A Maize Mutant Impaired in SL Biosynthesis (zmccd8) Shows a Lower Growth, an Altered Response to Nitrogen Starvation, and a Potential Secondary Effect on Drought Tolerance
Abstract
1. Introduction
2. Results
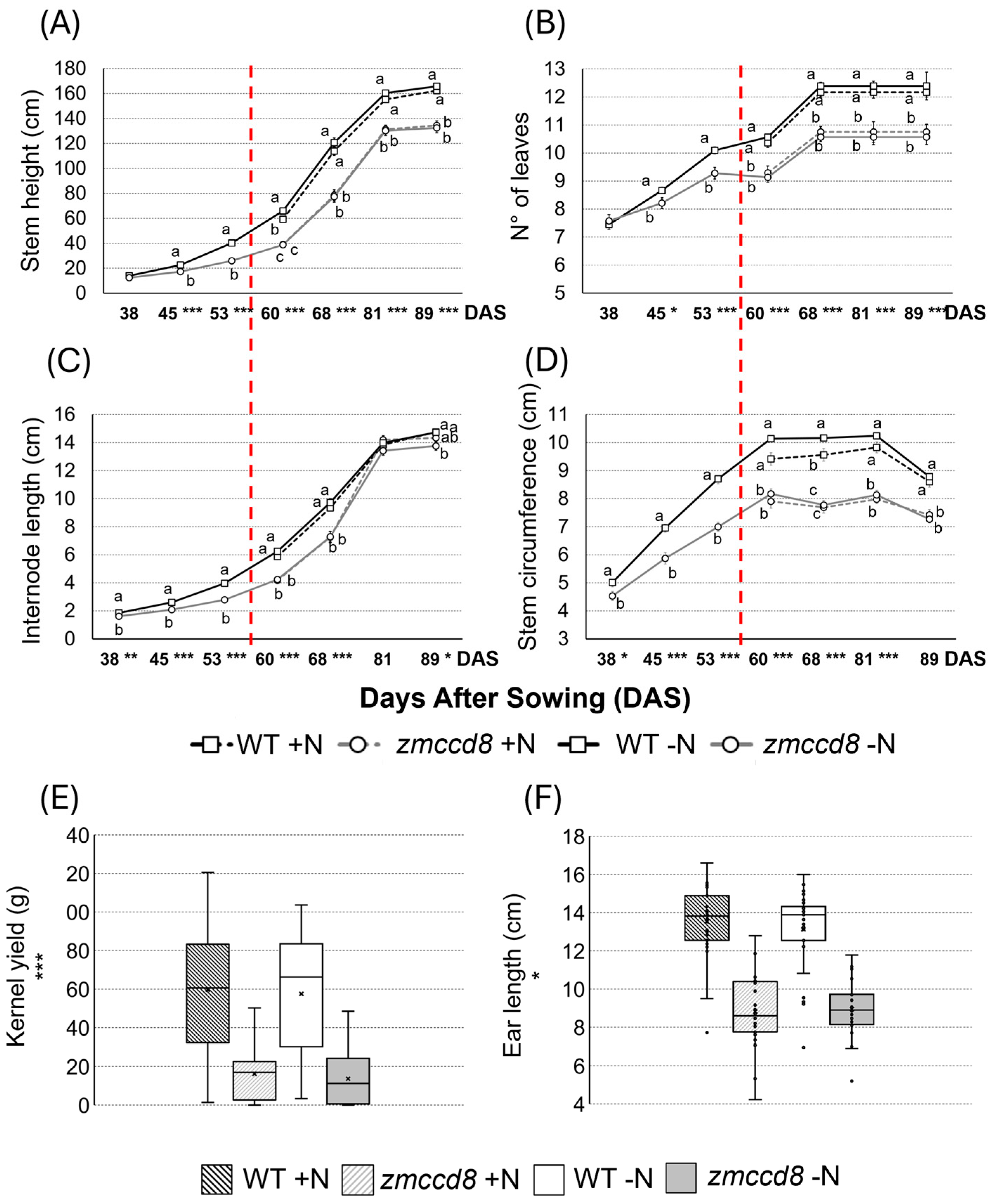
2.1. Phenotypic Analysis of Growth Traits
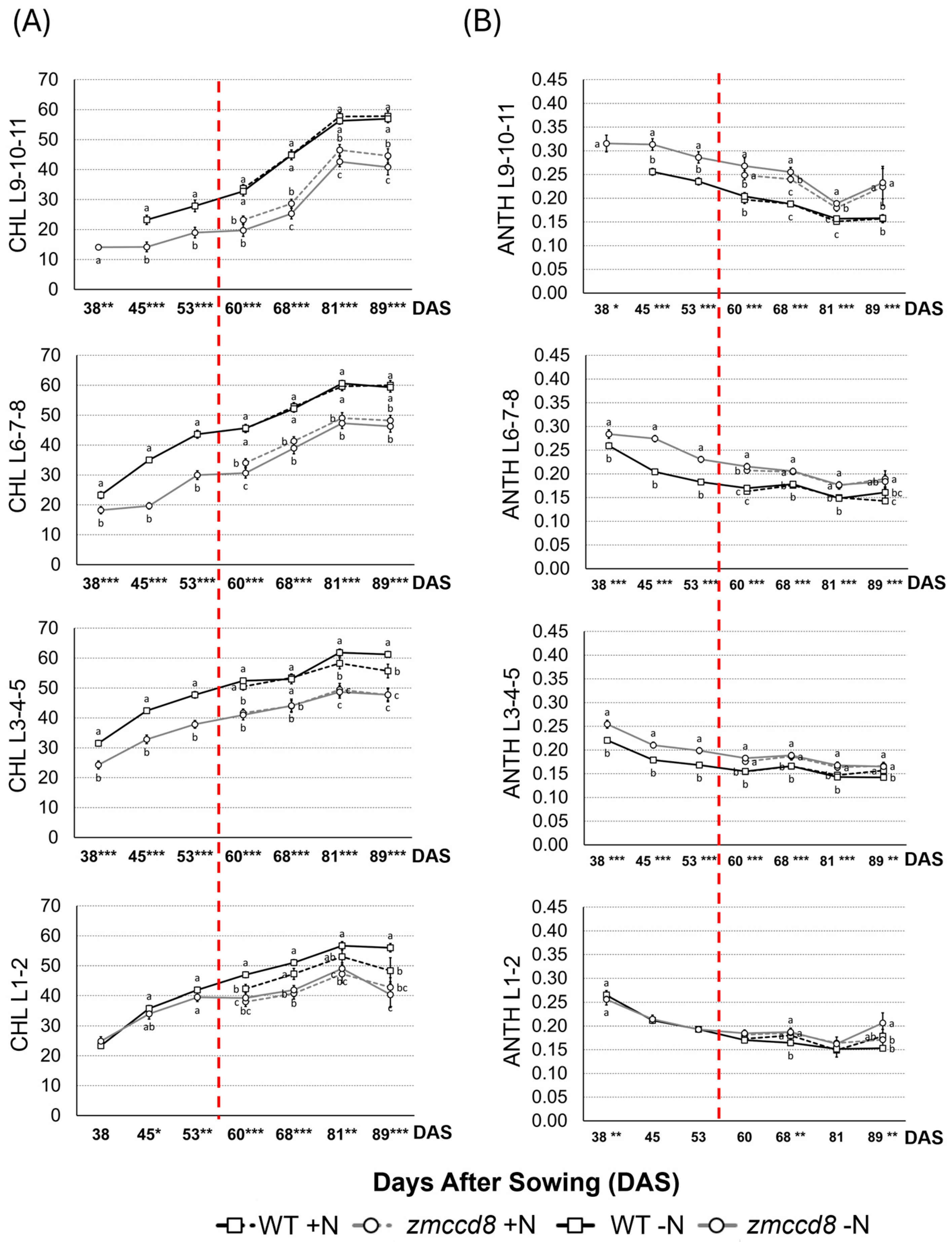
2.2. Assessment of Chlorophyl and Anthocyanin Contents in Leaves
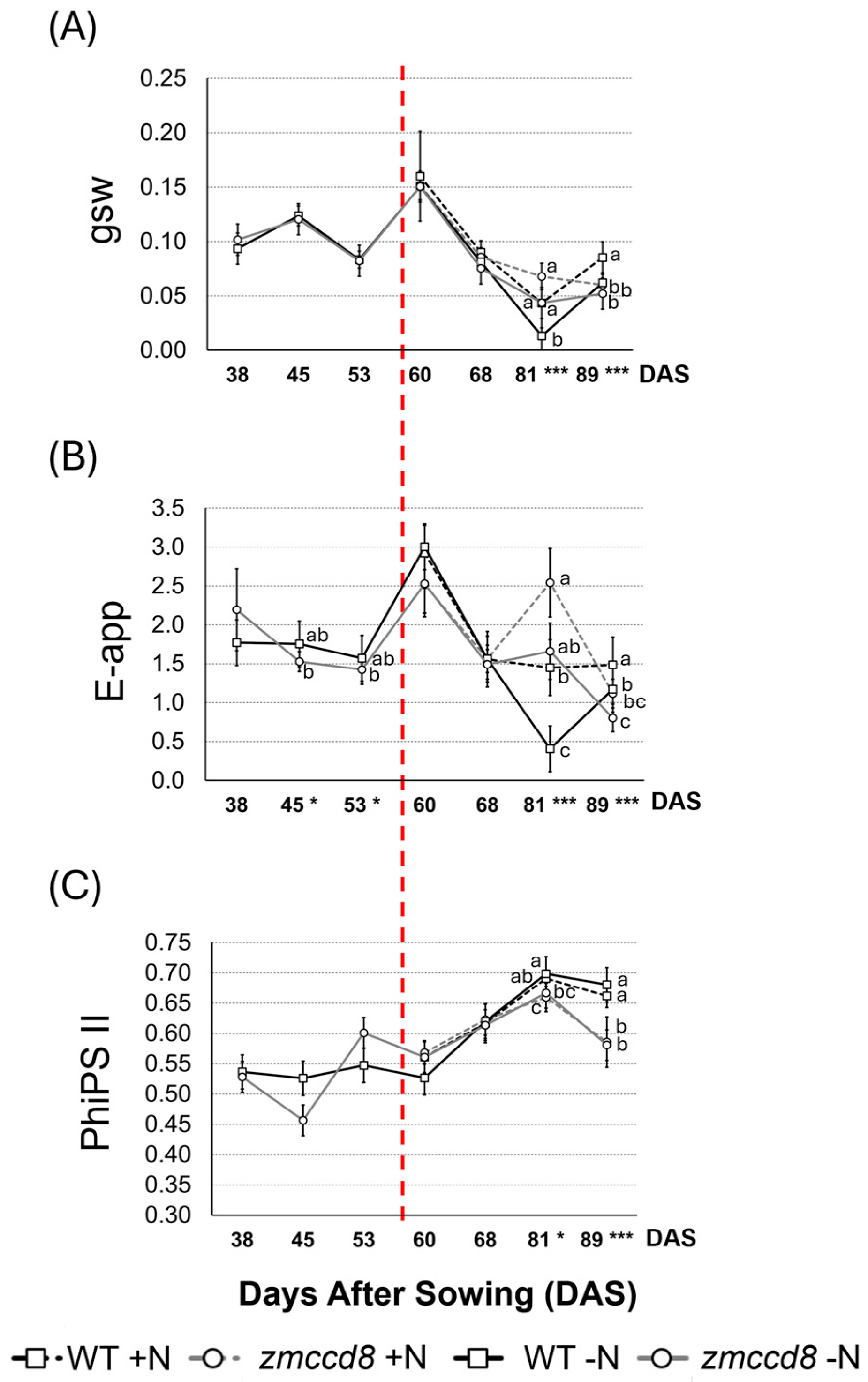
2.3. Assessment of Stomatal Conductance, Transpiration Rate, and Photosystem II Efficiency
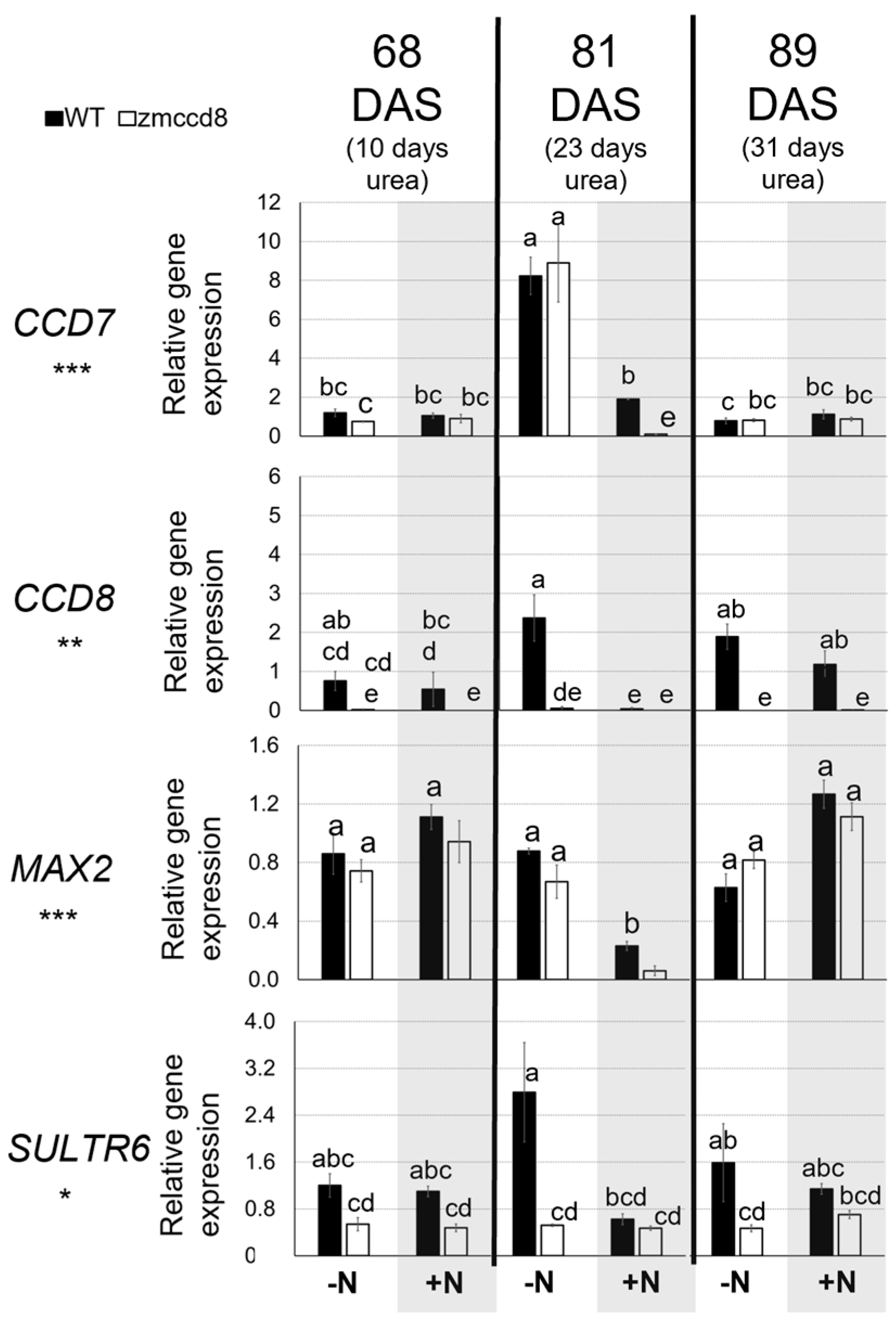
2.4. Molecular Analysis of Maize Gene Related to SL Biosynthesis and Signaling and Water Stress Response
3. Discussion
4. Materials and Methods
4.1. Maize Growth Conditions
4.2. Phenotypical Analysis
4.3. Optic Measurements of Chlorophyll and Anthocyanins in Leaves
4.4. Stomatal Conductance, Transpiration Rate, and Photosystem II Efficiency Leaf Analysis
4.5. RNA Extraction and cDNA Synthesis
4.6. Gene Selection for Gene Expression Analysis
4.7. Quantitative Reverse Transcription PCR (qRT-PCR)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Epstein, E.; Bloom, A. Mineral Nutrition of Plants: Principles and Perspectives, 2nd ed.; Sinauer Associates: Sunderland, MA, USA, 2005; p. 399. [Google Scholar]
- Ye, J.Y.; Tian, W.H.; Jin, C.W. Nitrogen in plants: From nutrition to the modulation of abiotic stress adaptation. Stress Biol. 2022, 2, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Anas, M.; Liao, F.; Verma, K.K.; Sarwar, M.A.; Mahmood, A.; Chen, Z.-L.; Li, Q.; Zeng, X.-P.; Li, Y.