Genome-Wide Identification, Bioinformatic Characterization, and Expression Profiling of Starch Synthase (SS) Genes in Foxtail Millet under Drought Condition
Abstract
1. Introduction
2. Results
2.1. Identification and Phylogenetic Analysis of SS in Millet
2.2. Gene Structure and Motif Analysis
2.3. Chromosomal Location, Gene Duplication of SiSSs and Synteny Analysis of SiSSs in Four Species
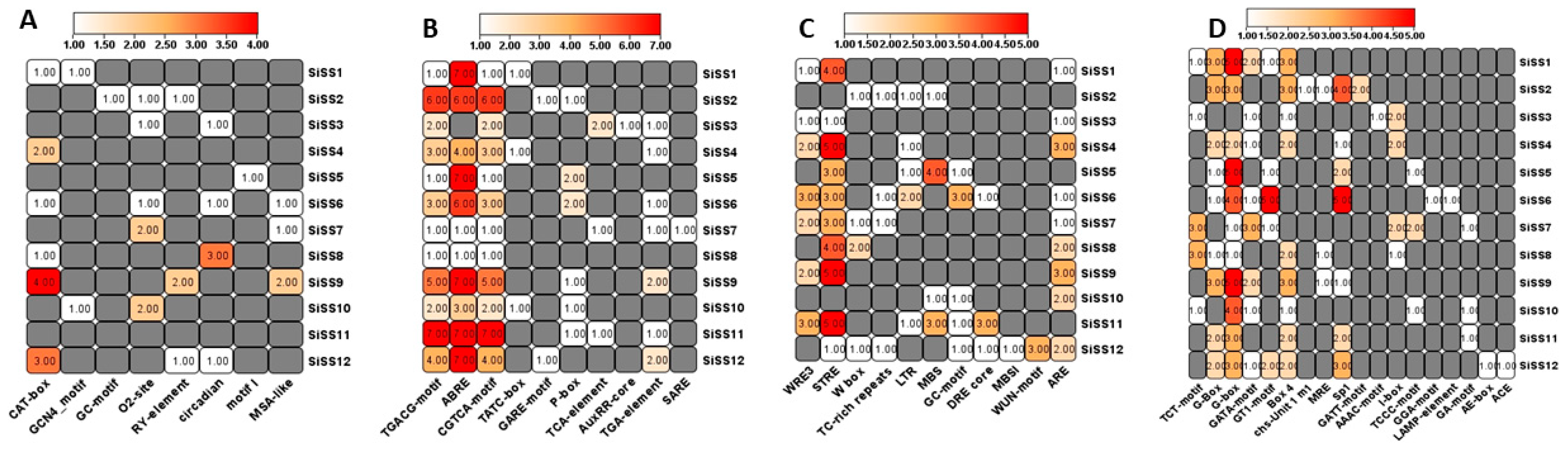
2.4. Promoter Analysis of SiSS Genes
2.5. Three-Dimensional Protein Structure of SiSSs
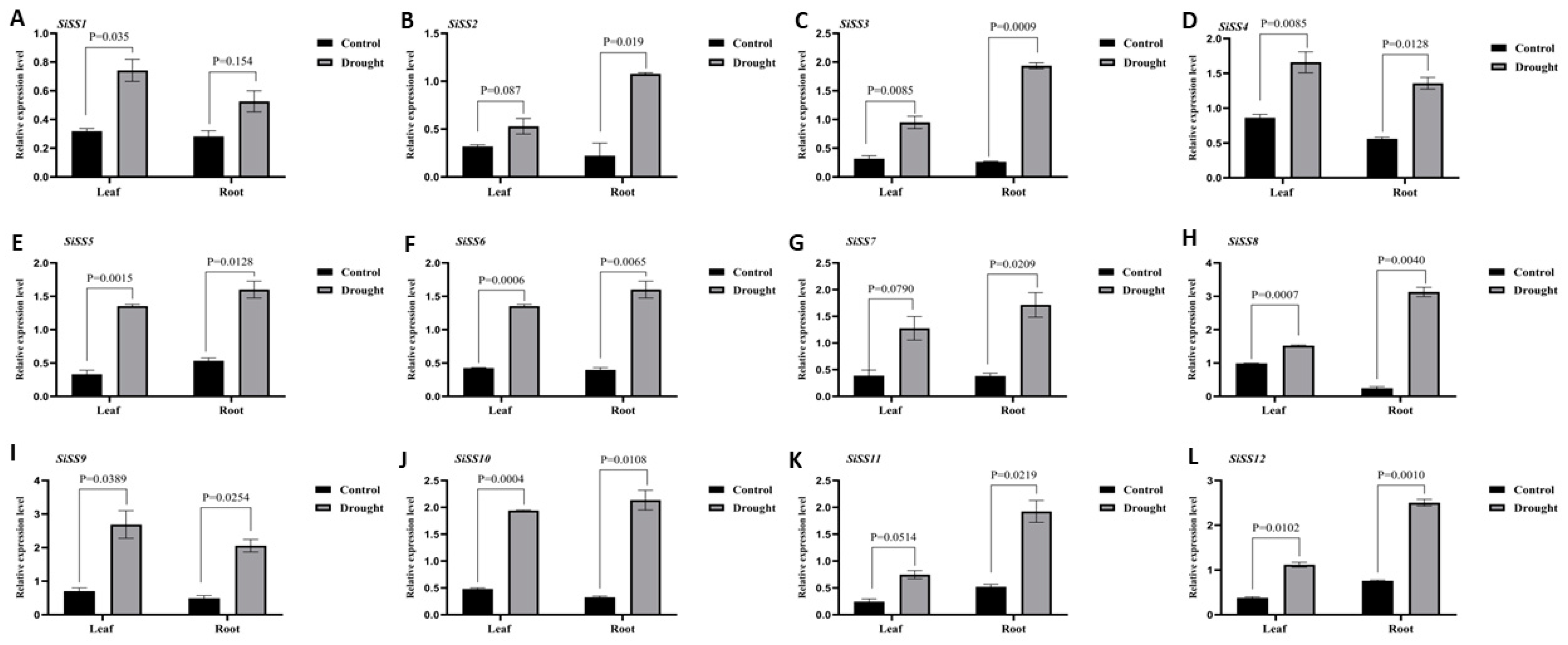
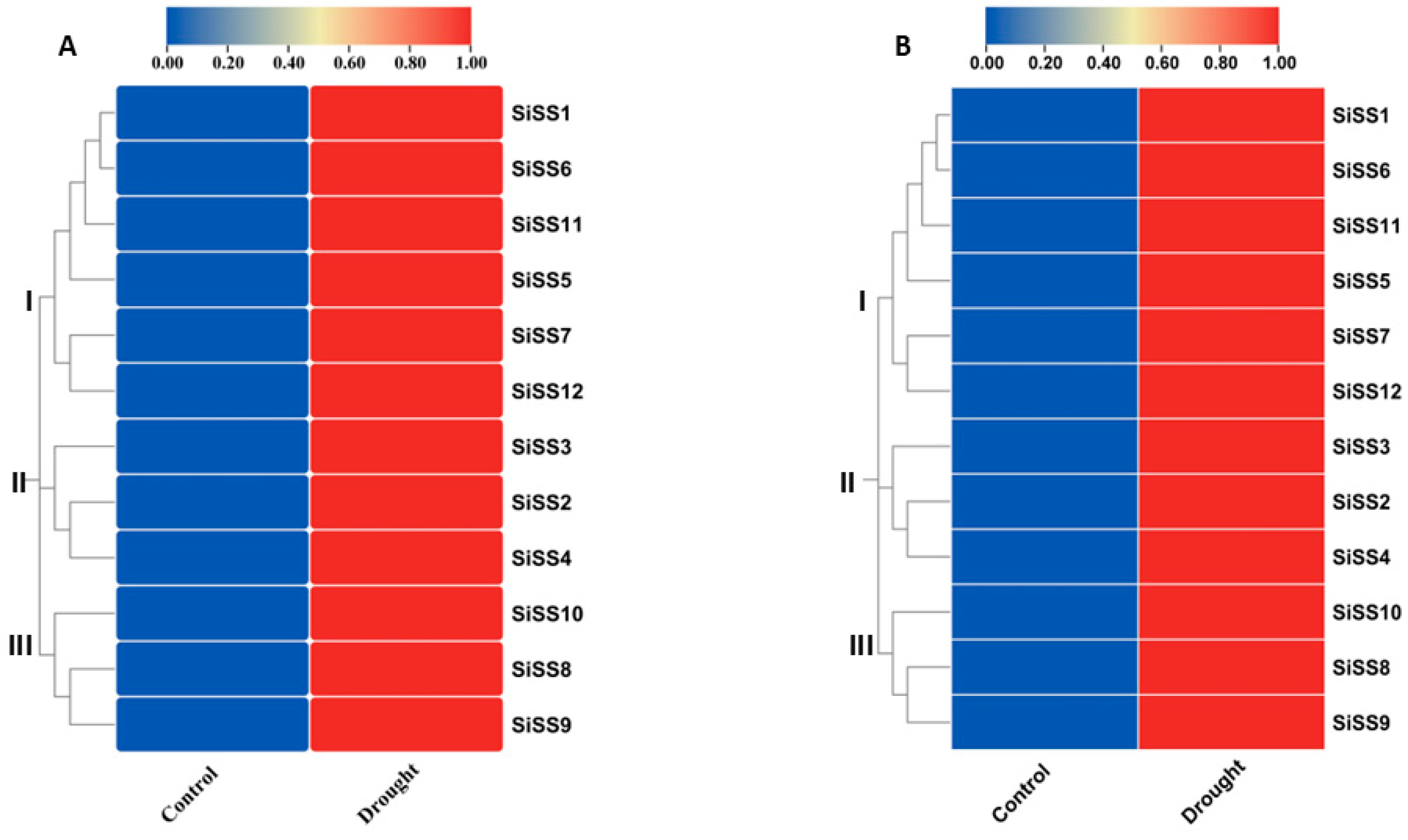
2.6. Expression Analysis of Putative SiSSs and Cluster Analysis under Drought Conditions
3. Discussion
4. Materials and Methods
4.1. Identification of the Starch Synthase (SS) Genes in Millet
4.2. Multiple Sequence Alignment, Gene Structure, Motifs, Gene Duplication, and Phylogenetic Analysis
4.3. Identification of Cis-Regulatory Elements and Prediction of Three-Dimensional Modeling
4.4. Plant Material and Growth Conditions
4.5. RNA Isolation and Gene Expression Analysis
4.6. Assay of Starch Content and Starch Synthase Activity
4.7. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Thalmann, M.; Santelia, D. Starch as a determinant of plant fitness under abiotic stress. New Phytol. 2017, 214, 943–951. [Google Scholar] [CrossRef]
- Georgelis, N.; Shaw, J.R.; Hannah, L.C. Phylogenetic analysis of ADP-glucose pyrophosphorylase subunits reveals a role of subunit interfaces in the allosteric properties of the enzyme. Plant Physiol. 2009, 151, 67–77. [Google Scholar] [CrossRef][Green Version]
- Qu, J.; Xu, S.; Zhang, Z.; Chen, G.; Zhong, Y.; Liu, L.; Zhang, R.; Xue, J.; Guo, D. Evolutionary, structural and expression analysis of core genes involved in starch synthesis. Sci. Rep. 2018, 8, 12736. [Google Scholar] [CrossRef] [PubMed]
- Cummings, J.H.; Englyst, H.N. Gastrointestinal effects of food carbohydrate. Am. J. Clin. Nutr. 1995, 61, 938–945. [Google Scholar] [CrossRef] [PubMed]
