Paenibacillus polymyxa A26 and Its Surfactant-Deficient Mutant Degradation of Polycyclic Aromatic Hydrocarbons
Abstract
1. Introduction
2. Material and Methods
2.1. Bacterial Growth, Culture Conditions and Chemicals
2.2. NAP Degradation in Culture Medium
2.3. Analysis of NAP Degradation Efficiency
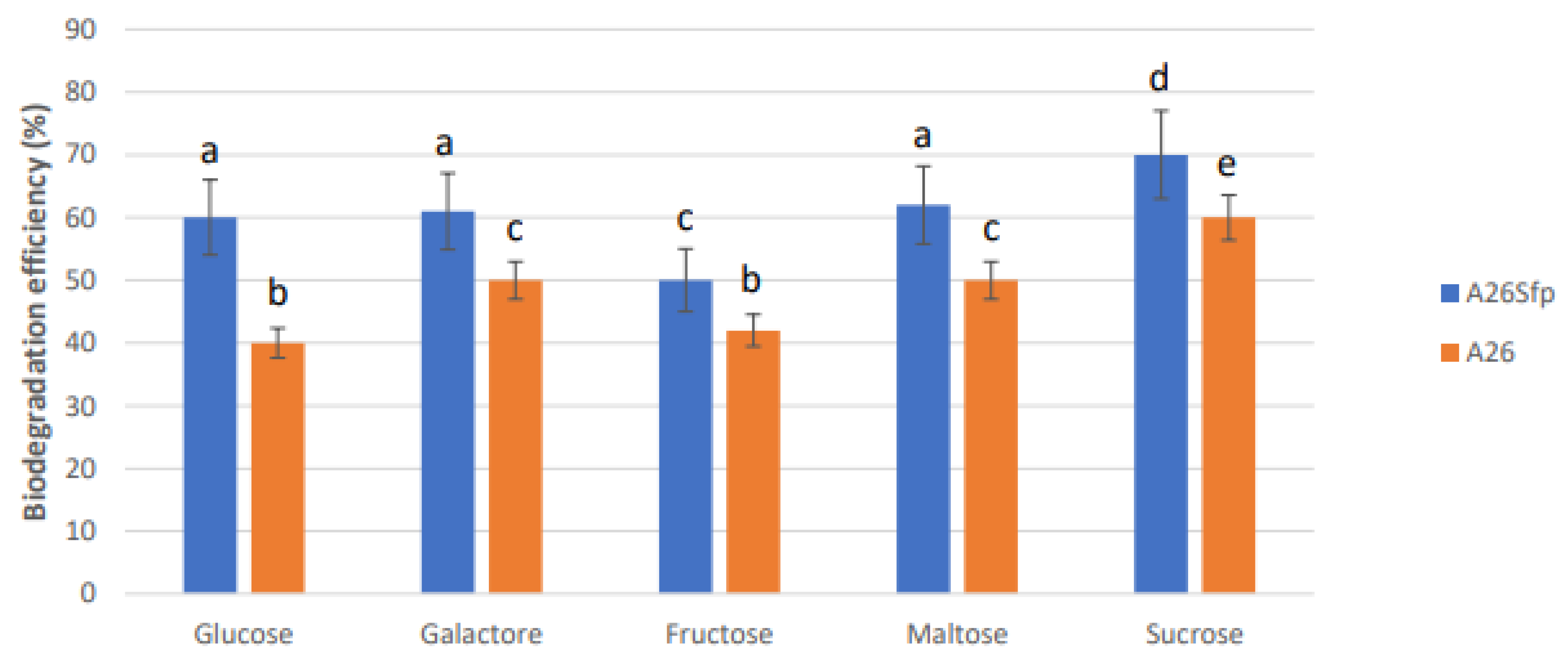
2.4. Effect of Different Carbon Sources on NAP Degradation
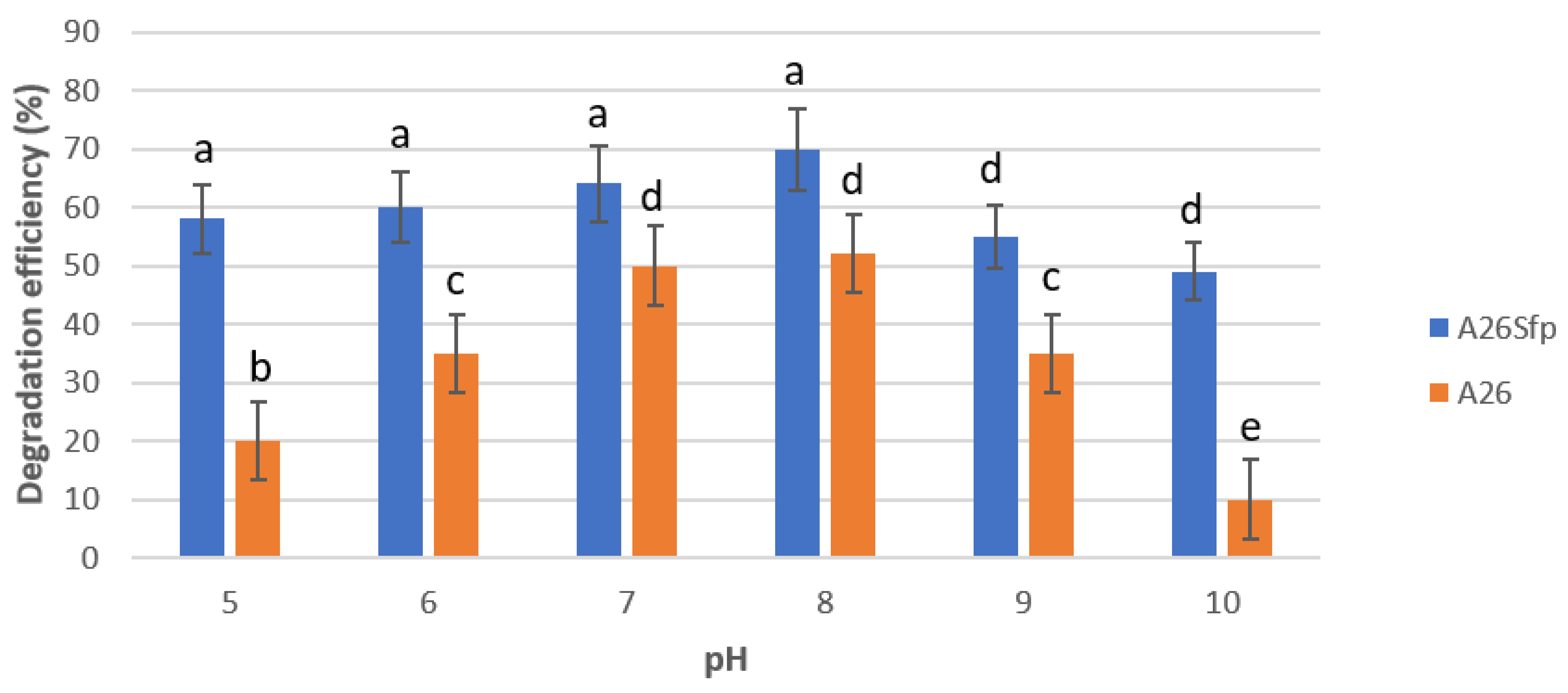
2.5. Effect of pH on NAP Degradation
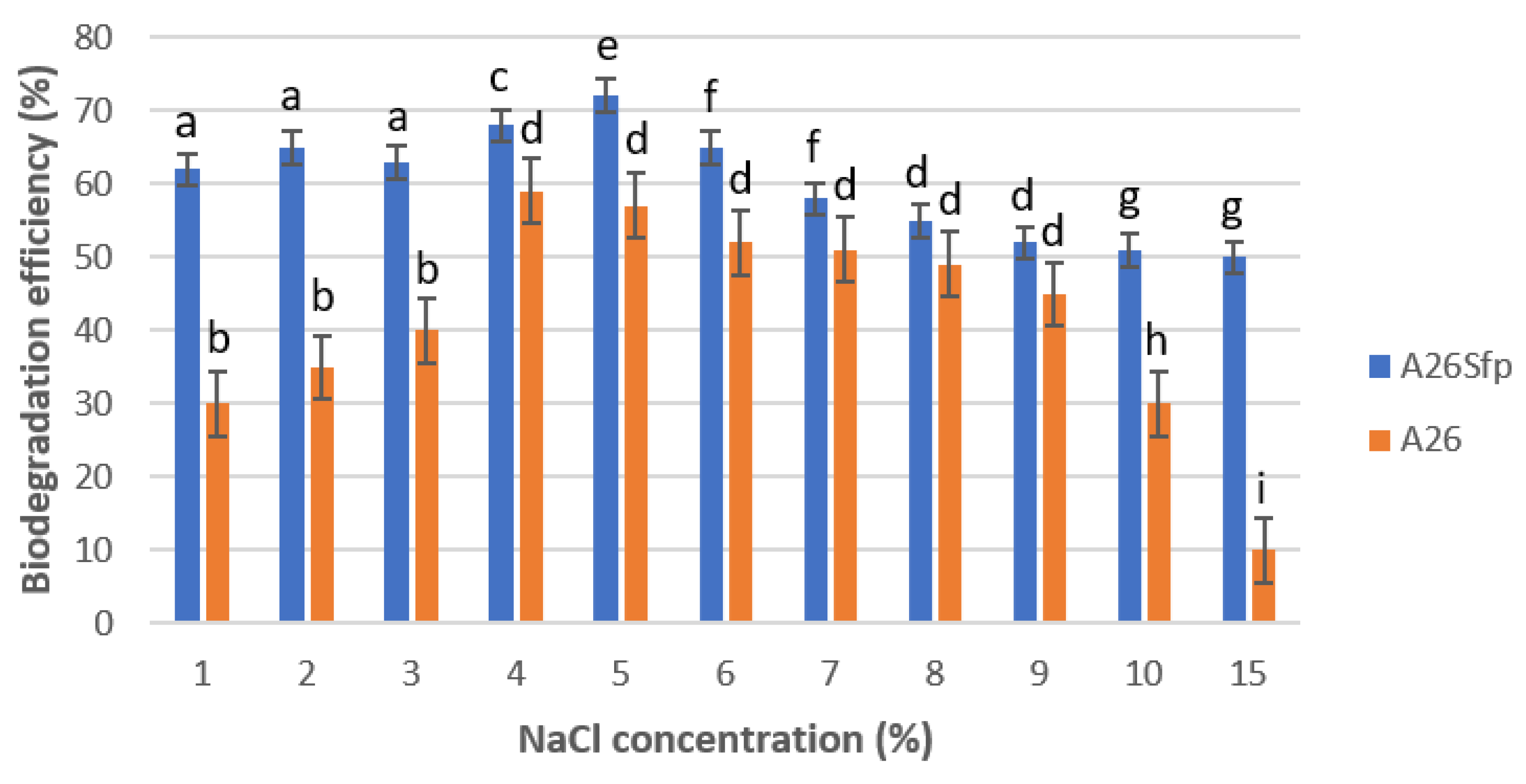
2.6. Effect of NaCl on NAP Degradation
2.7. EPS Extraction
2.8. Glucan Production Assay
2.9. Emulsification Assay
2.10. Drop-Collapse Test
2.11. Oil Spreading Assay
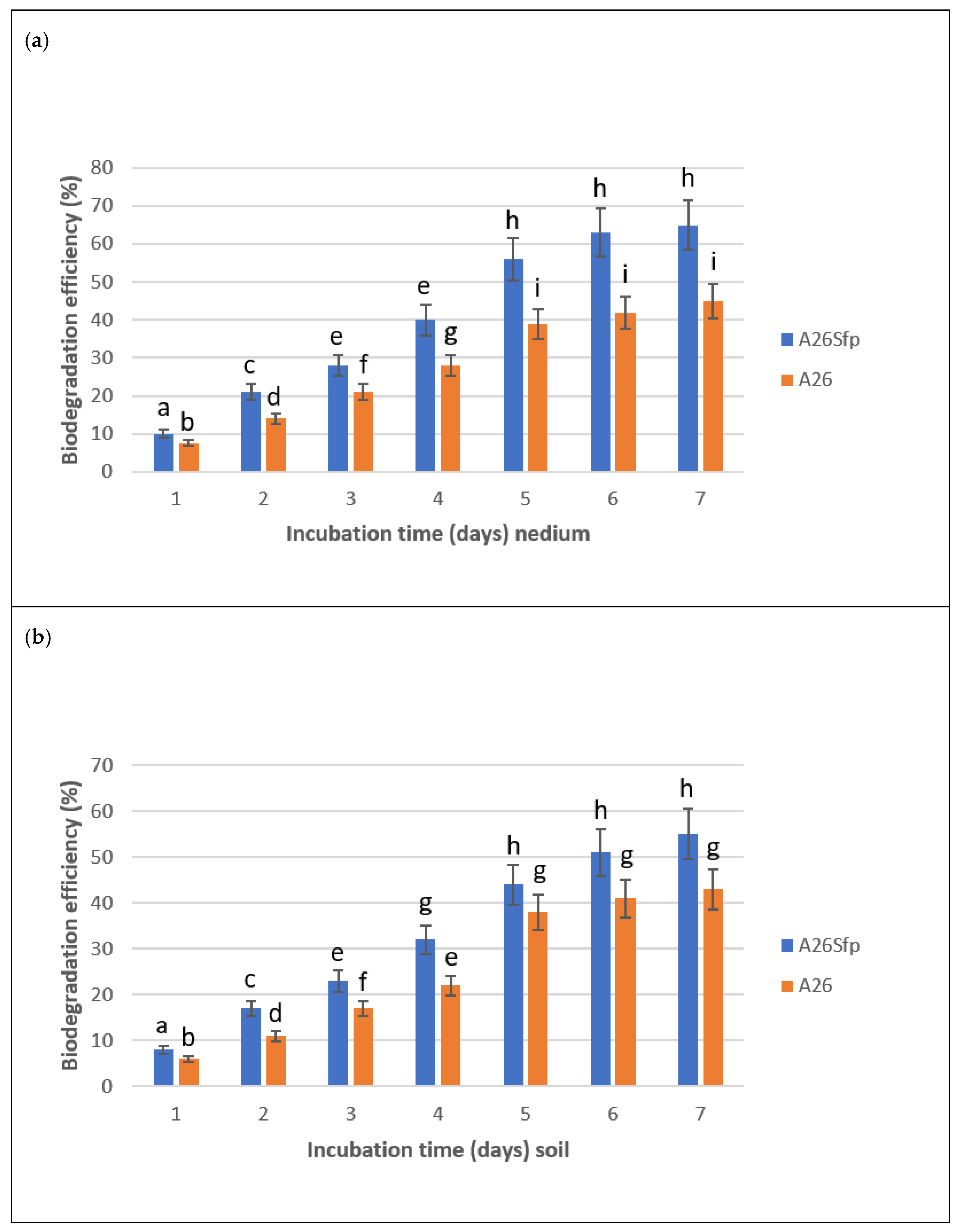
2.12. NAP Degradation in Soil
2.13. Data Confirmation and Validation
3. Results
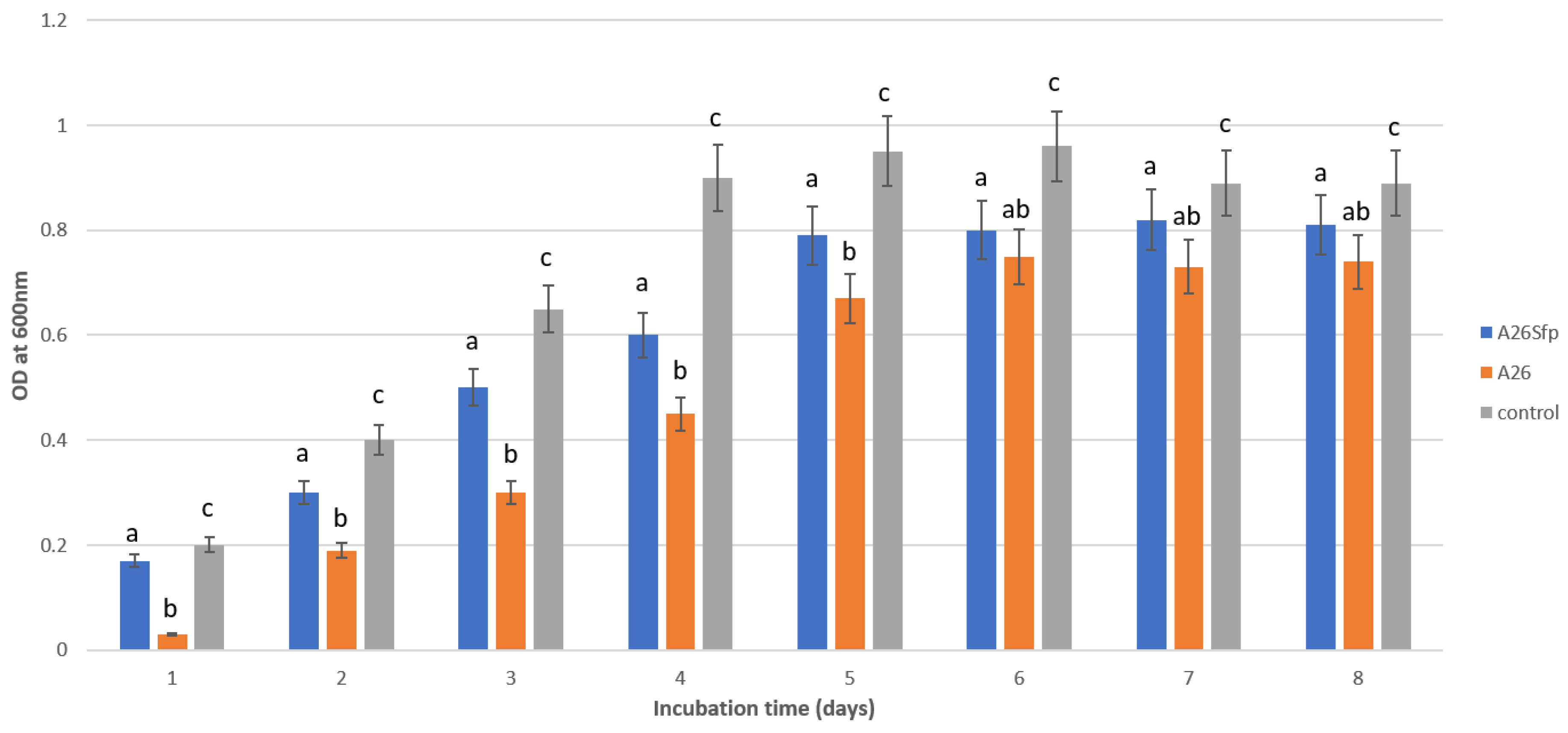
3.1. A26 and A26Sfp NAP Degradation Ability Is Dependent on the Medium pH, Salinity and C Source
3.2. A26Sfp Ability to Emulsify Kerosene and Hexadecane Is Improved Compared to the Wild Type
3.3. Exopolysaccharide Glucan Production of A26Sfp Is Improved Compared to That of the Wild Type
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Polli, J.R.; Rushing, B.R.; Lish, L.; Lewis, L.; Selim, M.I.; Pan, X. Quantitative analysis of PAH compounds in DWH crude oil and their effects on Caenorhabditis elegans germ cell apoptosis, associated with CYP450s upregulation. Sci. Total Environ. 2020, 745, 140639. [Google Scholar] [CrossRef] [PubMed]
- Fang, C.; Bo, J.; Zheng, R.; Hong, F.; Kuang, W.; Jiang, Y.; Chen, J.; Zhang, Y.; Segner, H. Biomonitoring of aromatic hydrocarbons in clam Meretrix meretrix from an emerging urbanization area, and implications for human health. Ecotoxicol. Environ. Saf. 2020, 192, 110271. [Google Scholar] [CrossRef] [PubMed]