-R. Fate of nitrogen in agriculture and environment: Agronomic, eco-physiological and molecular approaches to improve nitrogen use efficiency. Biol. Res. 2020, 53, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Bloom, A.J. The increasing importance of distinguishing among plant nitrogen sources. Curr. Opin. Plant Biol. 2015, 25, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Pinton, R.; Tomasi, N.; Zanin, L. Molecular and physiological interactions of urea and nitrate uptake in plants. Plant Signal. Behav. 2016, 11, e1076603. [Google Scholar] [CrossRef]
- Castaldelli, G.; Colombani, N.; Tamburini, E.; Vincenzi, F.; Mastrocicco, M. Soil type and microclimatic conditions as drivers of urea transformation kinetics in maize plots. CATENA 2018, 166, 200–208. [Google Scholar] [CrossRef]
- Muratore, C.; Espen, L.; Prinsi, B. Nitrogen Uptake in Plants: The Plasma Membrane Root Transport Systems from a Physiological and Proteomic Perspective. Plants 2021, 10, 681. [Google Scholar] [CrossRef]
- Forde, B.G. Nitrogen signalling pathways shaping root system architecture: An update. Curr. Opin. Plant Biol. 2014, 21, 30–36. [Google Scholar] [CrossRef]
- Sun, H.; Tao, J.; Liu, S.; Huang, S.; Chen, S.; Xie, X.; Yoneyama, K.; Zhang, Y.; Xu, G. Strigolactones are involved in phosphate- and nitrate-deficiency-induced root development and auxin transport in rice. J. Exp. Bot. 2014, 65, 6735–6746. [Google Scholar] [CrossRef]
- Ito, S.; Ito, K.; Abeta, N.; Takahashi, R.; Sasaki, Y.; Yajima, S. Effects of strigolactone signaling on Arabidopsis growth under nitrogen deficient stress condition. Plant Signal. Behav. 2016, 11, e1126031. [Google Scholar] [CrossRef]
