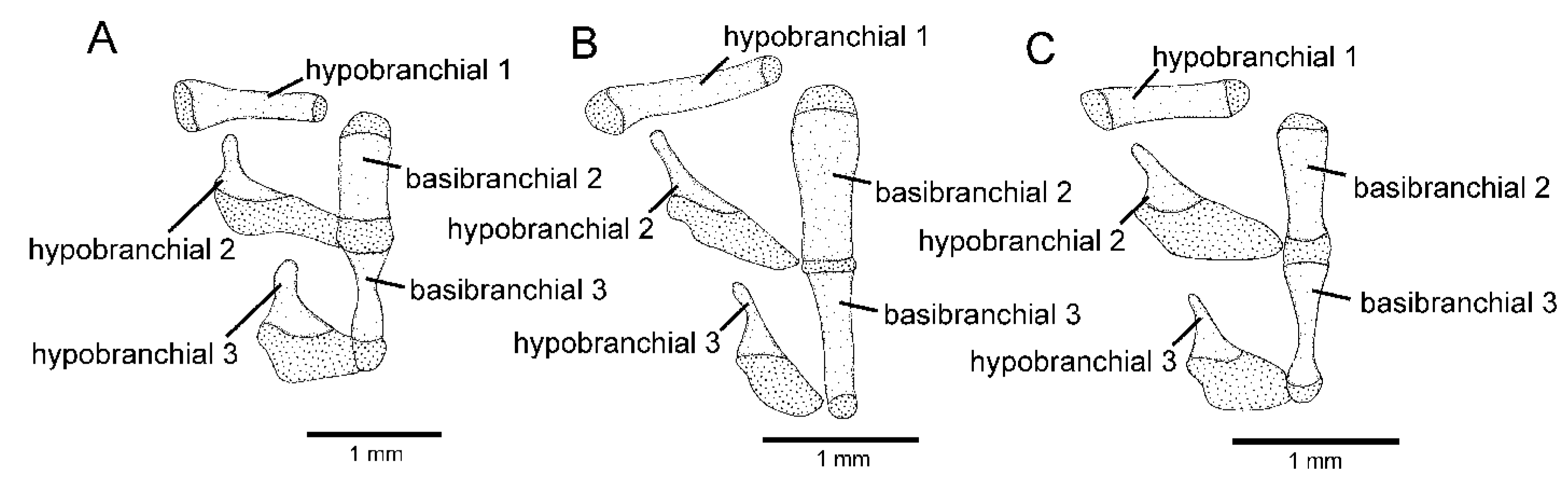
2.2. Morphological Data
2.3. DNA Extraction, Amplification, and Sequencing
2.4. Phylogenetic Analyses
3. Results
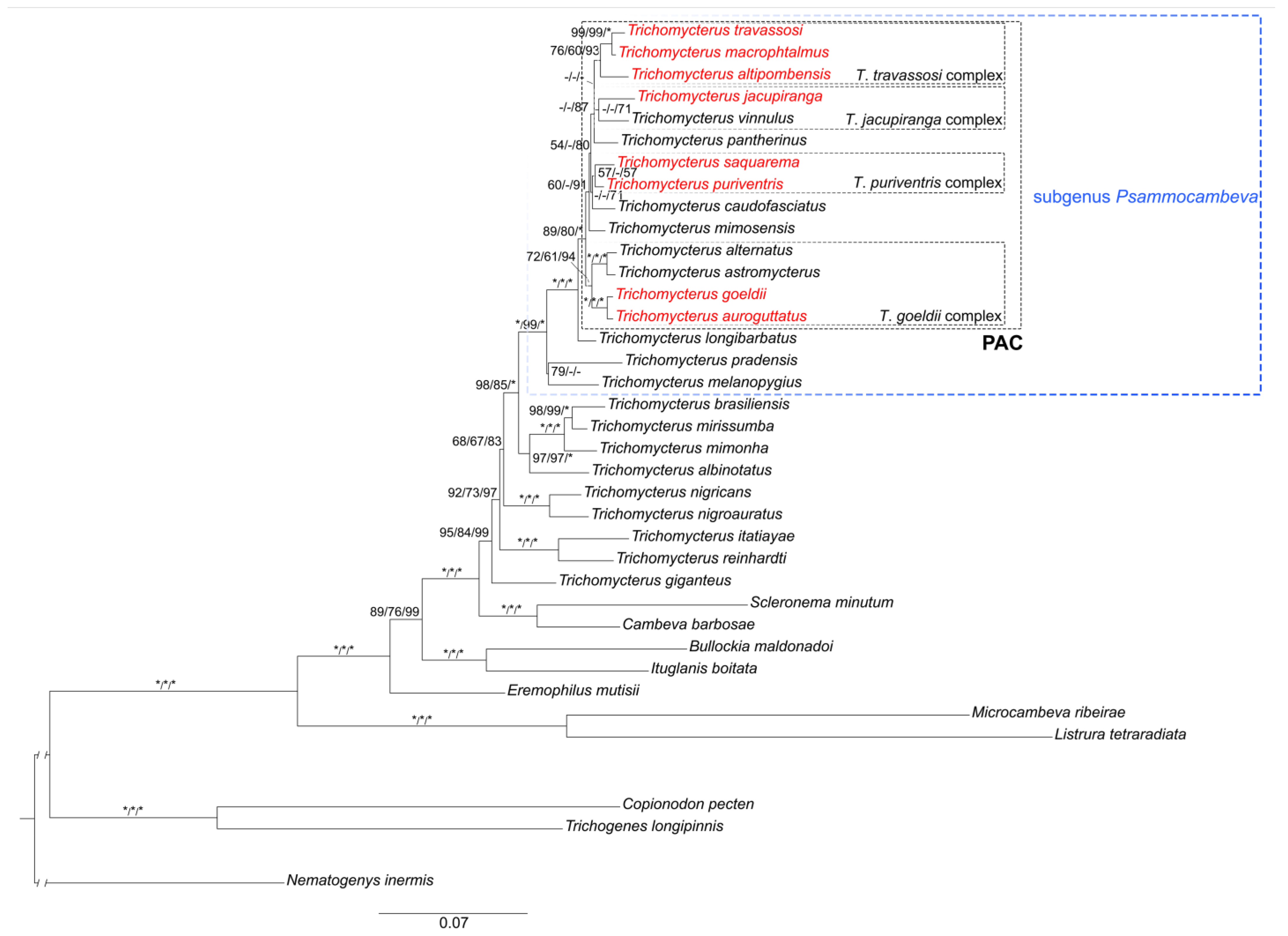
3.1. Phylogenetic Relationships
3.2. The Trichomycterus goeldii Complex
3.3. The Trichomycterus jacupiranga Complex
3.4. The Trichomycterus puriventris Complex
3.5. The Trichomycterus travassosi Complex
3.6. Key to Identification of Species of Psammocambeva from RPSA
4. Discussion
4.1. Psammocambeva Taxonomy
4.2. Perils of Underestimating Species Diversity
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
Appendix B
| COX1 | CYTB | MYH6 | RAG2 | |
|---|---|---|---|---|
| Nematogenys inermis | KY857952 | - | KY858107 | KY858182 |
| Copionodon pecten | KY857929 | - | KY858084 | KY858169 |
| Trichogenes longipinnis | MK123682 | MK123704 | MF431104 | MF431117 |
| Microcambeva ribeirae | MN385807 | OK334290 | MN385819 | MN385832 |
| Listrura tetraradiata | MN385804 | JQ231088 | MN385814.1 | MN385826.1 |
| Eremophilus mutisii | KY857931 | - | KY858086 | KY858171 |
| Ituglanis boitata | MK123684 | MK123706 | MF431105 | MK123758 |
| Bullockia maldonadoi | KY857926 | FJ772237 | KY858081 | KY858166 |
| Scleronema minutum | MK123685 | MK123707 | MK123735.1 | MK123759.1 |
| Cambeva barbosae | MK123689 | MK123713 | MK123740.1 | MN385820 |
| Trichomycterus itatiayae | MW671552 | MW679291 | KY858128 | KY858198 |
| Trichomycterus reinhardti | MK123698 | MK123727 | MF431106 | MF431119 |
| Trichomycterus giganteus | MK123693 | MK123720 | MK123746 | MT446426 |
| Trichomycterus nigricans | MN813005 | MK123723 | MK123750 | MK123765 |
| Trichomycterus nigroauratus | MK123696 | MK123724 | MK123751 | MK123766 |
| Trichomycterus albinotatus | MN813007 | MK123716 | MK123743 | MN812990 |
| Trichomycterus brasiliensis | MK123691 | MK123717 | MK123744 | MK123763 |
| Trichomycterus mimonha | MW196749 | MW196758 | MW196770 | MW196783 |
| Trichomycterus mirissumba | MW196752 | MW196761 | MW196773 | MW196786 |
| Trichomycterus caudofasciatus | MN812995 | MK123719 | MK123719 | MK123764 |
| Trichomycterus jacupiranga | MK123702 | MK123731.1 | MK123757 | OP688469 |
| Trichomycterus pantherinus | MK123697 | MK123725 | MK123752 | MN812989 |
| Trichomycterus pradensis | MN813003 | MK123726 | MK123753 | MN812988 |