- Saliu, F.; Meucci, E.; Allevi, C.; Savini, A.; Imiete, I.E.; Della Pergola, R. Evaluation of chitosan aggregates as pickering emulsifier for the remediation of marine sediments. Chemosphere 2021, 273, 129733. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Zhang, H.; Zhang, M.; Shen, X.; Zhang, X.; Wu, F.; Hu, J.; Wang, B.; Wang, X. Removal of PAHs at high concentrations in a soil washing solution containing TX-100 via simultaneous sorption and biodegradation processes by immobilized degrading bacteria in PVA-SA hydrogel beads. J. Hazard. Mater. 2021, 410, 124533. [Google Scholar] [CrossRef]
- Pichler, N.; de Souza, F.M.; dos Santos, V.F.; Martins, C.C. Polycyclic aromatic hydrocarbons (PAHs) in sediments of the amazon coast: Evidence for localized sources in contrast to massive regional biomass burning. Environ. Pollut. 2021, 268, 115958. [Google Scholar] [CrossRef]
- Huang, X.-D.; El-Alawi, Y.; Penrose, D.M.; Glick, B.R.; Greenberg, B.M. Responses of three grass species to creosote during phytoremediation. Environ. Pollut. 2004, 130, 453–463. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Li, X.; Liu, J.; Cheng, Y.; Zou, J.; Zhai, F.; Sun, Z.; Han, L. Soil microbial community succession and interactions during combined plant/white-rot fungus remediation of polycyclic aromatic hydrocarbons. Sci. Total. Environ. 2021, 752, 142224. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Lopez, C.; Posada-Baquero, R.; Garcia, J.L.; Castilla-Alcantara, J.C.; Cantos, M.; Ortega-Calvo, J.J. Root-mediated bacterial accessibility and cometabolism of pyrene in soil. Sci. Total Environ. 2021, 760, 143408. [Google Scholar] [CrossRef] [PubMed]
- de Almeida Couto, C.R.; Alvarez, V.M.; Marques, J.M.; de Azevedo Jurelevicius, D.; Seldin, L. Exploiting the aerobic endospore-forming bacterial diversity in saline and hypersaline environments for biosurfactant production. BMC Microbiol. 2015, 15, 240. [Google Scholar] [CrossRef]
