Abstract
Emerging fungal plant pathogens are significant biotic stresses to crops that threaten global food security, biodiversity, and agricultural sustainability. Historically, these pathogens cause devastating crop losses and continue to evolve rapidly due to climate change, international trade, and intensified farming practices. Recent advancements in diagnostic technologies, including remote sensing, sensor-based detection, and molecular techniques, are transforming disease monitoring and detection. These tools, when combined with data mining and big data analysis, facilitate real-time surveillance and early intervention strategies. There is a need for extension and digital advisory services to empower farmers with actionable insights for effective disease management. This manuscript presents an inclusive review of the socioeconomic and historical impacts of fungal plant diseases, the mechanisms driving the emergence of these pathogens, and the pressing need for global surveillance and reporting systems. By analyzing recent advancements and the challenges in the surveillance and diagnosis of fungal pathogens, this review advocates for an integrated, multidisciplinary approach to address the growing threats posed by these emerging fungal diseases. Fostering innovation, enhancing accessibility, and promoting collaboration at both national and international levels are crucial for the agricultural community to protect crops from these emerging biotic stresses, ensuring food security and supporting sustainable farming practices.
1. Introduction
The worldwide population is anticipated to surpass 10 billion by the year 2050, demanding a 60% increase in crop production [1]. However, biotic stresses from plant diseases present a major hurdle in achieving this increase, often leading to considerable pre- and post-harvest crop losses. Yield reductions can range from 10% to 40% [2]. Additionally, emerging plant pathogens, driven by intensified agricultural practices, global trade, and climate change, further complicate these issues due to their widespread outbreaks, varied host ranges, broad geographic distributions, and increased pathogenicity.
Fungal plant pathogens are significant biotic stressors in agriculture, causing widespread diseases that threaten global food security [3]. Over 8000 fungal and oomycete species have been linked to plant diseases, affecting various agricultural sectors, including horticulture, floriculture, and forest systems [4,5]. These microorganisms can infect plants at any developmental stage in natural environments, either independently or in synergy with other pathogens, leading to diseases such as blight, rot, mildew, and rust [6,7]. The consequences of fungal infections include reduced crop yield, quality, and economic value, presenting significant challenges for agricultural systems worldwide [8].
Emerging fungal pathogens, unlike endemic ones, often cause novel and intense outbreaks with devastating effects on crops and the livelihoods of farmers. Historical examples include the Irish Potato Famine (1840s), caused by Phytophthora infestans, and the Bengal Famine (1943), linked to Cochliobolus miyabeanus, both of which resulted in millions of deaths and massive displacement [9]. Recent cases, such as the 2016 wheat blast epidemic in Bangladesh, led to severe yield losses of up to 51% [10]. In Sub-Saharan Africa, maize farmers frequently suffer from Aspergillus flavus infection, which poses a threat to food safety and causes financial hardship [11]. In Bangladesh, smallholder vegetable farmers face challenges with emerging outbreaks of white mold, caused by Sclerotinia sclerotiorum, which leads to substantial yield reductions and financial hardships [12,13,14,15]. Globally, Fusarium oxysporum TR4 endangers banana production, while Hemileia vastatrix (coffee rust) has displaced hundreds of thousands of workers across Central America, exacerbating poverty and migration [16,17,18]. The wheat stem rust pathogen Puccinia graminis f. sp. tritici (Ug99) poses a significant threat to 37% of global wheat production, with outbreaks spreading from Africa to Asia [19]. These examples underscore the far-reaching socioeconomic impacts of emerging fungal diseases, especially in vulnerable farming systems.
The emergence of fungal plant pathogens is driven by complex interactions among anthropogenic activities, environmental changes, and genetic evolution. Human-mediated transport of infected plant materials, often referred to as “pathogen pollution,” has significantly expanded the global range of pathogens. Historical examples include the introduction of Cryphonectria parasitica to North America and the spread of Tilletia indica via seed trade, both of which caused severe ecological and economic damage [20,21]. Similarly, the 2016 wheat blast outbreak in Bangladesh likely originated from infected seeds imported from Brazil [10]. Natural aerial dispersal, especially of fungi with airborne spores, further facilitates emergence. Rust pathogens, for instance, Puccinia striiformis f. sp. tritici, exploit long-distance wind currents and benefit from crop genetic uniformity [22]. Climate events such as hurricanes have also contributed to the transcontinental movement of pathogens, as seen in the spread of soybean rust by Hurricane Ivan [9]. Pathogens adapt rapidly through evolutionary processes, including mutation, recombination, and host jumps. New virulent races, such as wheat stem rust race TTKTT in Ethiopia and Puccinia striiformis races in Europe, have emerged by overcoming host resistance genes [22,23]. Climate change exacerbates this process by expanding the habitats of pathogens, increasing sporulation rates, and shifting disease distributions. Modern monoculture-based agriculture creates dense, genetically uniform crop systems that favor pathogen specialization and adaptation, exemplified by the rise of Zymoseptoria tritici [24]. Genetic mechanisms, such as horizontal gene transfer, mobile chromosomes, and hybridization, also drive the evolution of new pathogenic strains, including wheat blast fungus (MoT) and Phytophthora hybrids [25,26,27].