| Trichomycterus goeldii | MT435136 | MT436453 | MT436451 | MT446427 |
| Trichomycterus alternatus | MK123690 | MK123715 | MK123742 | MN812991.1 |
| Trichomycterus melanopygius | KY857976 | KY858045 | KY858127 | KY858197 |
| Trichomycterus astromycterus | ON036881 | OK652453 | OK652451.1 | OK652448.1 |
| Trichomycterus saquarema | OP698258 | OP688464 | - | OP688470 |
| Trichomycterus macrophtalmus | OL741727 | OL752426 | - | OL752421 |
| Trichomycterus travassosi | MK123701 | MK123730 | MK123757 | OL752425 |
| Trichomycterus auroguttatus | MT435135 | MT436452 | MT436450 | OP699434 |
| Trichomycterus puriventris | OP698259 | OP688465 | OP688467 | OP688471 |
| Trichomycterus vinnulus | - | OK652452 | OK652450 | OK652449 |
| Trichomycterus altipombensis | OP698260 | OP688466 | OP688468 | OP688472 |
Appendix C
| Partition | Base pairs | Evolutive Model |
|---|---|---|
| COX1 1st | 174 | TIM + I + G |
| COX1 2nd | 174 | HKY + I |
| COX1 3rd | 174 | TRN + G |
| CYTB 1st | 363 | K80 + I + G |
| CYTB 2nd | 363 | TIM + I |
| CYTB 3rd | 362 | GTR + I + G |
| MYH6 1st | 181 | TRN + I + G |
| MYH6 2nd | 181 | TRN + I + G |
| MYH6 3rd | 181 | K80 + G |
| RAG2 1st | 274 | K81 + I + G |
| RAG2 2nd | 274 | TVM + G |
| RAG2 3rd | 273 | K80 + G |
Appendix D

References
- Katz, A.M.; Barbosa, M.A.; Mattos, J.L.O.; Costa, W.J.E.M. Multigene analysis of the catfish genus Trichomycterus and description of a new South American trichomycterine genus (Siluriformes, Trichomycteridae). Zoosystematics Evol. 2018, 94, 557–566. [Google Scholar] [CrossRef] [Green Version]
- Ochoa, L.E.; Datovo, A.; DoNascimiento, C.; Roxo, F.F.; Sabaj, M.H.; Chang, J.; Melo, B.F.; Silva Gabriel, S.C.; Foresti, F.; Alfaro, M.; et al. Phylogenomic analysis of trichomycterid catfshes (Teleostei: Siluriformes) inferred from ultraconserved elements. Sci. Rep. 2020, 10, 2697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fricke, R.; Eschmeyer, W.N.; Van der Laan, R. Eschmeyer’s Catalog of Fishes: Genera, Species, References. 2022. Available online: http://researcharchive.calacademy.org/research/ichthyology/catalog/fishcatmain.asp (accessed on 3 December 2022).
- Eigenmann, C.H. The Pygidiidae, a subfamily of South American catfishes. Mem. Carnegie Mus. 1918, 7, 259–398. [Google Scholar] [CrossRef]