- Yoneyama, K. How Do Strigolactones Ameliorate Nutrient Deficiencies in Plants? Cold Spring Harb. Perspect. Biol. 2019, 11, a034686. [Google Scholar] [CrossRef]
- Ravazzolo, L.; Trevisan, S.; Manoli, A.; Quaggiotti, S. The Control of Zealactone Biosynthesis and Exudation is Involved in the Response to Nitrogen in Maize Root. Plant Cell Physiol. 2019, 60, 2100–2112. [Google Scholar] [CrossRef] [PubMed]
- Ravazzolo, L.; Boutet-Mercey, S.; Perreau, F.; Forestan, C.; Varotto, S.; Ruperti, B.; Quaggiotti, S. Strigolactones and Auxin Cooperate to Regulate Maize Root Development and Response to Nitrate. Plant Cell Physiol. 2021, 62, 610–623. [Google Scholar] [CrossRef] [PubMed]
- Kapulnik, Y.; Koltai, H. Fine-tuning by strigolactones of root response to low phosphate. J. Integr. Plant Biol. 2016, 58, 203–212. [Google Scholar] [CrossRef] [PubMed]
- Shindo, M.; Shimomura, K.; Yamaguchi, S.; Umehara, M. Upregulation of DWARF27 is associated with increased strigolactone levels under sulfur deficiency in rice. Plant Direct 2018, 2, e00050. [Google Scholar] [CrossRef]
- Zhang, Y.; Lv, S.; Wang, G.; Zhang, Y.; Lv, S.; Wang, G. Strigolactones are common regulators in induction of stomatal closure in planta. Plant Signal. Behav. 2018, 13, e1444322. [Google Scholar] [CrossRef]
- Wang, L.; Wang, B.; Jiang, L.; Liu, X.; Li, X.; Lu, Z.; Meng, X.; Wang, Y.; Smith, S.M.; Li, J. Strigolactone Signaling in Arabidopsis Regulates Shoot Development by Targeting D53-Like SMXL Repressor Proteins for Ubiquitination and Degradation. Plant Cell 2015, 27, 3128–3142. [Google Scholar] [CrossRef]