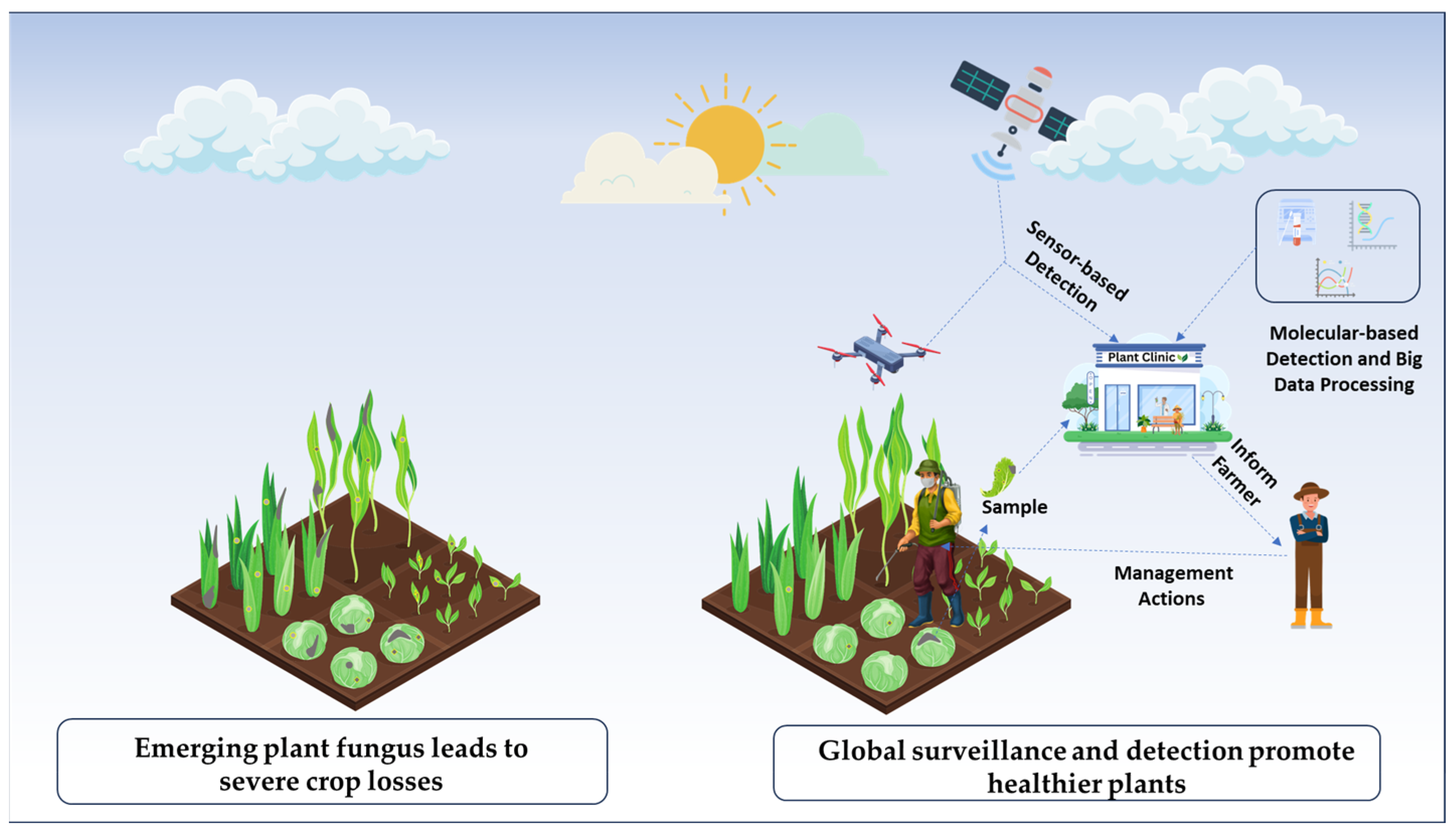
Embracing innovation and the application of advanced technologies are crucial for global surveillance and monitoring of emerging fungal diseases, which can help prevent crop losses. Equipped with multispectral and hyperspectral imagery, satellite and drone-based remote sensing technologies facilitate the early identification of plant diseases (Figure 1). By analyzing environmental and climatic conditions, this data, combined with AI-driven models, enables the prediction of disease outbreaks. The precise early-stage identification of fungal pathogens is facilitated by molecular diagnostics techniques such as polymerase chain reaction (PCR), next-generation sequencing (NGS), Loop-mediated isothermal amplification (LAMP), and sensor-based detection [28,29,30,31]. These methods, supported by metagenomics and bioinformatics resources such as GenBank and MycoBank, enhance the cataloging and analysis of fungal species, even at low concentrations of pathogen DNA, thereby advancing diagnostics and species delimitation [31]. Additionally, Internet of Things (IoT)-based monitoring and automated disease control, as part of smart farming technologies, help maximize agricultural practices by providing real-time data for early surveillance and intervention. However, implementing these technologies in resource-limited settings requires novel cost-effective frameworks.
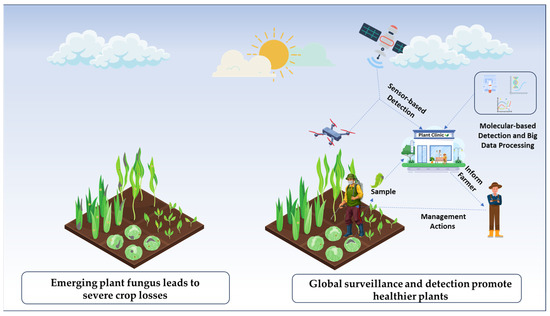
Figure 1.
Tech-driven approaches for surveillance and diagnostics of emerging fungal plant pathogens. Advanced technologies, such as remote sensing, molecular tools, and artificial intelligence, are utilized to collect environmental and genetic data. This data is then analyzed to identify fungal phytopathogens, enabling accurate prediction of the emerging diseases. This approach helps reduce losses and improve crop health.
Despite advancements in disease surveillance and detection technologies and tools, significant knowledge gaps remain regarding the comprehensive analyses of emerging fungal diseases, their broader socioeconomic effects, emergence mechanisms, and the effectiveness of current innovations in detecting and surveilling emerging fungal plant pathogens. Addressing these gaps is crucial for developing integrated, real-time surveillance systems to predict and manage fungal epidemics.
This review aims to deal with the current knowledge on emerging fungal plant diseases, assess their socioeconomic implications, and explore recent advancements in diagnostic and surveillance methods for emerging fungal plant pathogens. By providing insights into the integration of modern, innovative technological tools, this study is expected to contribute to the development of robust frameworks for plant disease management, safeguard food security, and maintain sustainable agricultural practices.
2. Recent Advances in Surveillance, Monitoring, and Detection of Emerging Fungal Diseases
Recent advancements in the surveillance and detection of emerging fungal diseases are transforming plant health management by enabling early identification and rapid response to pathogen threats. These technologies can be broadly categorized into four major domains: remote sensing systems, laboratory-based molecular diagnostics, field-deployable diagnostic tools, and data-driven analytical platforms (Table 1). Remote sensing technologies, including satellite imagery, unmanned aerial vehicles (UAVs), drones, and hyperspectral sensors, facilitate large-scale, non-invasive monitoring of crop health and enable the early detection of disease outbreaks across agricultural landscapes. Laboratory-based diagnostics utilize high-precision molecular techniques, including PCR, nucleic acid sequence-based amplification (NASBA), microarray analysis, and high-throughput sequencing (HTS), which enable the accurate identification and in-depth characterization of fungal pathogens. Field-applicable diagnostic tools, including LAMP, quantitative LAMP (qLAMP), and rolling circle amplification (RCA)-based assays, offer rapid, sensitive, and cost-effective detection directly in field conditions. In parallel, data-driven platforms using artificial intelligence (AI), natural language processing (NLP), geoparsing, social media mining, and metagenomic approaches support real-time surveillance, risk forecasting, and predictive modeling. Collectively, the integration of these complementary tools is revolutionizing plant disease surveillance by improving diagnostic accuracy, shortening response time, and guiding more effective disease management strategies.

Table 1.
Techniques and tools for surveillance and detection in agriculture and their benefits.