- Hayes, M.M.; Paz, H.J.; Stout, C.C.; Werneke, D.C.; Armbruster, J.W. A hotspot atop: Rivers of the Guyana Highlands hold high diversity of endemic pencil catfish (Teleostei: Ostariophysi: Siluriformes). Biol. J. Linn. Soc. 2020, 129, 862–874. [Google Scholar] [CrossRef]
- Costa, W.J.E.M.; Mattos, J.L.O.; Katz, A.M. Two new catfish species from central Brazil comprising a new clade supported by molecular phylogeny and comparative osteology (Siluriformes: Trichomycteridae). Zool. Anz. 2021, 293, 124–137. [Google Scholar] [CrossRef]
- Costa, W.J.E.M. Comparative osteology, phylogeny and classification of the eastern South American catfish genus Trichomycterus (Siluriformes: Trichomycteridae). Taxonomy 2021, 1, 160–191. [Google Scholar] [CrossRef]
- Costa, W.J.E.M.; Mattos, J.L.O.; Katz, A.M. Phylogenetic position of Trichomycterus payaya and examination of osteological characters diagnosing the Neotropical catfish genus Ituglanis (Siluriformes: Trichomycteridae). Zool. Stud. 2021, 60, 43. [Google Scholar] [CrossRef]
- Costa, W.J.E.M.; Sampaio, W.M.S.; Giongo, P.; de Almeida, F.B.; Azevedo-Santos, V.M.; Katz, A.M. An enigmatic interstitial trichomycterine catfish from south-eastern Brazil found at about 1000 km away from its sister group (Siluriformes: Trichomycteridae). Zool. Anz. 2022, 297, 85–96. [Google Scholar] [CrossRef]
- Vilardo, P.J.; Katz, A.M.; Costa, W.J.E.M. Phylogenetic position of Trichomycterus astromycterus (Siluriformes: Trichomycteridae), an enigmatic trichomycterid from eastern Brazil, inferred from molecular data. J. Fish Biol. 2022, 100, 1093–1096. [Google Scholar] [CrossRef]
- Costa, W.J.E.M.; Mattos, J.L.O.; Amorim, P.F.; Vilardo, P.J.; Katz, A.M. Relationships of a new species support multiple origin of melanism in Trichomycterus from the Atlantic Forest of south-eastern Brazil (Siluriformes: Trichomycteridae). Zool. Anz. 2020, 288, 74–83. [Google Scholar] [CrossRef]
- Myers, N.; Mittermeir, R.A.; Mittermeir, C.G.; da Fonseca, G.A.B.; Kent, J. Biodiversity hotspots for conservation priorities. Nature 2000, 403, 853–858. [Google Scholar] [CrossRef] [PubMed]
- Costa, W.J.E.M.; Katz, A.M. A new catfish of the genus Trichomycterus from the Rio Paraíba do Sul Basin, south-eastern Brazil, a supposedly migrating species (Siluriformes, Trichomycteridae). Zoosystematics Evol. 2022, 98, 13–21. [Google Scholar] [CrossRef]