- Ruyter-Spira, C.; Kohlen, W.; Charnikhova, T.; van Zeijl, A.; van Bezouwen, L.; de Ruijter, N.; Cardoso, C.; Lopez-Raez, J.A.; Matusova, R.; Bours, R.; et al. Physiological Effects of the Synthetic Strigolactone Analog GR24 on Root System Architecture in Arabidopsis: Another Belowground Role for Strigolactones? Plant Physiol. 2010, 155, 721–734. [Google Scholar] [CrossRef]
- Villaécija-Aguilar, J.A.; Hamon-Josse, M.; Carbonnel, S.; Kretschmar, A.; Schmidt, C.; Dawid, C.; Bennett, T.; Gutjahr, C. SMAX1/SMXL2 regulate root and root hair development downstream of KAI2-mediated signalling in Arabidopsis. PLOS Genet. 2019, 15, e1008327. [Google Scholar] [CrossRef]
- Quaggiotti, S.; Buzzicotti, L.; Koch, K.E.; Guan, J.C.; Trevisan, S.; Varotto, S.; Ruperti, B.; Ravazzolo, L. Strigolactone roles in maize tolerance to low nitrogen involve shifts in acquisition and partitioning of protein, sulfur, and iron. Plant Soil 2024, 1–24. [Google Scholar] [CrossRef]
- Visentin, I.; Ferigolo, L.F.; Russo, G.; Krukowski, P.K.; Capezzali, C.; Tarkowská, D.; Gresta, F.; Deva, E.; Nogueira, F.T.S.; Schubert, A.; et al. Strigolactones promote flowering by inducing the miR319-LA-SFT module in tomato. Proc. Natl. Acad. Sci. USA 2024, 121. [Google Scholar] [CrossRef]
- Ueda, H.; Kusaba, M. Strigolactone Regulates Leaf Senescence in Concert with Ethylene in Arabidopsis. Plant Physiol. 2015, 169, 138–147. [Google Scholar] [CrossRef] [PubMed]
- Végh, A.; Incze, N.; Fábián, A.; Huo, H.; Bradford, K.J.; Balázs, E.; Soós, V. Comprehensive Analysis of DWARF14-LIKE2 (DLK2) Reveals Its Functional Divergence from Strigolactone-Related Paralogs. Front. Plant Sci. 2017, 8, 1641. [Google Scholar] [CrossRef] [PubMed]