- Keenan, M.J.; Zhou, J.; Hegsted, M.; Pelkman, C.; Durham, H.A.; Coulon, D.B.; Martin, R.J. Role of resistant starch in improving gut health, adiposity, and insulin resistance. Adv. Nutr. 2015, 6, 198–205. [Google Scholar] [CrossRef] [PubMed]
- Annor, G.A.; Tyl, C.; Marcone, M.; Ragaee, S.; Marti, A. Why do millets have slower starch and protein digestibility than other cereals? Trends Food Sci. Technol. 2017, 66, 73–83. [Google Scholar] [CrossRef]
- Dhaka, A.; Muthamilarasan, M.; Prasad, M. A comprehensive study on core enzymes involved in starch metabolism in the model nutricereal, foxtail millet (Setaria italica L.). J. Cereal Sci. 2021, 97, 103153. [Google Scholar] [CrossRef]
- Muthamilarasan, M.; Dhaka, A.; Yadav, R.; Prasad, M. Exploration of millet models for developing nutrient rich graminaceous crops. Plant Sci. 2016, 242, 89–97. [Google Scholar] [CrossRef]
- Saleh, A.S.; Zhang, Q.; Chen, J.; Shen, Q. Millet grains: Nutritional quality, processing, and potential health benefits. Compr. Rev. Food Sci. Food Saf. 2013, 12, 281–295. [Google Scholar] [CrossRef]
- Chandra, A.K.; Chandora, R.; Sood, S.; Malhotra, N. Global production, demand, and supply. In Millets and Pseudo Cereals; Elsevier: Amsterdam, The Netherlands, 2021; pp. 7–18. [Google Scholar]
- Muthamilarasan, M.; Singh, N.K.; Prasad, M. Chapter One—Multi-omics approaches for strategic improvement of stress tolerance in underutilized crop species: A climate change perspective. In Advances in Genetics; Kumar, D., Ed.; Academic Press: Cambridge, MA, USA, 2019; pp. 1–38. [Google Scholar]
- Derbyshire, M.C.; Batley, J.; Edwards, D. Use of multiple ‘omics techniques to accelerate the breeding of abiotic stress tolerant crops. Curr. Plant Biol. 2022, 32, 100262. [Google Scholar] [CrossRef]
- Hirose, T.; Terao, T. A comprehensive expression analysis of the starch synthase gene family in rice (Oryza sativa L.). Planta 2004, 220, 9–16. [Google Scholar] [CrossRef]
- Yang, Z.; Wang, Y.; Xu, S.; Xu, C.; Yan, C. Molecular evolution and functional divergence of soluble starch synthase genes in cassava (Manihot esculenta crantz). Evol. Bioinform. Online 2013, 9, 239–249. [Google Scholar] [CrossRef]