- Sanjad, N. Emílio Goeldi (1859–1917): A ventura de um naturalista entre a Europa e o Brasil; EMC Edições: Rio de Janeiro, Brazil, 2009; 232p. [Google Scholar]
- Boulenger, G.A. Description of a new siluroid fish from the Organ Mountains, Brazil. Ann. Mag. Nat. Hist. 1896, 18, 154. [Google Scholar] [CrossRef] [Green Version]
- Miranda Ribeiro, P. Duas novas espécies de peixes na coleção ictiológica do Museu Nacional (Pisces, Callichthyidae et Pygidiidae). Rev. Bras. De Biol. 1949, 9, 143–145. [Google Scholar]
- Costa, W.J.E.M. Description de huit nouvelles espèces du genre Trichomycterus (Siluriformes: Trichomycteridae), du Brésil oriental. Rev. Française D’aquariologie Et Herpetol. 1992, 18, 101–110. [Google Scholar]
- Barbosa, M.A.; Costa, W.J.E.M. Trichomycterus macrophthalmus (Teleostei: Siluriformes: Trichomycteridae), a new species of catfish from the rio Paraiba do Sul basin, southeastern Brazil. Vertebr. Zool. 2012, 62, 79–82. [Google Scholar]
- Barbosa, M.A.; Costa, W.J.E.M. Trichomycterus puriventris (Teleostei: Siluriformes: Trichomycteridae), a new species of catfish from the rio Paraiba do Sul basin, southeastern Brazil. Vertebr. Zool. 2012, 62, 155–160. [Google Scholar]
- Reis, V.J.C.; de Pinna, M.C.C. The type specimens of Trichomycterus alternatus (Eigenmann, 1917) and Trichomycterus zonatus (Eigenmann, 1918), with elements for future revisionary work (Teleostei: Siluriformes: Trichomycteridae). Zootaxa 2019, 4585, 100–120. [Google Scholar] [CrossRef]
- Reis, V.J.C.; de Pinna, M.C.C. Diversity and systematics of Trichomycterus Valenciennes 1832 (Siluriformes: Trichomycteridae) in the Rio Doce Basin: Iterating DNA, phylogeny and classical taxonomy. Zool. J. Linn. Soc. 2022. [Google Scholar] [CrossRef]
- Vilardo, P.J.; Katz, A.M.; Costa, W.J.E.M. Relationships and description of a new species of Trichomycterus (Siluriformes: Trichomycteridae) from the Rio Paraíba do Sul basin, south-eastern Brazil. Zool. Stud. 2020, 59, 53. [Google Scholar] [CrossRef]
- Leary, S.; Underwood, W.; Anthony, R.; Cartner, S.; Corey, D.; Grandin, T.; Greenacre, C.; Gwaltney-Brant, S.; McCrackin, M.; Meyer, R.; et al. AVMA Guidelines for the Euthanasia of Animals: 2020 Edition. 2020. Available online: http://www.avma.org/sites/default/files/2020-02/Guidelines-on-Euthanasia-2020.pdf (accessed on 3 December 2022).