- Guan, J.C.; Koch, K.E.; Suzuki, M.; Wu, S.; Latshaw, S.; Petruff, T.; Goulet, C.; Klee, H.J.; McCarty, D.R. Diverse Roles of Strigolactone Signaling in Maize Architecture and the Uncoupling of a Branching-Specific Subnetwork. Plant Physiol. 2012, 160, 1303–1317. [Google Scholar] [CrossRef]
- Nomura, T.; Seto, Y.; Kyozuka, J. Unveiling the complexity of strigolactones: Exploring structural diversity, biosynthesis pathways, and signaling mechanisms. J. Exp. Bot. 2023, 75, 1134–1147. [Google Scholar] [CrossRef]
- A Dun, E.; Brewer, P.B.; Gillam, E.M.J.; A Beveridge, C. Strigolactones and Shoot Branching: What Is the Real Hormone and How Does It Work? Plant Cell Physiol. 2023, 64, 967–983. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Li, W.; Burritt, D.J.; Tian, H.; Zhang, H.; Liang, X.; Miao, Y.; Mostofa, M.G.; Tran, L.-S.P. Strigolactones interact with other phytohormones to modulate plant root growth and development. Crop. J. 2022, 10, 1517–1527. [Google Scholar] [CrossRef]
- Jing, Y.; Yang, Z.; Yang, Z.; Bai, W.; Yang, R.; Zhang, Y.; Zhang, K.; Zhang, Y.; Sun, J. Sequential activation of strigolactone and salicylate biosynthesis promotes leaf senescence. New Phytol. 2024, 242, 2524–2540. [Google Scholar] [CrossRef]
- Buelbuel, S.; Sakuraba, Y.; Sedaghatmehr, M.; Watanabe, M.; Hoefgen, R.; Balazadeh, S.; Mueller-Roeber, B. Arabidopsis BBX14 negatively regulates nitrogen starvation- and dark-induced leaf senescence. Plant J. 2023, 116, 251–268. [Google Scholar] [CrossRef]
- Guo, Y.; Ren, G.; Zhang, K.; Li, Z.; Miao, Y.; Guo, H. Leaf senescence: Progression, regulation, and application. Mol. Hortic. 2021, 1, 1–25. [Google Scholar] [CrossRef]