- Alvarez, V.M.; Jurelevicius, D.; Marques, J.M.; de Souza, P.M.; de Araújo, L.V.; Barros, T.G.; de Souza, R.O.M.A.; Freire, D.M.G.; Seldin, L. Bacillus amyloliquefaciens TSBSO 3.8, a biosurfactant-producing strain with biotechnological potential for microbial enhanced oil recovery. Colloids Surfaces B Biointerfaces 2015, 136, 14–21. [Google Scholar] [CrossRef] [PubMed]
- Nevo, E.; Fu, Y.B.; Pavlicek, T.; Khalifa, S.; Tavasi, M.; Beiles, A. Evolution of wild cereals during 28 years of global warming in Israel. Proc. Natl. Acad. Sci. USA 2012, 109, 3412–3415. [Google Scholar] [CrossRef]
- Timmusk, S.; Kim, S.-B.; Nevo, E.; El-Daim, I.A.; Ek, B.; Bergquist, J.; Behers, L. Sfp-type PPTase inactivation promotes bacterial biofilm formation and ability to enhance wheat drought tolerance. Front. Microbiol. 2015, 6, 387. [Google Scholar] [CrossRef] [PubMed]
- Timmusk, S.; Nevo, E.; Ayele, F.; Noe, S.; Niinemets, Y. Fighting Fusarium Pathogens in the Era of Climate Change: A Conceptual Approach. Pathogens 2020, 9, 419. [Google Scholar] [CrossRef] [PubMed]
- Timmusk, S.; Copolovici, D.; Copolovici, L.; Teder, T.; Nevo, E.; Behers, L. Paenibacillus polymyxa biofilm polysaccharides antagonise Fusarium graminearum. Sci. Rep. 2019, 9, 662. [Google Scholar] [CrossRef] [PubMed]
- Timmusk, S.; Seisenbaeva, G.A.; Behers, L. Titania (TiO2) nanoparticles enhance the performance of growth-promoting rhizobacteria. Sci. Rep. 2018, 8, 617. [Google Scholar] [CrossRef] [PubMed]
- Timmusk, S.; Conrad, J.; Niinemets, Y.; Nevo, E.; Behers, L.; Bergqvist, J.; Noe, S. Managing Plant Stress in the Era of Climate Change: Realising the Global Sustainable Development Goals. Available online: http://www.global-engage.com/agricultural-biotechnology/managing-plant-stress-in-the-era-of-climate-change-realising-global-sustainable-development-goals/ (accessed on 22 September 2021).