- Close, B.; Banister, K.; Baumans, V.; Bernoth, E.M.; Bromage, N.; Bunyan, J.; Erhardt, W.; Flecknell, P.; Gregory, N.; Hackbarth, H.; et al. Recommendations for euthanasia of experimental animals: Part 1. Lab. Anim. 1996, 30, 293–316. [Google Scholar] [CrossRef] [PubMed]
- Close, B.; Banister, K.; Baumans, V.; Bernoth, E.M.; Bromage, N.; Bunyan, J.; Erhardt, W.; Flecknell, P.; Gregory, N.; Hackbarth, H.; et al. Recommendations for euthanasia of experimental animals: Part 2. Lab. Anim. 1997, 31, 1–32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Costa, W.J.E.M.; Katz, A.M.; Mattos, J.L.O.; Amorim, P.F.; Mesquita, B.O.; Vilardo, P.J.; Barbosa, M.A. Historical review and redescription of three poorly known species of the catfish genus Trichomycterus from south-eastern Brazil (Siluriformes: Trichomycteridae). J. Nat. Hist. 2020, 53, 2905–2928. [Google Scholar] [CrossRef]
- Bockmann, F.A.; Sazima, I. Trichomycterus maracaya, a new catfish from the upper rio Paraná, southeastern Brazil (Siluriformes: Trichomycteridae), with notes on the T. brasiliensis species-complex. Neotrop. Ichthyol. 2004, 2, 61–74. [Google Scholar] [CrossRef]
- Taylor, W.R.; Van Dyke, G.C. Revised procedures for staining and clearing small fishes and other vertebrates for bone and cartilage study. Cybium 1985, 9, 107–119. [Google Scholar]
- Arratia, G.; Huaquin, L. Morphology of the lateral line system and of the skin of diplomystic and certain primitive loricarioid catfishes and systematic and ecological considerations. Bonn Zool. Monogr. 1995, 36, 1–110. [Google Scholar]
- Villa-Verde, L.; Lazzarotto, H.; Lima, S.Q.M. A new glanapterygine catfish of the genus Listrura (Siluriformes: Trichomycteridae) from southeastern Brazil, corroborated by morphological and molecular data. Neotrop. Ichthyol. 2012, 10, 527–538. [Google Scholar] [CrossRef] [Green Version]
- Unmack, P.J.; Bennin, A.P.; Habi, E.M.; Victoriano, P.F.; Johnson, J.B. Impact of ocean barriers, topography, and glaciation on the phylogeography of the catfish Trichomycterus areolatus (Teleostei: Trichomycteridae) in Chile. Biol. J. Linn. Soc. 2009, 97, 876–892. [Google Scholar] [CrossRef] [Green Version]
- Barros, L.C.; Santos, U.; Cioffi, M.D.B.; Dergam, J.A. Evolutionary divergence among Oligosarcus spp. Ostariophysi, Characidae from the São Francisco and Doce River Basins: Oligosarcus solitarius Menezes, 1987 shows the highest rates of chromosomal evolution in the Neotropical region. Zebrafish 2015, 12, 102–110. [Google Scholar] [CrossRef]