- Hajibarat, Z.; Saidi, A. Senescence-associated proteins and nitrogen remobilization in grain filling under drought stress condition. J. Genet. Eng. Biotechnol. 2022, 20, 101–114. [Google Scholar] [CrossRef]
- Mu, X.; Chen, Y. The physiological response of photosynthesis to nitrogen deficiency. Plant Physiol. Biochem. 2020, 158, 76–82. [Google Scholar] [CrossRef]
- Sakuraba, Y. Molecular basis of nitrogen starvation-induced leaf senescence. Front. Plant Sci. 2022, 13, 1013304. [Google Scholar] [CrossRef]
- Shi, L.; Li, X.; Fu, Y.; Li, C. Environmental Stimuli and Phytohormones in Anthocyanin Biosynthesis: A Comprehensive Review. Int. J. Mol. Sci. 2023, 24, 16415. [Google Scholar] [CrossRef]
- Jezek, M.; Allan, A.C.; Jones, J.J.; Geilfus, C. Why do plants blush when they are hungry? New Phytol. 2023, 239, 494–505. [Google Scholar] [CrossRef]
- Peng, M.; Hudson, D.; Schofield, A.; Tsao, R.; Yang, R.; Gu, H.; Bi, Y.-M.; Rothstein, S.J. Adaptation of Arabidopsis to nitrogen limitation involves induction of anthocyanin synthesis which is controlled by the NLA gene. J. Exp. Bot. 2008, 59, 2933–2944. [Google Scholar] [CrossRef]
- Gonzalez-Dugo, V.; Durand, J.-L.; Gastal, F. Water deficit and nitrogen nutrition of crops. A review. Agron. Sustain. Dev. 2010, 30, 529–544. [Google Scholar] [CrossRef]
- Xu, Z.; Zhou, G.; Shimizu, H. Plant responses to drought and rewatering. Plant Signal. Behav. 2010, 5, 649–654. [Google Scholar] [CrossRef]