2.1. Remote Sensing Technologies
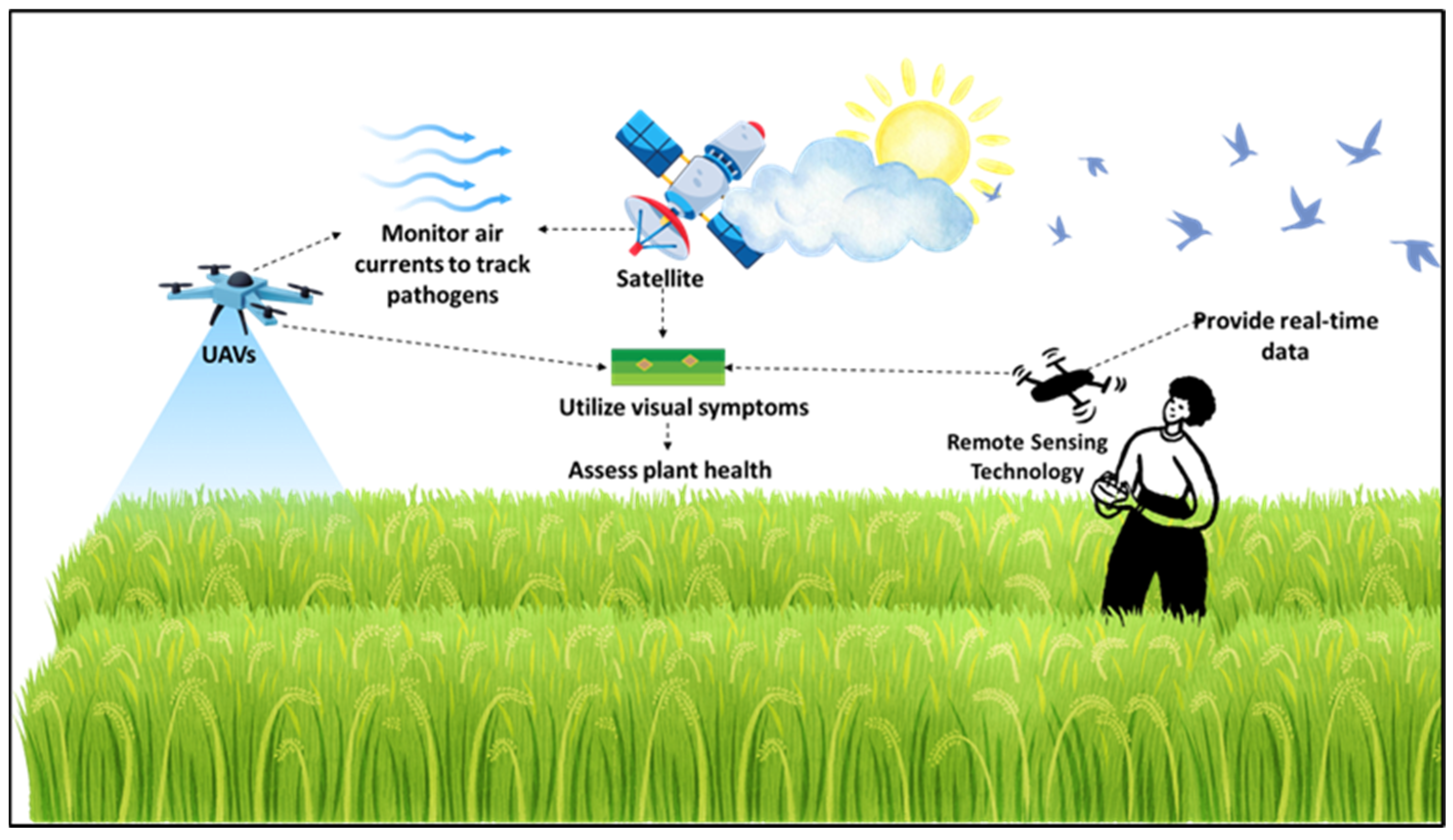
The application of remote sensing technologies in plant disease surveillance has advanced considerably, especially with the integration of satellite imagery, hyperspectral sensors, and UAVs (Figure 2). The principle behind these technologies lies in detecting changes in plant reflectance or spectral signatures associated with stress or infection, which can be captured across various spatial and spectral resolutions. Each of these tools contributes uniquely to disease monitoring, offering specific benefits and limitations (Figure 2). While remote sensing technology requires a significant initial investment in UAVs and satellite imagery, its long-term benefits for large-scale farms, including reduced labor costs and targeted crop interventions, outweigh the costs. However, smallholder farmers may struggle with the financial burden without government support or collaborative farming initiatives.
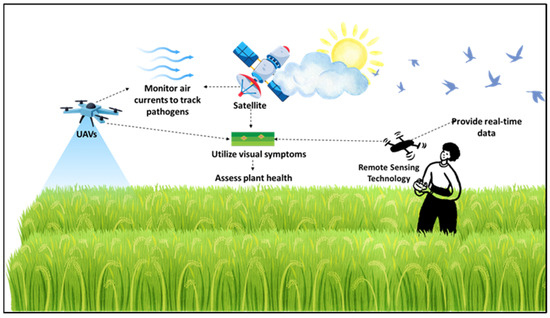
Figure 2.
Application of remote sensing technologies in monitoring and surveillance of emerging fungal diseases. This approach integrates satellite imagery and unmanned aerial vehicles (UAVs) or drones to assess crop health and detect plant diseases, particularly fungal infections. Satellites monitor environmental conditions and track the atmospheric movement of airborne pathogens. UAVs capture high-resolution data, allowing for the identification of visible symptoms of diseases such as blight and wilt. Meanwhile, hyperspectral imaging, utilized through both UAVs and satellites, enhances the early detection of plant stress and disease occurrence. This multi-scale remote sensing approach facilitates real-time monitoring, targeted interventions, and improved disease management strategies across extensive agricultural landscapes.
Satellite imagery is currently being widely used in agriculture for various purposes. It operates by capturing reflectance data over large areas, often using multispectral or high-resolution sensors to detect vegetation stress patterns that may be associated with disease symptoms. Its key strength lies in broad-scale coverage and long-term monitoring, making it ideal for regional or national-scale disease surveillance. However, quantifying disease prevalence in entire plants using satellite images remains a challenge due to the difficulty of identifying specific diseases. Moreover, it faces limitations in resolution and weather-dependence (e.g., cloud cover) [41].
Hyperspectral imaging is a promising solution to address the limitations of satellite imagery [41]. Hyperspectral sensors can capture a broad spectrum of wavelengths, providing detailed data that can help in detecting early signs of plant disease. This technology works by capturing the reflectance signatures of plants associated with stress or infection, often before visible symptoms appear. This method offers high sensitivity and early detection potential but comes with drawbacks such as complex data processing and limited utility for small-scale farmers [41]. Franke and Menz [42] demonstrated the effectiveness of high-resolution multispectral and hyperspectral remote sensing in monitoring powdery mildew and leaf rust in a 6-hectare winter wheat field. By utilizing QuickBird imagery and HyMap hyperspectral data, along with ground-truth validation from 54 sampled points, they successfully distinguished between healthy and infected plants, particularly during the later growth stages.