- Timmusk, S.; Wagner, E.G. The plant-growth-promoting rhizobacterium Paenibacillus polymyxa induces changes in Arabidopsis thaliana gene expression: A possible connection between biotic and abiotic stress responses. Mol. Plant Microbe Interact. 1999, 12, 951–959. [Google Scholar] [CrossRef] [PubMed]
- Timmusk, S.; Grantcharova, N.; Wagner, E.G. Paenibacillus polymyxa invades plant roots and forms biofilms. Appl. Environ. Microbiol. 2005, 71, 7292–7300. [Google Scholar] [CrossRef]
- Timmusk, S. Biofilm forming Paenibacillus polymyxa antagonises oomycete plant pathogens. J. Biotechnol. 2008, 136, 265–266. [Google Scholar] [CrossRef]
- Timmusk, S.; van West, P.; Gow, N.A.; Huffstutler, R.P. Paenibacillus polymyxa antagonizes oomycete plant pathogens Phytophthora palmivora and Pythium aphanidermatum. J. Appl. Microbiol. 2009, 106, 1473–1481. [Google Scholar] [CrossRef]
- Timmusk, S.; Behers, L. Rhizobacterial application for sustainable water management on the areas of limited water resources. Irrig. Drain. Syst. Eng. 2012, 12. [Google Scholar] [CrossRef]
- Timmusk, S.; Timmusk, K.; Behers, L. Rhizobacterial plant drought stress tolerance enhancement. J. Food Secur. 2013, 1, 10–16. [Google Scholar]
- Timmusk, S.; Behers, L.; Muthoni, J.; Muraya, A.; Aronsson, A.-C. Perspectives and Challenges of Microbial Application for Crop Improvement. Front. Plant Sci. 2017, 8, 49. [Google Scholar] [CrossRef] [PubMed]
- Fetsiukh, A.; Conrad, J.; Bergquist, J.; Timmusk, S. Silica particles trigger the exopolysaccharide production of harsh environment isolates of growth-promoting rhizobacteria and increase their ability to enhance wheat biomass in drought-stressed soils. Int. J. Mol. Sci. 2021, 22, 6201. [Google Scholar] [CrossRef]