- Agami, R.A.; Alamri, S.A.; Abd El-Mageed, T.A.; Abousekken, M.; Hashem, M. Role of exogenous nitrogen supply in alleviating the deficit irrigation stress in wheat plants. Agric. Water Manag. 2018, 210, 261–270. [Google Scholar] [CrossRef]
- Cramer, G.R.; Urano, K.; Delrot, S.; Pezzotti, M.; Shinozaki, K. Effects of abiotic stress on plants: A systems biology perspective. BMC Plant Biol. 2011, 11, 163. [Google Scholar] [CrossRef]
- Luqman, M.; Shahbaz, M.; Maqsood, M.F.; Farhat, F.; Zulfiqar, U.; Siddiqui, M.H.; Masood, A.; Aqeel, M.; Haider, F.U. Effect of strigolactone on growth, photosynthetic efficiency, antioxidant activity, and osmolytes accumulation in different maize (Zea mays L.) hybrids grown under drought stress. Plant Signal. Behav. 2023, 18, 2262795. [Google Scholar] [CrossRef]
- Korek, M.; Marzec, M. Strigolactones and abscisic acid interactions affect plant development and response to abiotic stresses. BMC Plant Biol. 2023, 23, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Dong, L.; Durairaj, J.; Guan, J.-C.; Yoshimura, M.; Quinodoz, P.; Horber, R.; Gaus, K.; Li, J.; Setotaw, Y.B.; et al. Maize resistance to witchweed through changes in strigolactone biosynthesis. Science 2023, 379, 94–99. [Google Scholar] [CrossRef] [PubMed]
- Forestan, C.; Farinati, S.; Zambelli, F.; Pavesi, G.; Rossi, V.; Varotto, S. Epigenetic signatures of stress adaptation and flowering regulation in response to extended drought and recovery in Zea mays. Plant Cell Environ. 2019, 43, 55–75. [Google Scholar] [CrossRef] [PubMed]
- Posit team. 2023. RStudio: Integrated Development Environment for R. Posit Software, PBC, Boston, MA. March. Available online: https://www.posit.co/ (accessed on 19 March 2024).