- Zhang, H.; Jang, S.G.; Lar, S.M.; Lee, A.R.; Cao, F.Y.; Seo, J.; Kwon, S.W. Genome-Wide Identification and Genetic Variations of the Starch Synthase Gene Family in Rice. Plants 2021, 10, 1154. [Google Scholar] [CrossRef] [PubMed]
- Brosius, J. Gene duplication and other evolutionary strategies: From the RNA world to the future. J. Struct. Funct. Genom. 2003, 3, 1–17. [Google Scholar] [CrossRef]
- Chen, H.-J.; Chen, J.-Y.; Wang, S.-J. Molecular regulation of starch accumulation in rice seedling leaves in response to salt stress. Acta Physiol. Plant. 2008, 30, 135–142. [Google Scholar] [CrossRef]
- Wei, L.; Wang, L.; Yang, Y.; Wang, P.; Guo, T.; Kang, G. Abscisic acid increases leaf starch content of polyethylene glycol-treated wheat seedlings by temporally increasing transcripts of genes encoding starch synthesis enzymes. Acta Physiol. Plant. 2015, 37, 206. [Google Scholar] [CrossRef]
- Thalmann, M.; Pazmino, D.; Seung, D.; Horrer, D.; Nigro, A.; Meier, T.; Kölling, K.; Pfeifhofer, H.W.; Zeeman, S.C.; Santelia, D. Regulation of leaf starch degradation by abscisic acid is important for osmotic stress tolerance in plants. Plant Cell 2016, 28, 1860–1878. [Google Scholar] [CrossRef] [PubMed]
- Thitisaksakul, M.; Jiménez, R.C.; Arias, M.C.; Beckles, D.M. Effects of environmental factors on cereal starch biosynthesis and composition. J. Cereal Sci. 2012, 56, 67–80. [Google Scholar] [CrossRef]
- Cho, Y.-G.; Kang, K.-K. Functional Analysis of Starch Metabolism in Plants. Plants 2020, 9, 1152. [Google Scholar] [CrossRef]
- Wang, S.-J.; Yeh, K.-W.; Tsai, C.-Y. Regulation of starch granule-bound starch synthase I gene expression by circadian clock and sucrose in the source tissue of sweet potato. Plant Sci. 2001, 161, 635–644. [Google Scholar] [CrossRef]
- Prathap, V.; Ali, K.; Singh, A.; Vishwakarma, C.; Krishnan, V.; Chinnusamy, V.; Tyagi, A. Starch accumulation in rice grains subjected to drought during grain filling stage. Plant Physiol. Biochem. 2019, 142, 440–451. [Google Scholar]