- Timmusk, S.; Paalme, V.; Pavlicek, T.; Bergquist, J.; Vangala, A.; Danilas, T.; Nevo, E. Bacterial distribution in the rhizosphere of wild barley under contrasting microclimates. PLoS ONE 2011, 6, e17968. [Google Scholar] [CrossRef]
- Copp, J.N.; Neilan, B.A. The Phosphopantetheinyl Transferase Superfamily: Phylogenetic Analysis and Functional Implications in Cyanobacteria. Appl. Environ. Microbiol. 2006, 72, 2298–2305. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kim, S.-B.; Timmusk, S. A simplified method for Paenibacillus polymyxa gene knockout and insertional screening. PLoS ONE 2013, 8, e68092. [Google Scholar] [CrossRef]
- Abbas, M.; Adil, M.; Ehtisham-Ul-Haque, S.; Munir, B.; Yameen, M.; Ghaffar, A.; Shar, G.A.; Asif Tahir, M.; Iqbal, M. Vibrio fischeri bioluminescence inhibition assay for ecotoxicity assessment: A review. Sci. Total Environ. 2018, 626, 1295–1309. [Google Scholar] [CrossRef]
- Nascimento, F.X.; Glick, B.R.; Rossi, M.J. Isolation and characterization of novel soil- and plant-associated bacteria with multiple phytohormone-degrading activities using a targeted methodology. Access Microbiol. 2019, 1, e000053. [Google Scholar] [CrossRef] [PubMed]
- Rutering, M.; Schmid, J.; Ruhmann, B.; Schilling, M.; Sieber, V. Controlled production of polysaccharides-exploiting nutrient supply for levan and heteropolysaccharide formation in Paenibacillus sp. Carbohydr. Polym. 2016, 148, 326–334. [Google Scholar] [CrossRef] [PubMed]
- McIntosh, M.; Stone, B.A.; Stanisich, V.A. Curdlan and other bacterial (1-->3)-beta-D-glucans. Appl. Microbiol. Biotechnol. 2005, 68, 163. [Google Scholar] [CrossRef]
- Cooper, D.G.; Goldenberg, B.G. Surface-active agents from two bacillus species. Appl. Environ. Microbiol. 1987, 53, 224–229. [Google Scholar] [CrossRef] [PubMed]