Tracking the atmospheric movement of phytopathogens is crucial for implementing effective quarantine strategies and predicting the long-distance dispersal of airborne pathogens [32]. In this context, UAVs, also known as drones, are effective tools for monitoring the movement of pathogens across large agricultural landscapes. Drones can operate at altitudes ranging from tens to hundreds of meters above the ground, providing real-time data on the spread of plant diseases. The key benefits of using UAVs or drones include their flexibility, high-resolution, and low-altitude imaging capabilities. They are particularly effective for detecting visible disease symptoms, monitoring disease progression, and mapping affected zones. However, dependence on trained personnel for operation and data interpretation is a major challenge.
The use of drones or UAVs has been assessed and found to be effective in identifying fungal diseases that exhibit clear visual signs. Blight and wilt were the primary diseases analyzed utilizing drone data due to their prominent symptoms in the imagery [43]. Sporangia of Phytophthora infestans in potato fields were detected at elevations of 90 m, showcasing the potential of drones for air sampling of pathogens [32]. In addition to pathogen monitoring, drones equipped with remote sensing technology are being utilized to assess plant health, such as monitoring water stress and tracking disease progression. This capability is significant for identifying field areas at risk of disease development, allowing for targeted intervention [41]. On the other hand, the application of drone-mounted hyperspectral imaging to assess wheat stem rust severity enables farmers and breeders to manage diseases more effectively and select resistant varieties [44]. With advancements in technology and artificial intelligence for precise fertilizer, fungicide, and crop management applications, this drone technology will emerge as an essential component of a farmer’s precision equipment arsenal.
As the technology behind remote sensing continues to evolve, higher-resolution satellite imagery and UAV-based systems will make it increasingly feasible to detect diseases at the individual plant or field level. The combination of remote sensing data, such as satellite imagery and hyperspectral data, with predictive modeling holds the potential to enhance the surveillance of emerging fungal diseases. By integrating information on pathogen movement, predictive models, environmental conditions, and plant health, this system can help forecast disease spread and enable targeted interventions [32,45].
2.2. Lab-Based Detection Techniques
2.2.1. PCR-Based Techniques
PCR has revolutionized the molecular identification of fungal pathogens and remains a cornerstone of plant disease diagnostics [46]. Various PCR variants offer trade-offs in sensitivity, specificity, and cost, making them suitable for different diagnostic contexts (Table 2).

Table 2.
Advanced PCR techniques, their mode of action, and applications in fungal plant pathogen detection.
End-point PCR operates by amplifying DNA using universal or species-specific primers, followed by gel electrophoresis or sequencing for identification. Its principle lies in DNA amplification until reaction saturation without real-time monitoring. Pros include affordability, simplicity, and suitability for routine diagnostics. Cons involve lower sensitivity and the need for post-PCR processing and sequencing. It has been used to detect Sphaeropsis pyriputrescens and Phacidiopycnis washingtonensis in apples, with species-specific primers providing accurate identification [47].
Nested PCR improves end-point PCR by using two sets of primers in successive amplification rounds to enhance sensitivity and specificity. Its principle involves a second PCR using the product of the first as a template. Pros include high sensitivity and specificity; cons include increased time, cost, and risk of contamination due to multiple handling steps. It has been used to detect Pilidiella granati, the causal agent of twig blight in pomegranate, down to 10 pg of DNA [48].
Multiplex PCR enables the simultaneous detection of multiple pathogens in a single reaction using multiple primer sets. The principle of this method lies in the concurrent amplification of distinct targets. Its pros include efficiency, time, and reagent savings, while its cons are complex primer design and challenges related to cross-reactivity. It has successfully identified 12 fungal pathogens associated with cranberry fruit rot [49] and detected Fusarium oxysporum and Phytophthora spp. in cacti [50].
Quantitative PCR (qPCR) enables the real-time monitoring of DNA amplification and the quantification of pathogen load. The principle is based on fluorescence detection during amplification. Pros include high sensitivity, specificity, speed, and quantification ability; cons involve high initial equipment cost and technical expertise. qPCR has detected Cryphonectria parasitica in chestnuts at single-spore sensitivity [51], Ramularia collo-cygni in barley [52], and Verticillium longisporum in UK crops [56].
BIO-PCR enhances sensitivity by combining a short pre-enrichment (incubation) phase with traditional PCR. Its principle involves pathogen biomass increase before DNA extraction. The pros include improved detection for low-titer pathogens and cost-effectiveness; the cons are longer processing times and delayed results. It has been used for detecting Colletotrichum lupini in lupin seeds [53] and Alternaria alternata in seedborne diseases [54].
Magnetic Capture Hybridization PCR (MCH-PCR) utilizes magnetic beads with specific probes to isolate target DNA before amplification, thereby minimizing the effects of inhibitors. The principle is targeted DNA capture followed by amplification. Pros include improved accuracy in complex samples; cons include higher cost and reliance on specialized reagents. This technique has detected Acidovorax avenae and Didymella bryoniae in cucurbit seeds [55]. The continued evolution of PCR technology is enhancing the precision and efficiency of fungal disease diagnostics, ultimately leading to more effective management strategies for plant diseases.