- Manoli, A.; Sturaro, A.; Trevisan, S.; Quaggiotti, S.; Nonis, A. Evaluation of candidate reference genes for qPCR in maize. J. Plant Physiol. 2012, 169, 807–815. [Google Scholar] [CrossRef]
- Pan, X.; Zheng, H.; Zhao, J.; Xu, Y.; Li, X. ZmCCD7/ZpCCD7 encodes a carotenoid cleavage dioxygenase mediating shoot branching. Planta 2016, 243, 1407–1418. [Google Scholar] [CrossRef]
- Guan, J.-C.; Li, C.; Flint-Garcia, S.; Suzuki, M.; Wu, S.; Saunders, J.W.; Dong, L.; Bouwmeester, H.J.; McCarty, D.R.; Koch, K.E. Maize domestication phenotypes reveal strigolactone networks coordinating grain size evolution with kernel-bearing cupule architecture. Plant Cell 2022, 35, 1013–1037. [Google Scholar] [CrossRef]
- Huang, Q.; Wang, M.; Xia, Z. The SULTR gene family in maize (Zea mays L.): Gene cloning and expression analyses under sulfate starvation and abiotic stress. J. Plant Physiol. 2018, 220, 24–33. [Google Scholar] [CrossRef]
- Nonis, A.; Ruperti, B.; Falchi, R.; Casatta, E.; Enferadi, S.T.; Vizzotto, G. Differential expression and regulation of a neutral invertase encoding gene from peach (Prunus persica): Evidence for a role in fruit development. Physiol. Plant. 2007, 129, 436–446. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Rozen, S.; Skaletsky, H. Primer3 on the WWW for general users and for biologist programmers. Bioinform. Methods Protoc. 2000, 132, 365–386. [Google Scholar]
| Gene ID | Maize GDB ID | Functions | Primers | References |
|---|---|---|---|---|
| CCD7 | Zm00001eb074640 | Carotenoid cleavage dioxygenase 7, involved in SL biosynthesis | TCCGGCTCGCGCAGATTC | [12,13,43,47] |
| CTGCCCAGAACCCATGGA | ||||
| CCD8 | Zm00001eb153000 | Carotenoid cleavage dioxygenase 8, involved in SL biosynthesis | AGAAAGGTGTCTCTGCTGCT | [12,13,24,43] |
| CTATGGGCTCGCTCACATGA | ||||
| MAX2 | Zm00001eb376660 | Encoding F-box protein MAX2 involved in SL signaling | GAACAAGACCGGCATCCAAC | [48] |
| TTAACTCGTCAGGCCTCCAG | ||||
| SULTR6 | Zm00001eb154590 | Sulfate Transporter 6, mediates the uptake and translocation of sulfate | TAGGCGTCTTCAGGTTAGGG | [20,43,44,49] |
| GAGGTCTGTCTTTGGCGTGA | ||||
| MPE | Zm00001eb257640 | Housekeeping gene, encoding the membrane protein PB1A10.07c | TGTACTCGGCAATGCTCTTG | [46] |
| TTTGATGCTCCAGGCTTACC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ravazzolo, L.; Chichi, A.; Meggio, F.; Buzzicotti, L.; Ruperti, B.; Varotto, S.; Malagoli, M.; Quaggiotti, S. A Maize Mutant Impaired in SL Biosynthesis (zmccd8) Shows a Lower Growth, an Altered Response to Nitrogen Starvation, and a Potential Secondary Effect on Drought Tolerance. Stresses 2024, 4, 614-626. https://doi.org/10.3390/stresses4040039
Ravazzolo L, Chichi A, Meggio F, Buzzicotti L, Ruperti B, Varotto S, Malagoli M, Quaggiotti S. A Maize Mutant Impaired in SL Biosynthesis (zmccd8) Shows a Lower Growth, an Altered Response to Nitrogen Starvation, and a Potential Secondary Effect on Drought Tolerance. Stresses. 2024; 4(4):614-626. https://doi.org/10.3390/stresses4040039
Chicago/Turabian StyleRavazzolo, Laura, Andrea Chichi, Franco Meggio, Leonardo Buzzicotti, Benedetto Ruperti, Serena Varotto, Mario Malagoli, and Silvia Quaggiotti. 2024. "A Maize Mutant Impaired in SL Biosynthesis (zmccd8) Shows a Lower Growth, an Altered Response to Nitrogen Starvation, and a Potential Secondary Effect on Drought Tolerance" Stresses 4, no. 4: 614-626. https://doi.org/10.3390/stresses4040039
APA StyleRavazzolo, L., Chichi, A., Meggio, F., Buzzicotti, L., Ruperti, B., Varotto, S., Malagoli, M., & Quaggiotti, S. (2024). A Maize Mutant Impaired in SL Biosynthesis (zmccd8) Shows a Lower Growth, an Altered Response to Nitrogen Starvation, and a Potential Secondary Effect on Drought Tolerance. Stresses, 4(4), 614-626. https://doi.org/10.3390/stresses4040039