- Jain, D.; Collins-Thompson, D.; Lee, H. A drop-collapsing test for screening surfactant producing microorganisms. J. Microbiol. Meth. 1991, 13, 271–279. [Google Scholar] [CrossRef]
- Abd El-Daim, I.; Haggblom, P.; Karlsson, M.; Stenstrom, E.; Timmusk, S. Paenibacillus polymyxa A26 Sfp-type PPTase inactivation limits bacterial antagonism against Fusarium graminearum but not of F. culmorum in kernel assay. Front. Plant Sci. 2015, 6, 368. [Google Scholar] [CrossRef]
- Boonchan, S.; Britz, M.L.; Stanley, G.A. Degradation and mineralization of high-molecular-weight polycyclic aromatic hydrocarbons by defined fungal-bacterial cocultures. Appl. Environ. Microbiol. 2000, 66, 1007–1019. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Ji, M.; Zhai, H.; Zhao, J. Organic matter composition, BaP biodegradation and microbial communities at sites near and far from the bioanode in a soil microbial fuel cell. Sci. Total Environ. 2021, 772, 144919. [Google Scholar] [CrossRef]
- Huang, X.D.; El-Alawi, Y.; Penrose, D.M.; Glick, B.R.; Greenberg, B.M. A multi-process phytoremediation system for removal of polycyclic aromatic hydrocarbons from contaminated soils. Environ. Pollut. 2004, 130, 465–476. [Google Scholar] [CrossRef] [PubMed]
- Lominchar, M.A.; Santos, A.; de Miguel, E.; Romero, A. Remediation of aged diesel contaminated soil by alkaline activated persulfate. Sci. Total Environ. 2018, 622–623, 41–48. [Google Scholar] [CrossRef]
- Wang, F.; Wu, C.; Li, Q. Treatment of refractory organics in strongly alkaline dinitrodiazophenol wastewater with microwave irradiation-activated persulfate. Chemosphere 2020, 254, 126773. [Google Scholar] [CrossRef]
- Kang, H.J.; Jung, Y.; Kwon, J.H. Changes in ecotoxicity of naphthalene and alkylated naphthalenes during photodegradation in water. Chemosphere 2019, 222, 656–664. [Google Scholar] [CrossRef]