- Ward, R.D.; Zemlak, T.S.; Innes, B.H.; Last, P.R.; Hebert, P.D. DNA barcoding Australia’s fish species. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2005, 360, 1847–1857. [Google Scholar] [CrossRef]
- Hardman, M.; Page, L.M. Phylogenetic relationships among bullhead catfishes of the genus Ameiurus (Siluriformes: Ictaluridae). Copeia 2003, 2003, 20–33. [Google Scholar] [CrossRef]
- Costa, W.J.E.M.; Henschel, E.; Katz, A.M. Multigene phylogeny reveals convergent evolution in small interstitial catfishes from the Amazon and Atlantic forests (Siluriformes: Trichomycteridae). Zool. Scr. 2020, 49, 159–173. [Google Scholar] [CrossRef]
- Cramer, C.A.; Bonatto, S.L.; Reis, R.E. Molecular phylogeny of the Neoplecostominae and Hypoptopomatinae (Siluriformes: Loricariidae) using multiple genes. Mol. Phylogenetics Evol. 2011, 59, 43–52. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Ortí, G.; Zhang, G.; Lu, G. A practical approach to phylogenomics: The phylogeny of ray-finned fish (Actinopterygii) as a case study. BMC Evol. Biol. 2007, 7, 44. [Google Scholar] [CrossRef] [Green Version]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Chenna, R.; Sugawara, H.; Koike, T.; Lopez, R.; Gibson, T.J.; Higgins, D.G.; Thompson, J.D. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003, 31, 3497–3500. [Google Scholar] [CrossRef]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. PartitionFinder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 2017, 34, 772–773. [Google Scholar] [CrossRef] [Green Version]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [Green Version]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior summarisation in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Minh, B.Q.; Nguyen, M.A.T.; von Haeseler, A. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 2013, 30, 1188–1195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Costa, W.J.E.M. Feeding habits of a fish community in a tropical coastal stream, Rio Mato Grosso, Brazil. Stud. Neotrop. Fauna Environ. 1987, 22, 145–153. [Google Scholar] [CrossRef]
- Donin, L.M.; Ferrer, J.; Carvalho, T.P. Taxonomical study of Trichomycterus (Siluriformes: Trichomycteridae) from the Ribeira de Iguape River basin reveals a new species recorded in the early 20th century. J. Fish Biol. 2020, 96, 886–904. [Google Scholar] [CrossRef] [PubMed]