- Lu, H.; Hu, Y.; Wang, C.; Liu, W.; Ma, G.; Han, Q.; Ma, D. Effects of High Temperature and Drought Stress on the Expression of Gene Encoding Enzymes and the Activity of Key Enzymes Involved in Starch Biosynthesis in Wheat Grains. Front. Plant Sci. 2019, 10, 1414. [Google Scholar] [CrossRef]
- Van Harsselaar, J.K.; Lorenz, J.; Senning, M.; Sonnewald, U.; Sonnewald, S. Genome-wide analysis of starch metabolism genes in potato (Solanum tuberosum L.). BMC Genom. 2017, 18, 37. [Google Scholar] [CrossRef]
- Huang, T.; Luo, X.; Fan, Z.; Yang, Y.; Wan, W. Genome-wide identification and analysis of the sucrose synthase gene family in cassava (Manihot esculenta Crantz). Gene 2021, 769, 145191. [Google Scholar] [CrossRef]
- Dong, S.; Beckles, D.M. Dynamic changes in the starch-sugar interconversion within plant source and sink tissues promote a better abiotic stress response. J. Plant Physiol. 2019, 234, 80–93. [Google Scholar] [CrossRef]
- Vrinten, P.L.; Nakamura, T. Wheat granule-bound starch synthase I and II are encoded by separate genes that are expressed in different tissues. Plant Physiol. 2000, 122, 255–264. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Li, H.; Liu, J.; Liu, A.; Cao, X.; Liu, C.; Cheng, D.; Zhao, Z.; Song, J. Genome-Wide Identification and Expression Profiling of Starch-Biosynthetic Genes in Common Wheat. Russ. J. Genet. 2020, 56, 1445–1456. [Google Scholar] [CrossRef]
- Schwarte, S.; Brust, H.; Steup, M.; Tiedemann, R. Intraspecific sequence variation and differential expression in starch synthase genes of Arabidopsis thaliana. BMC Res. Notes 2013, 6, 84. [Google Scholar] [CrossRef] [PubMed]
- Patron, N.J.; Smith, A.M.; Fahy, B.F.; Hylton, C.M.; Naldrett, M.J.; Rossnagel, B.G.; Denyer, K. The altered pattern of amylose accumulation in the endosperm of low-amylose barley cultivars is attributable to a single mutant allele of granule-bound starch synthase I with a deletion in the 5′-non-coding region. Plant Physiol. 2002, 130, 190–198. [Google Scholar] [CrossRef]
- Prathap, V.; Aruna, T. Correlation between expression and activity of ADP glucose pyrophosphorylase and starch synthase and their role in starch accumulation during grain filling under drought stress in rice. Plant Physiol. Biochem. 2020, 157, 239–243. [Google Scholar]