2.2.2. DNA Microarray Technology
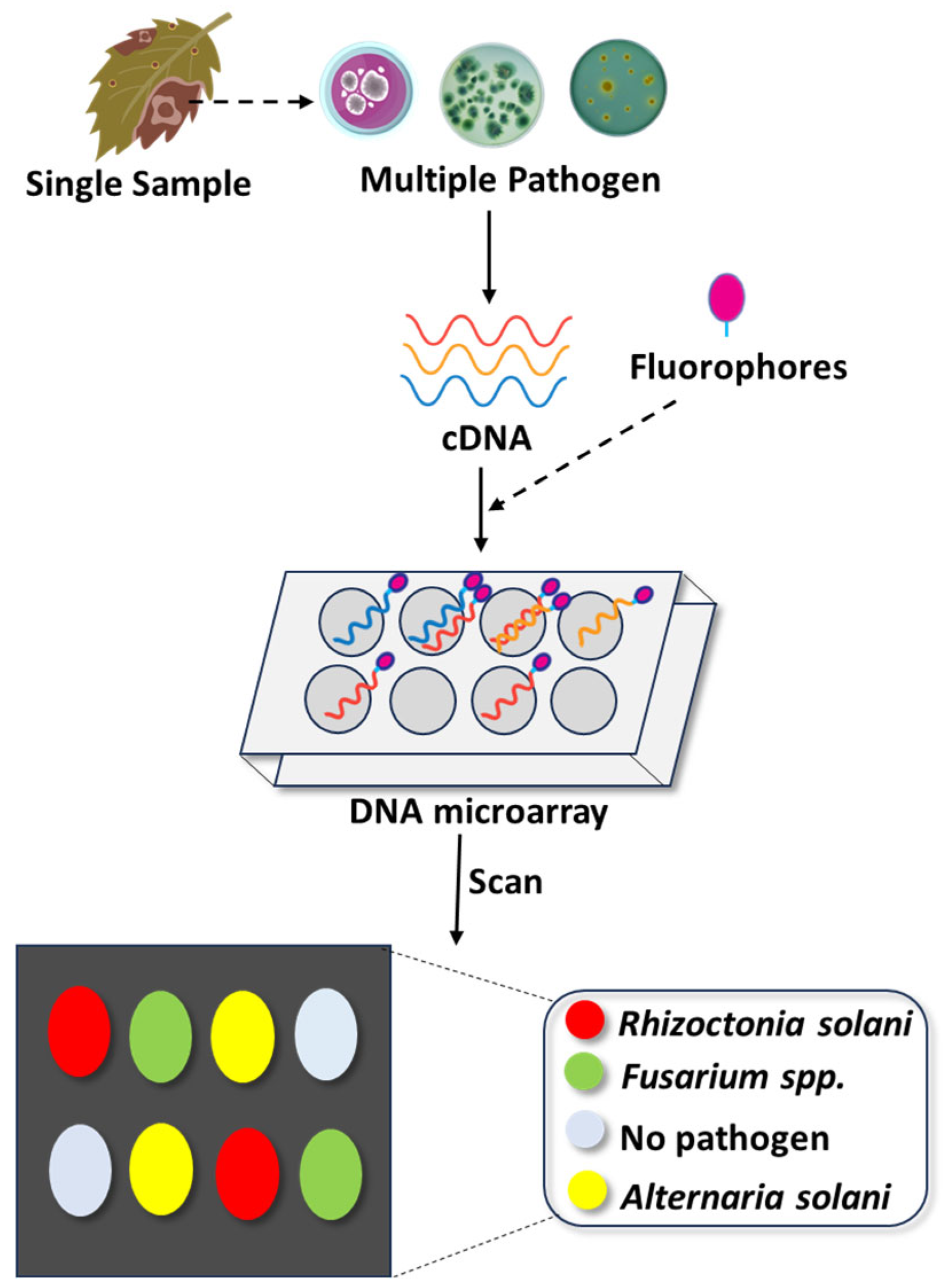
DNA microarray technology, also known as DNA chips, gene chips, or biochips, is a high-throughput molecular diagnostic method that enables the simultaneous detection and identification of numerous pathogens, including fungi, based on nucleic acid hybridization. It involves a solid substrate, typically glass or silicon, to which thousands of specific DNA sequences (probes) are attached in predefined locations (Figure 3). These probes hybridize with labelled target cDNA from the sample, and hybridization events are detected through fluorescence, chemiluminescence, or silver staining.
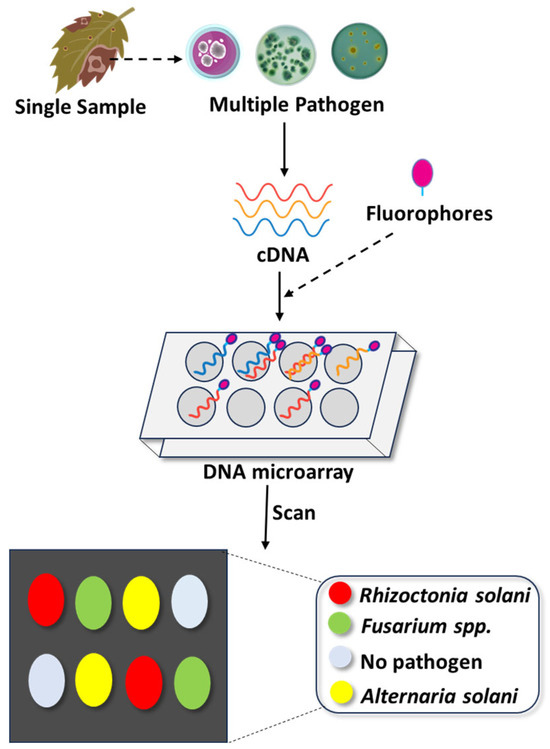
Figure 3.
Multiplex detection of emerging fungal pathogens using DNA microarray technology. This schematic illustrates the workflow for DNA microarray-based diagnostics, which simultaneously detects multiple fungal pathogens from a single plant sample. After sample collection from infected tissues, total RNA is extracted and then reverse-transcribed into complementary DNA (cDNA). The resulting cDNA is fluorescently labeled and hybridized to specific oligonucleotide probes immobilized on a microarray chip. Each probe corresponds to a unique fungal pathogen (e.g., Rhizoctonia solani, Fusarium spp., Alternaria solani). Hybridization is visualized through distinct fluorophore signals, with the resulting fluorescence pattern interpreted pathogens. This high-throughput, sensitive technique significantly enhances diagnostic accuracy and efficiency in plant disease surveillance.
Application cases demonstrate the utility of DNA microarrays in plant pathology. For example, Nikitin et al. [57] used a qPCR microarray system on 48-well silicon chips to identify multiple fungal pathogens, including Rhizoctonia solani, Spongospora subterranea, Alternaria solani, Fusarium spp., and Colletotrichum coccodes. In a separate study, the Array-Tube microarray system, which utilizes 42 species-specific probes targeting the ITS, tef1, and 16S rDNA regions, effectively detected various root rot pathogens, including Aphanomyces cochlioides, Botrytis cinerea, and Penicillium expansum, in artificially infected sugar beet tissues. [58]. This low-density microarray correctly identified pathogens in 23 out of 25 diseased samples, demonstrating its accuracy and efficiency for the simultaneous detection of multiple fungal pathogens. Wöhrle et al. [59] further highlighted that DNA microarrays can be manufactured cost-effectively and reproducibly, contributing to their practical adoption in diagnostic workflows.