- Girvan, M.S.; Campbell, C.D.; Killham, K.; Prosser, J.I.; Glover, L.A. Bacterial diversity promotes community stability and functional resilience after perturbation. Environ. Microbiol. 2005, 7, 301–313. [Google Scholar] [CrossRef]
- Mahmood, S.; Paton, G.I.; Prosser, J.I. Cultivation-independent in situ molecular analysis of bacteria involved in degradation of pentachlorophenol in soil. Environ. Microbiol. 2005, 7, 1349–1360. [Google Scholar] [CrossRef] [PubMed]
- de-Bashan, L.E.; Bashan, Y. Immobilized microalgae for removing pollutants: Review of practical aspects. Bioresour. Technol. 2010, 101, 1611–1627. [Google Scholar] [CrossRef] [PubMed]
- Jami, M.; Lai, Q.; Ghanbari, M.; Moghadam, M.S.; Kneifel, W.; Domig, K.J. Celeribacter persicus sp. nov., a polycyclic-aromatic-hydrocarbon-degrading bacterium isolated from mangrove soil. Int. J. Syst. Evol. Microbiol. 2016, 66, 1875–1880. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Van Hamme, J.D.; Ward, O.P. Surfactants in microbiology and biotechnology: Part 2. Application aspects. Biotechnol. Adv. 2007, 25, 99–121. [Google Scholar] [CrossRef] [PubMed]
- Milliasseau, D.; Jeftic, J.; Pessel, F.; Plusquellec, D.; Benvegnu, T. Transformation of Pectins into Non-Ionic or Anionic Surfactants Using a One-Pot and Cascade Mode Process. Molecules 2021, 26, 1956. [Google Scholar] [CrossRef]
- Talvenmaki, H.; Saartama, N.; Haukka, A.; Lepikko, K.; Pajunen, V.; Punkari, M.; Yan, G.; Sinkkonen, A.; Piepponen, T.; Silvennoinen, H.; et al. In situ bioremediation of Fenton’s reaction-treated oil spill site, with a soil inoculum, slow release additives, and methyl-beta-cyclodextrin. Environ. Sci. Pollut. Res. Int. 2021, 28, 20273–20289. [Google Scholar] [CrossRef] [PubMed]
| Name | Origin | Publications |
|---|---|---|