- Liu, H.; Yu, G.; Wei, B.; Wang, Y.; Zhang, J.; Hu, Y.; Liu, Y.; Yu, G.; Zhang, H.; Huang, Y. Identification and phylogenetic analysis of a novel starch synthase in maize. Front. Plant Sci. 2015, 6, 1013. [Google Scholar] [CrossRef] [PubMed]
- Senoura, T.; Isono, N.; Yoshikawa, M.; Asao, A.; Hamada, S.; Watanabe, K.; Ito, H.; Matsui, H. Characterization of starch synthase I and II expressed in early developing seeds of kidney bean (Phaseolus vulgaris L.). Biosci. Biotechnol. Biochem. 2004, 68, 1949–1960. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Dai, M.; Yang, X.; Chen, Q.; Bai, Z. Comprehensive genomic identification of cotton starch synthase genes reveals that GhSS9 regulates drought tolerance. Front. Plant Sci. 2023, 14, 1163041. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Zhao, Q.; Chen, L.; Yao, X.; Zhang, W.; Zhang, B.; Xie, F. Effect of drought stress on sugar metabolism in leaves and roots of soybean seedlings. Plant Physiol. Biochem. 2020, 146, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Peng, L.; Gong, J.; Wang, J.; Wu, C.; Sui, X.; Tian, Y.; Hu, M.; Li, C.; He, X.; et al. Effects of water stress on starch synthesis and accumulation of two rice cultivars at different growth stages. Front. Plant Sci. 2023, 14, 1133524. [Google Scholar] [CrossRef] [PubMed]
- Yi, B.; Zhou, Y.-f.; Gao, M.-y.; Zhang, Z.; Han, Y.; Yang, G.-d.; Xu, W.; Huang, R.-d. Effect of drought stress during flowering stage on starch accumulation and starch synthesis enzymes in sorghum grains. J. Integr. Agric. 2014, 13, 2399–2406. [Google Scholar] [CrossRef]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J.; et al. Pfam: The protein families database. Nucleic Acids Res. 2014, 42, D222–D230. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein identification and analysis tools on the ExPASy server. In The Proteomics Protocols Handbook; Springer: Totowa, NJ, USA, 2005. [Google Scholar]