2.2.3. Next-Generation or High-Throughput Sequencing
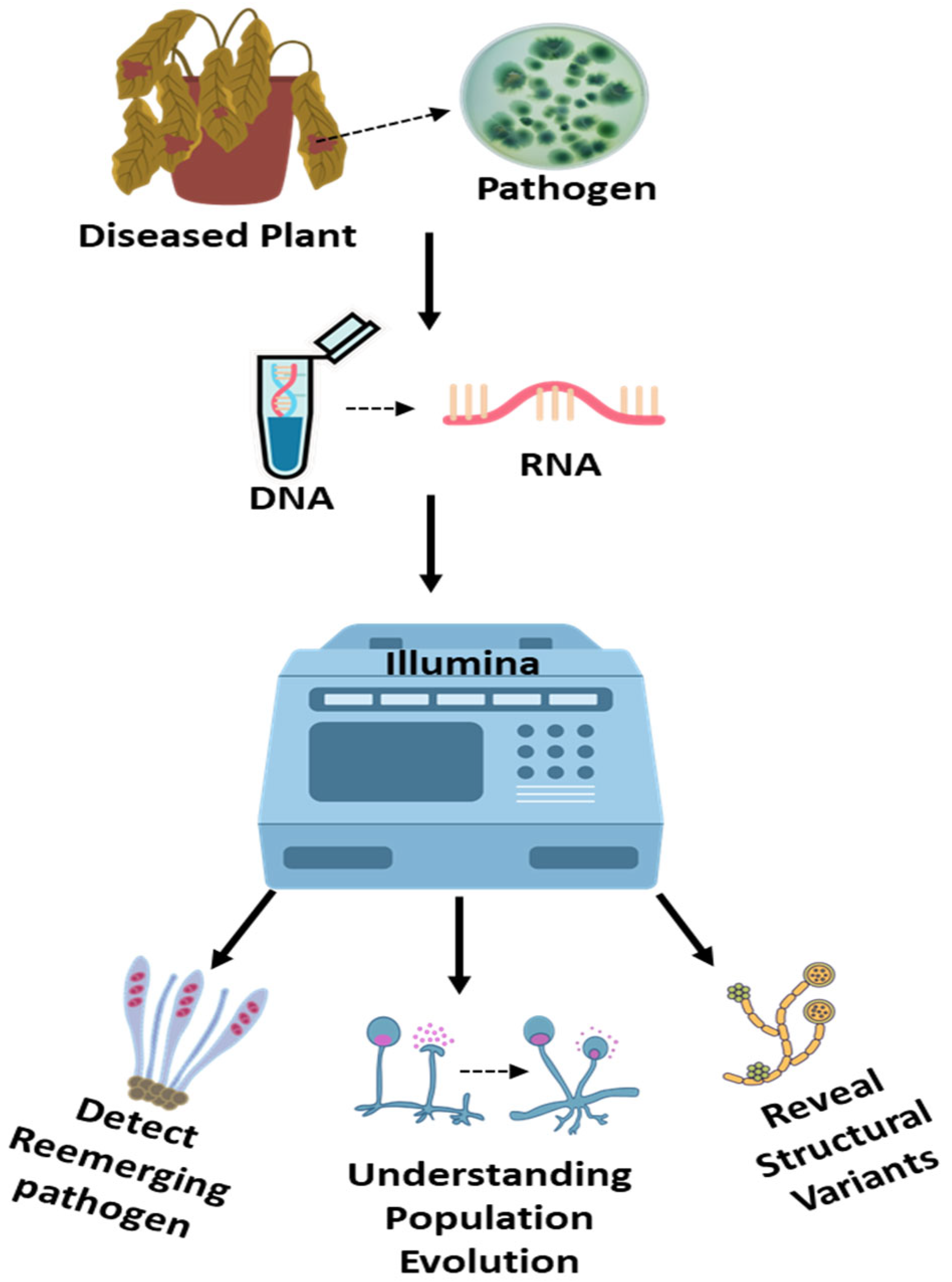
NGS, or HTS, is a powerful molecular approach that has advanced significantly, providing methods for comprehensive genomic analysis that can accurately detect fungal pathogens [60]. It enables comprehensive genomic analysis through massively parallel sequencing of DNA or RNA. The principle involves fragmenting nucleic acids, attaching adaptors, and simultaneously sequencing millions of short reads, which are then assembled and analyzed computationally (Figure 4). RNA-Seq, a form of NGS, targets messenger RNA to assess gene expression and identify active pathogens. RNA-Seq is proving invaluable for rapidly identifying fungal infections causing emerging plant diseases. This unbiased method can detect both known and novel fungal pathogens directly from infected plant tissues without prior knowledge of the target organism [60].
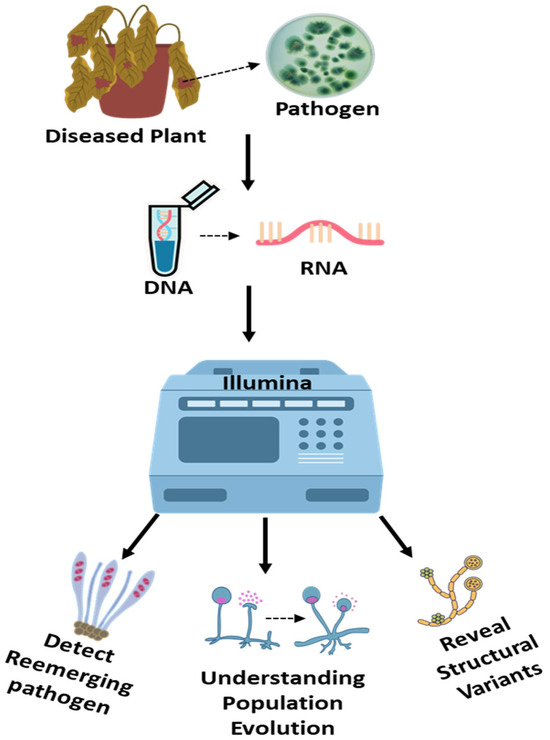
Figure 4.
Next-Generation Sequencing (NGS) for detection and characterization of fungal plant pathogens. This schematic illustrates the workflow of high-throughput next-generation sequencing (e.g., Illumina platform) for identifying and analyzing fungal pathogens from diseased plant tissues. The process begins with the collection of symptomatic plant samples, followed by nucleic acid extraction and library preparation. Sequencing generates massive datasets that enable precise identification of fungal species, including unculturable and cryptic taxa. NGS also facilitates in-depth genomic investigations, such as population diversity, evolutionary dynamics, virulence factor profiling, and detection of structural variants. This powerful tool enhances our understanding of emerging fungal threats and supports the development of targeted disease management strategies.
The advantages of NGS include high sensitivity and specificity, the ability to detect mixed infections, novel pathogens, and genomic variants, as well as its utility in both diagnostics and pathogen population studies. NGS allows for in-depth characterization of fungal evolution and emergence patterns [61]. Despite its advantages, NGS faces several challenges and limitations in field applications. One primary limitation is the requirement for sophisticated laboratory infrastructure and highly trained personnel, which restricts its accessibility in resource-limited settings [62]. The Portable Oxford Nanopore MinION has attempted to address this issue; however, the device still requires stable power sources, computational resources for data analysis, and careful handling to ensure sequencing accuracy [63]. Another challenge is the high cost associated with NGS, including sample preparation, sequencing reagents, and bioinformatics pipelines, making routine field deployment economically unfeasible in many cases [64].