| Paenibacillus polymyxa A26 | Wild barley rhizosphere, the Evolution Canyon, Haifa, Israel | [25] |
| Paenibacillus polymyxa A26Sfp | Wild barley rhizosphere, the Evolution Canyon, Haifa, Israel | [12,27] |
| Strains | Emulsion Index % Hexadecane (day) | Emulsion Index % Kerosene (day) | DC 1 | OS 2 | Glucan Titre (mg/L) | EPSTitre (mg/L) | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 3 | 7 | 1 | 3 | 7 | |||||
| A26Sfp | 60 ± 4.2 a | 63 ± 5.2 a | 60 ± 5.2 a | 63 ± 5.2 a | 59 ± 5.2 a | 60 ± 4.1 a | + | + | 1.2 ± 0.1 | 15 ± 0.2 |
| A26 | 42 ± 5.2 b | 44 ± 5.2 b | 45 ± 3.2 b | 43 ± 5.2 b | 45 ± 2.2 b | 43 ± 3.2 b | + | + | 0.62 ± 0.1 | 10 ± 0.2 |
| EPS A26Sfp | 41 ± 2.2 b | 45 ± 2.2 b | 44 ± 2.2 b | 43 ± 2.2 b | 44 ± 3.2 b | 45 ± 2.2 b | + | + | ND | ND |
| Glucan A26Sfp | 20 ± 2.0 c | 20 ± 2.2 c | 21 ± 2.2 c | 21 ± 2.2 c | 21 ± 2.1 c | 23 ± 2.2 c | + | + | ND | ND |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Timmusk, S.; Teder, T.; Behers, L. Paenibacillus polymyxa A26 and Its Surfactant-Deficient Mutant Degradation of Polycyclic Aromatic Hydrocarbons. Stresses 2021, 1, 266-276. https://doi.org/10.3390/stresses1040019
Timmusk S, Teder T, Behers L. Paenibacillus polymyxa A26 and Its Surfactant-Deficient Mutant Degradation of Polycyclic Aromatic Hydrocarbons. Stresses. 2021; 1(4):266-276. https://doi.org/10.3390/stresses1040019
Chicago/Turabian StyleTimmusk, Salme, Tiiu Teder, and Lawrence Behers. 2021. "Paenibacillus polymyxa A26 and Its Surfactant-Deficient Mutant Degradation of Polycyclic Aromatic Hydrocarbons" Stresses 1, no. 4: 266-276. https://doi.org/10.3390/stresses1040019
APA StyleTimmusk, S., Teder, T., & Behers, L. (2021). Paenibacillus polymyxa A26 and Its Surfactant-Deficient Mutant Degradation of Polycyclic Aromatic Hydrocarbons. Stresses, 1(4), 266-276. https://doi.org/10.3390/stresses1040019