- Yu, C.S.; Chen, Y.C.; Lu, C.H.; Hwang, J.K. Prediction of protein subcellular localization. Proteins Struct. Funct. Bioinform. 2006, 64, 643–651. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. In Nucleic Acids Symposium Series; Oxford University Press: Oxford, UK, 1999. [Google Scholar]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Bailey, T.L.; Williams, N.; Misleh, C.; Li, W.W. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006, 34 (Suppl. 2), W369–W373. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Koch, M.A.; Haubold, B.; Mitchell-Olds, T. Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis, and related genera (Brassicaceae). Mol. Biol. Evol. 2000, 17, 1483–1498. [Google Scholar] [CrossRef]
- Li, H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 2011, 27, 2987–2993. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef]
- Amoah, J.N.; Adu-Gyamfi, M.O.; Kwarteng, A.O. Effect of drought acclimation on antioxidant system and polyphenolic content of Foxtail Millet (Setaria italica L.). Physiol. Mol. Biol. Plants 2023, 29, 1577–1589. [Google Scholar] [CrossRef]
- Zadoks, J.C.; Chang, T.T.; Konzak, C.F. A decimal code for the growth stages of cereals. Weed Res. 1974, 14, 415–421. [Google Scholar] [CrossRef]
- Amoah, J.N.; Seo, Y.W. Effect of progressive drought stress on physio-biochemical responses and gene expression patterns in wheat. 3 Biotech 2021, 11, 440. [Google Scholar] [CrossRef]






| Gene Locus | Gene Name | Chrom ID | CDS (bp) | Peptide(aa) | MW(kDa) | pI | Instability Index | GRAVY | Aliphatic Index |
|---|---|---|---|---|---|---|---|---|---|
| Seita.1G318200.1 | SiSS1 | 1 | 2121 | 707 | 76.58 | 6.08 | 41.92 | −0.264 | 80.62 |
| Seita.1G359000.1 | SiSS2 | 1 | 2115 | 705 | 79.17 | 6.74 | 51.62 | −0.317 | 91.69 |
| Seita.3G167300.1 | SiSS3 | 3 | 2733 | 911 | 102.92 | 5.91 | 41.41 | −0.399 | 85.05 |
| Seita.4G007700.1 | SiSS4 | 4 | 1803 | 601 | 67.63 | 5.94 | 45.76 | −0.134 | 99.58 |
| Seita.4G065500.1 | SiSS5 | 4 | 1929 | 643 | 71.08 | 5.53 | 40.46 | −0.29 | 79.61 |
| Seita.4G099700.1 | SiSS6 | 4 | 2322 | 744 | 84.06 | 6.2 | 44.58 | −0.337 | 77.02 |