Furthermore, data analysis in NGS is computationally intensive, requiring robust bioinformatics tools and high-performance computing capabilities, which may not be readily available in field settings. Additionally, sequencing errors, particularly with third-generation sequencing platforms, can impact the reliability of results, necessitating thorough validation and cross-referencing with existing genomic databases [65]. Contamination risks also pose significant challenges, as metagenomic sequencing often includes host and environmental DNA, complicating pathogen identification [66]. Addressing these limitations will require continued advancements in portable sequencing technologies, cost-effective workflows, user-friendly bioinformatics tools, and contamination control measures to enhance the feasibility of NGS in field applications.
Several application cases illustrate the diagnostic potential of NGS. For example, Illumina MiSeq technology was utilized to sequence the genome of Calonectria pseudonaviculata, the pathogen of Sarcococca blight in ornamental plants, leading to the detection of novel fungal species [67]. Population genomic studies widely use NGS to reveal variations, including structural variants and single-nucleotide polymorphisms (SNPs), enhancing the understanding of fungal pathogen populations and their evolution [61]. NGS is particularly applicable in detecting re-emerging pathogens. A study on the wheat yellow rust fungus, Puccinia striiformis f. sp. tritici, used RNA-Seq to track changes in the fungal population in the UK, revealing novel and exotic lineages [68]. Similarly, NGS techniques have been employed to develop molecular diagnostics for the cucurbit downy mildew pathogen Pseudoperonospora cubensis by comparing the genomes of associated species [69]. The resulting diagnostic markers reliably identified P. cubensis across various hosts, enabling the accurate differentiation of this species from closely related oomycetes.
2.2.4. Nucleic Acid Sequence-Based Amplification (NASBA)
NASBA is an isothermal amplification technique designed explicitly for RNA targets. Operating at a constant temperature of approximately 41 °C, NASBA amplifies nucleic acids without the need for a thermal cycler [70]. The procedure involves converting a single-stranded RNA (ssRNA) template into complementary DNA (cDNA) using reverse transcriptase. This is then amplified using RNase H and T7 RNA polymerase. This combination of enzymes enables NASBA to produce over 108 copies of the target nucleic acid sequence in approximately 30 min, making it both rapid and efficient.
The main advantages of NASBA include its rapid amplification (within 30–90 min), isothermal operation (no thermal cycler required), and high sensitivity, capable of detecting as few as 10,000 rRNA molecules or 100 colony-forming units (cfu) per reaction. Its RNA specificity also allows detection of actively transcribing pathogens, potentially distinguishing live from dead cells. Furthermore, NASBA’s compatibility with real-time detection methods (e.g., molecular beacons) supports the development of quantitative diagnostics.
However, NASBA has certain limitations. It requires high-quality RNA extraction and is more sensitive to RNA degradation compared to DNA-based methods. The method also relies on multiple enzymes, increasing complexity and cost. Additionally, despite its success in clinical and bacterial plant pathogen diagnostics, the application of this method in fungal plant pathogen detection remains underexplored, likely due to the dominance of DNA-based methods in this area.
In terms of applications, NASBA has been effectively used to detect plant bacterial pathogens. For instance, Van Beckhoven et al. [71] employed NASBA with AmpliDet RNA to detect Clavibacter michiganensis subsp. sepedonicus (Cms), the causal agent of bacterial ring rot in potatoes. The method enabled real-time detection using molecular beacons and achieved a sensitivity of 10,000 rRNA molecules or 100 cfu per reaction. Notably, a specific primer pair could distinguish live from dead Cms cells, demonstrating the precision and utility of NASBA in complex plant tissue samples.
Although its use in fungal plant pathology is still limited, the isothermal and RNA-targeting nature of NASBA presents strong potential for developing portable, field-deployable diagnostic tools for rapid detection of fungal pathogens in agricultural systems [72].
2.2.5. DNA or RNA Probe-Based Techniques
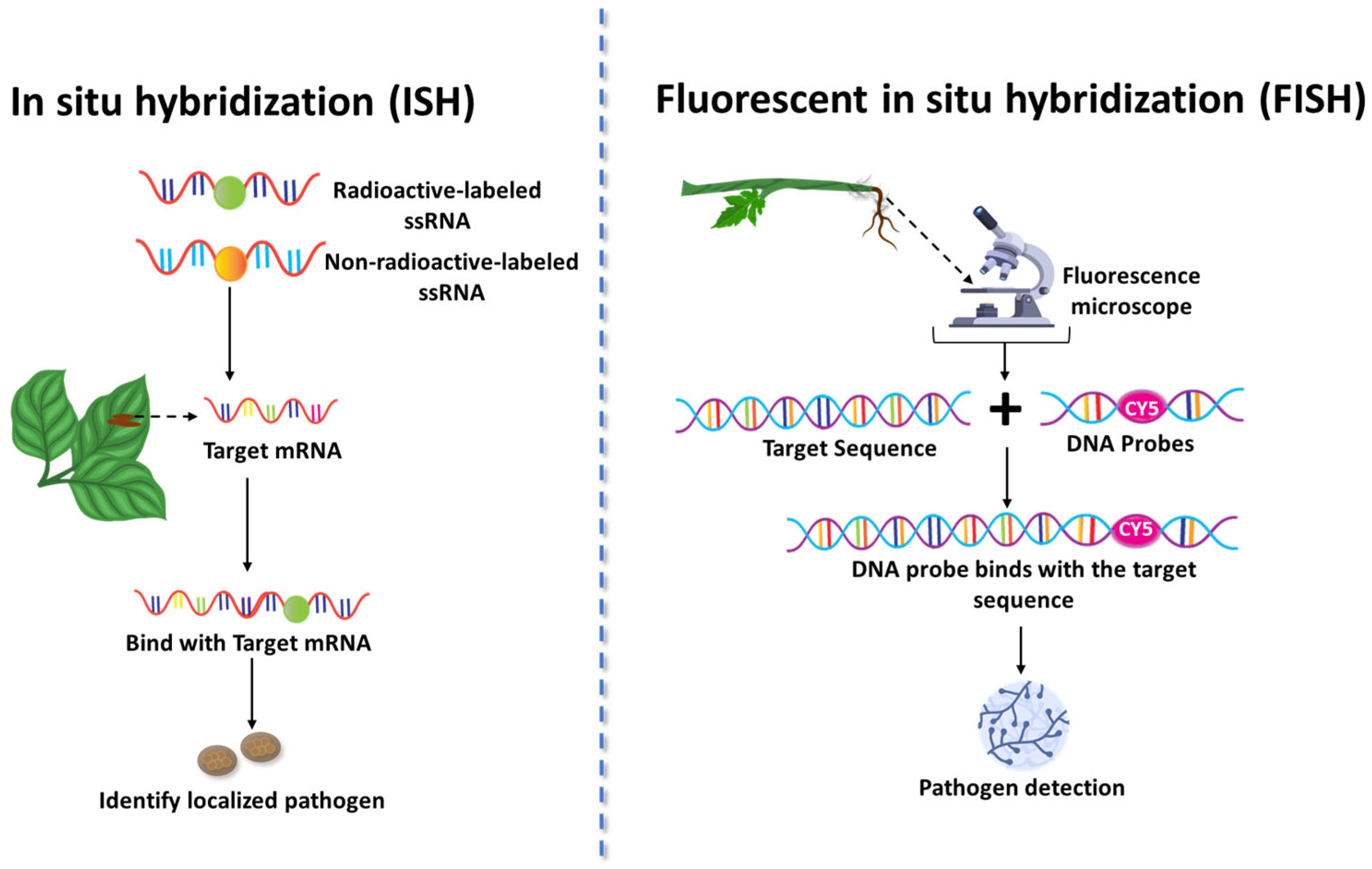
Assays based on DNA or RNA probes, such as fluorescent in situ hybridization (FISH) and in situ hybridization (ISH), have appeared as powerful tools for diagonizing fungal pathogens in plant tissues. These methods utilize the specificity of nucleic acid sequences to identify and localize pathogens within plant samples, offering advantages in both sensitivity and specificity. ISH involves designing antisense single-stranded RNA (ssRNA) probes that bind specifically to the target mRNA of interest (Figure 5). These probes are typically radioactive, although non-radioactive labels, such as biotin, digoxigenin, and alkaline phosphatase, are also employed [73]. Advantages of ISH include its high specificity for target gene expression and ability to provide spatial localization of pathogens within tissue sections. However, limitations include a complex optimization process, potential RNA degradation, and generally lower sensitivity compared to other nucleic acid detection methods.