- Lima, S.M.Q.; Berbel-Filho, W.M.; Vilasboa, A.; Lazoski, C.; Volpi, T.A.; Lazzarotto, H.; Russo, C.A.M.; Tatarenkov, A.; Avise, J.C.; Solé-Cava, A.M. Rio de Janeiro and other palaeodrainages evidenced by the genetic structure of an Atlantic Forest catfish. J. Biogeogr. 2021, 48, 1475–1488. [Google Scholar] [CrossRef]
- Wosiacki, W.B.; Oyakawa, O.T. Two new species of the catfish genus Trichomycterus (Siluriformes: Trichomycteridae) from the rio Ribeira de Iguape Basin, Southeastern Brazil. Neotrop. Ichthyol. 2005, 3, 465–472. [Google Scholar] [CrossRef]
- Bizerril, C.R.S.F.; Primo, P.B.S. Peixes de águas interiores do estado do Rio de Janeiro; Secretaria de Estado de Meio Ambiente e Desenvolvimento Sustentável; Fundação de Estudos do Mar: Rio de Janeiro, Brazil, 2001; 417p. [Google Scholar]
- Bizerril, C.R.S.F.; Araujo, L.M.N.; Tosin, P.C. (Eds.) Contribuição ao conhecimento da bacia do Rio Paraíba do Sul: Coletânea de estudos; Agência Nacional de Energia Elétrica: Brasília, Brazil, 1998; 113p. [Google Scholar]
- Polaz, C.N.M.; Bataus, Y.S.L.; Desbiez, A.; Reis, M.L. Plano de ação nacional para a conservação das espécies aquáticas ameaçadas de extinção da bacia do Rio Paraíba do Sul. Série Espécies Ameaçadas 16; Instituto Chico Mendes de Conservação da Biodiversidade: Brasília, Brazil, 2011; 140p. [Google Scholar]
- Costa, W.J.E.M.; Katz, A.M. Integrative taxonomy supports high species diversity of south-eastern Brazilian mountain catfishes of the T. reinhardti group (Siluriformes: Trichomycteridae). Syst. Biodivers. 2021, 19, 601–621. [Google Scholar] [CrossRef]





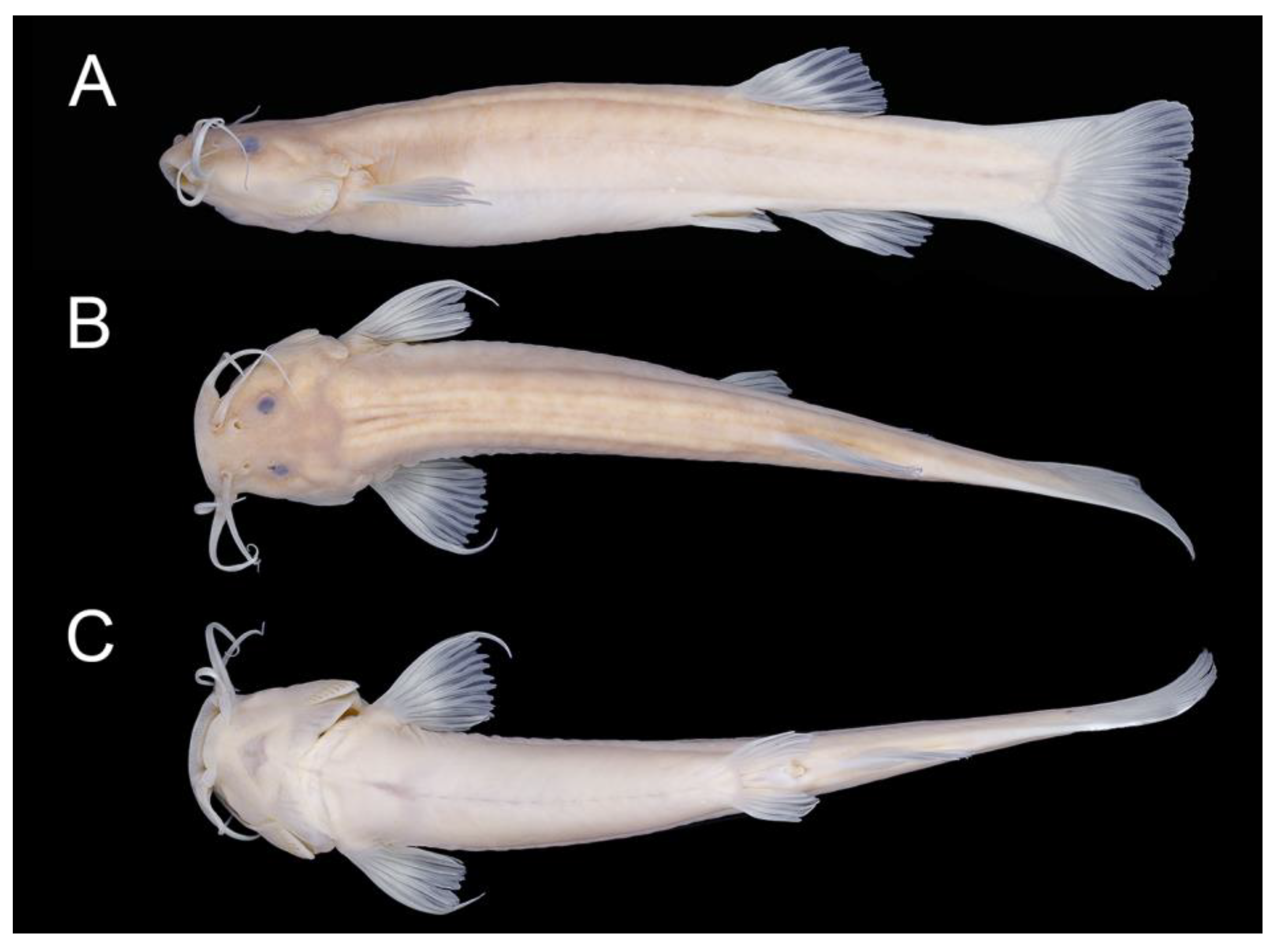



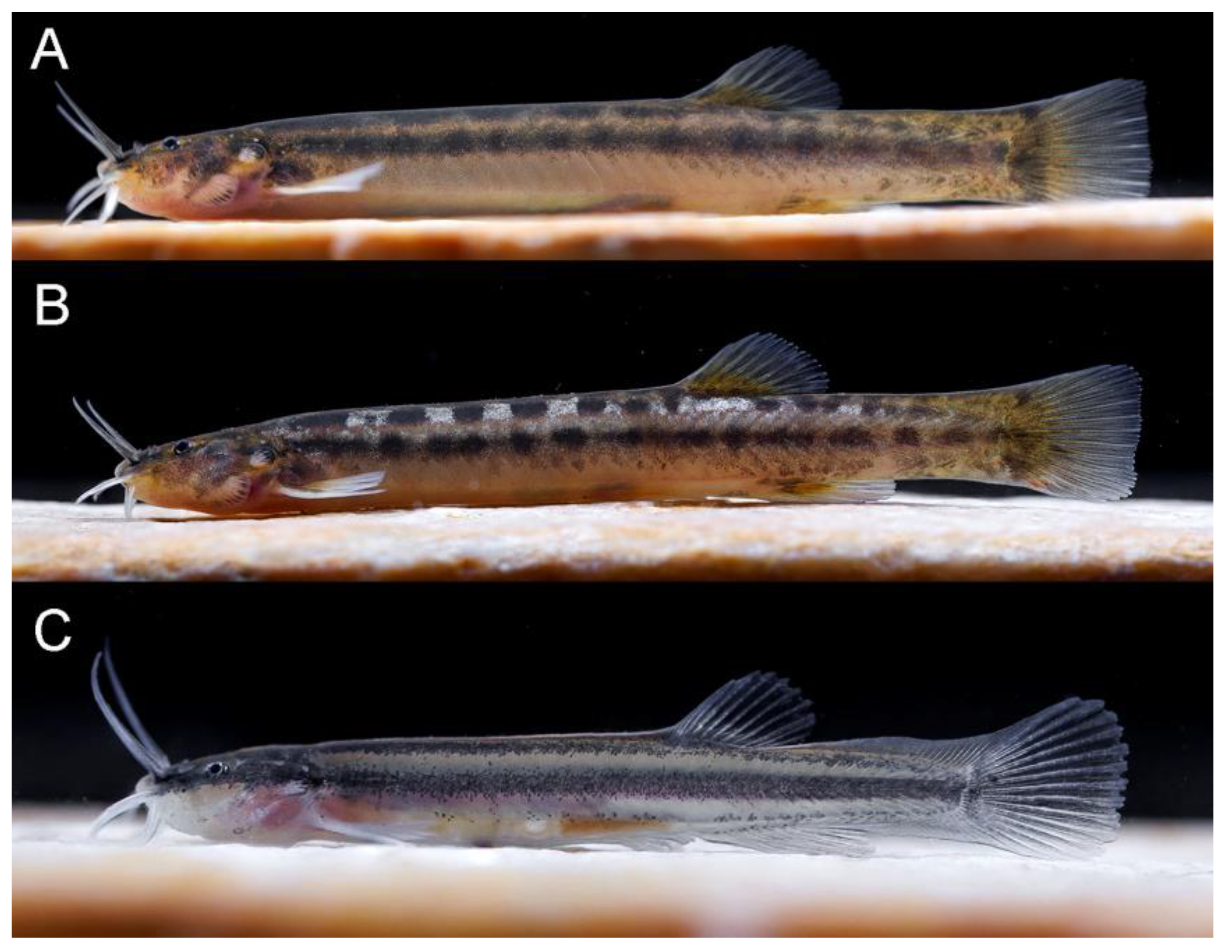





| Holotype | Paratypes (n = 6) | |
|---|---|---|
| Standard length (SL) | 53.1 | 45.8–56.7 |
| Percentage of standard length | ||
| Body depth | 13.3 | 12.0–14.3 |
| Caudal peduncle depth | 11.3 | 9.1–11.6 |
| Body width | 11.2 | 9.5–11.7 |
| Caudal peduncle width | 3.2 | 2.9–5.4 |
| Pre-dorsal length | 61.9 | 60.0–65.4 |
| Pre-pelvic length | 57.9 | 55.9–58.6 |
| Dorsal-fin base length | 12.6 | 10.4–11.8 |
| Anal-fin base length | 10.0 | 8.4–10.6 |
| Caudal-fin length | 15.9 | 15.0–17.0 |
| Pectoral-fin length | 14.4 | 12.7–13.8 |
| Pelvic-fin length | 10.7 | 9.6–11.1 |
| Head length | 20.8 | 20.3–21.3 |
| Percentage of head length | ||
| Head depth | 46.5 | 43.3–47.9 |
| Head width | 87.0 | 83.4–90.2 |