| Seita.5G098100.1 | SiSS7 | 5 | 1314 | 438 | 49.20 | 6.85 | 38.44 | −0.096 | 88.58 |
| Seita.6G036700.1 | SiSS8 | 6 | 4629 | 1543 | 174.78 | 5.21 | 47.18 | −0.431 | 81.67 |
| Seita.6G036800.1 | SiSS9 | 6 | 4245 | 1415 | 159.95 | 5.02 | 47.63 | −0.531 | 77.69 |
| Seita.7G243900.1 | SiSS10 | 7 | 3462 | 1154 | 130.65 | 5.39 | 49.59 | −0.582 | 49.59 |
| Seita.9G243600.1 | SiSS11 | 9 | 2568 | 856 | 95.23 | 5.9 | 44.48 | −0.319 | 44.48 |
| Seita.9G456900.1 | SiSS12 | 9 | 1242 | 414 | 46.42 | 8.96 | 44.82 | −0.25 | 83.39 |
| Seq_1 | Seq_2 | Ka | Ks | Ka/Ks | Time (Mya) | Duplication | Purification |
|---|---|---|---|---|---|---|---|
| SiSS1 | SiSS6 | 0.258 | 0.680 | 0.379 | 19.632 | Segmental | Yes |
| SiSS7 | SiSS12 | 0.135 | 2.095 | 0.065 | 10.313 | Segmental | Yes |
| SiSS2 | SiSS4 | 0.030 | 0.045 | 0.681 | 2.313 | Segmental | Yes |
| SiSS8 | SiSS9 | 0.147 | 0.320 | 0.459 | 11.174 | Tandem | Yes |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Amoah, J.N.; Adu-Gyamfi, M.O.; Kwarteng, A.O. Genome-Wide Identification, Bioinformatic Characterization, and Expression Profiling of Starch Synthase (SS) Genes in Foxtail Millet under Drought Condition. Stresses 2024, 4, 518-533. https://doi.org/10.3390/stresses4030033
Amoah JN, Adu-Gyamfi MO, Kwarteng AO. Genome-Wide Identification, Bioinformatic Characterization, and Expression Profiling of Starch Synthase (SS) Genes in Foxtail Millet under Drought Condition. Stresses. 2024; 4(3):518-533. https://doi.org/10.3390/stresses4030033
Chicago/Turabian StyleAmoah, Joseph N., Monica Ode Adu-Gyamfi, and Albert Owusu Kwarteng. 2024. "Genome-Wide Identification, Bioinformatic Characterization, and Expression Profiling of Starch Synthase (SS) Genes in Foxtail Millet under Drought Condition" Stresses 4, no. 3: 518-533. https://doi.org/10.3390/stresses4030033
APA StyleAmoah, J. N., Adu-Gyamfi, M. O., & Kwarteng, A. O. (2024). Genome-Wide Identification, Bioinformatic Characterization, and Expression Profiling of Starch Synthase (SS) Genes in Foxtail Millet under Drought Condition. Stresses, 4(3), 518-533. https://doi.org/10.3390/stresses4030033