| Snout length | 40.8 | 38.7–40.1 |
| Interorbital width | 29.6 | 28.7–31.0 |
| Preorbital length | 11.0 | 10.1–12.0 |
| Eye diameter | 10.9 | 11.0–13.5 |
| Holotype | Paratypes (n = 10) | |
|---|---|---|
| Standard length (SL) | 53.5 | 44.3–70.8 |
| Percentage of standard length | ||
| Body depth | 14.9 | 14.7–15.7 |
| Caudal peduncle depth | 11.5 | 10.5–12.5 |
| Body width | 13.2 | 11.7–14.5 |
| Caudal peduncle width | 4.1 | 3.3–4.6 |
| Pre-dorsal length | 60.4 | 56.6–62.6 |
| Pre-pelvic length | 54.8 | 51.3–57.1 |
| Dorsal-fin base length | 9.7 | 11.8–13.0 |
| Anal-fin base length | 8.4 | 7.3–9.7 |
| Caudal-fin length | 16.7 | 14.9–18.0 |
| Pectoral-fin length | 14.3 | 13.3–15.8 |
| Pelvic-fin length | 11.4 | 10.4–12.4 |
| Head length | 22.2 | 20.2–23.7 |
| Percentage of head length | ||
| Head depth | 43.6 | 42.7–50.6 |
| Head width | 87.2 | 82.0–90.8 |
| Snout length | 46.1 | 41.4–49.0 |
| Interorbital width | 26.5 | 23.5–27.5 |
| Preorbital length | 14.2 | 12.5–16.8 |
| Eye diameter | 14.9 | 14.5–16.5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Costa, W.J.E.M.; Mattos, J.L.; Vilardo, P.J.; Amorim, P.F.; Katz, A.M. Perils of Underestimating Species Diversity: Revisiting Systematics of Psammocambeva Catfishes (Siluriformes: Trichomycteridae) from the Rio Paraíba do Sul Basin, South-Eastern Brazil. Taxonomy 2022, 2, 491-523. https://doi.org/10.3390/taxonomy2040032
Costa WJEM, Mattos JL, Vilardo PJ, Amorim PF, Katz AM. Perils of Underestimating Species Diversity: Revisiting Systematics of Psammocambeva Catfishes (Siluriformes: Trichomycteridae) from the Rio Paraíba do Sul Basin, South-Eastern Brazil. Taxonomy. 2022; 2(4):491-523. https://doi.org/10.3390/taxonomy2040032
Chicago/Turabian StyleCosta, Wilson J. E. M., José Leonardo Mattos, Paulo J. Vilardo, Pedro F. Amorim, and Axel M. Katz. 2022. "Perils of Underestimating Species Diversity: Revisiting Systematics of Psammocambeva Catfishes (Siluriformes: Trichomycteridae) from the Rio Paraíba do Sul Basin, South-Eastern Brazil" Taxonomy 2, no. 4: 491-523. https://doi.org/10.3390/taxonomy2040032
APA StyleCosta, W. J. E. M., Mattos, J. L., Vilardo, P. J., Amorim, P. F., & Katz, A. M. (2022). Perils of Underestimating Species Diversity: Revisiting Systematics of Psammocambeva Catfishes (Siluriformes: Trichomycteridae) from the Rio Paraíba do Sul Basin, South-Eastern Brazil. Taxonomy, 2(4), 491-523. https://doi.org/10.3390/taxonomy2040032