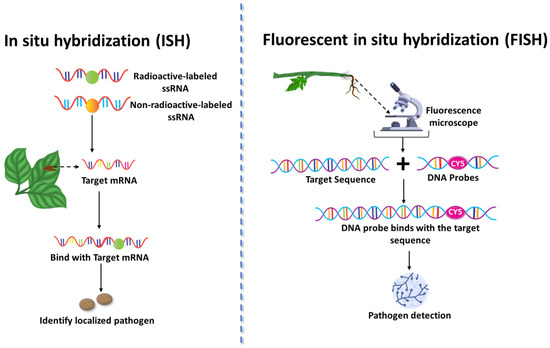
Figure 5.
In Situ Hybridization (ISH) and Fluorescent In Situ Hybridization (FISH) techniques for the detection of fungal pathogens in plant tissues. This figure illustrates two nucleic acid probe-based diagnostic approaches—In Situ Hybridization (ISH) and Fluorescent In Situ Hybridization (FISH)—used to detect and localize fungal pathogens directly within infected plant tissues. ISH involves the use of labeled single-stranded RNA (ssRNA) or DNA probes, either radioactive or non-radioactive, which hybridize with complementary pathogen mRNA or DNA sequences, allowing visualization of infection sites. FISH employs fluorophore-tagged DNA probes that bind specifically to fungal nucleic acids, enabling high-resolution detection through fluorescence microscopy. These techniques provide spatial and molecular insights into fungal colonization patterns and gene expression in planta.
ISH has been successfully used to identify rust pathogens, such as Puccinia horiana (Chrysanthemum rust), Uromyces transversalis (Gladiolus rust), and Phakopsora pachyrhizi (soybean rust), in paraffin-fixed plant tissues [74]. In this study, key optimizations of the ISH included proteinase K treatment (10 µg/mL) and fine-tuning of formamide concentration and wash temperatures. The validated protocol effectively distinguished rust fungi from plant tissues without background staining in healthy samples. Strong signals were observed in U. transversalis, while P. pachyrhizi showed weaker signals, suggesting a need for further refinement. This optimized ISH method provides a valuable tool for visualizing rust fungi, studying their morphology and interactions with host tissues, and elucidating their life cycles.
FISH operates on a similar hybridization principle but uses DNA or RNA probes labelled with fluorescent dyes (e.g., Cy3, Cy5) to detect target sequences directly under fluorescence microscopy [75]. The fluorochrome-labelled DNA or RNA probes emit fluorescence when hybridized with the target sequence. This method directly visualizes pathogens within cells or tissues using fluorescence microscopy [75]. FISH offers high sensitivity, specificity, speed, and detailed information on pathogen morphology, even in mixed species samples. The primary drawbacks of FISH are the requirement for expensive fluorescence equipment, potential interference from autofluorescence, and limitations in multiplexing when a large number of probes are needed.
FISH has been used efficiently to detect Sclerotium rolfsii, which causes southern blight in tomatoes, by targeting rRNA sequences of the pathogen. The technique identified soil-borne S. rolfsii at concentrations as low as 0.06 pg µL−1 using cyanine dye Cy3- and Cy5-labelled probes [76].
2.3. Field-Applicable
2.3.1. Loop-Mediated Isothermal Amplification (LAMP)
LAMP has recently become an essential tool for diagnosing fungal infections due to its practical application in the field [35]. This technique combines rapid nucleic acid extraction using microneedles with DNA amplification at a constant temperature. It employs a strand-displacing DNA polymerase and a set of four to six primers that target specific regions of the DNA sequence [77]. Unlike PCR methods, LAMP does not require thermal cycling, making it particularly suitable for field diagnostics and providing a more accessible alternative. The advantages of LAMP include high specificity, rapid amplification, which is often achieved within 30 min, and tolerance to PCR inhibitors commonly found in plant tissues. It can also work with crude samples, minimizing the need for extensive purification. These features make LAMP especially effective for on-site applications in plant pathology.
Despite its advantages, LAMP has some limitations and challenges that impact its field application. One significant drawback is the risk of cross-contamination, which can lead to false-positive results, as the high efficiency of the LAMP reaction can amplify even minute amounts of target DNA from previous assays if proper precautions are not taken [78]. Closed-tube detection methods, such as fluorescence-based readouts, can mitigate this issue; however, they require additional equipment, potentially limiting their field applicability [37]. Another challenge is primer design, as LAMP requires four to six primers that recognize distinct regions of the target DNA. This complexity makes assay development more intricate than conventional PCR and can lead to non-specific amplification if not carefully optimized [79]. Additionally, while LAMP can be performed using portable and battery-operated devices, maintaining precise temperature control in field conditions remains a challenge, especially in regions with extreme environmental fluctuations. The stability of the reaction mixture and the shelf life of the reagents also require further improvement to ensure long-term usability in remote locations without reliable cold-chain storage. Finally, while LAMP has demonstrated effectiveness in detecting single pathogens, its multiplexing capabilities remain limited compared to PCR, which restricts its application in scenarios requiring the simultaneous detection of multiple pathogens. Addressing these limitations through advancements in assay design, reagent stabilization, and contamination control strategies will be crucial for enhancing the field utility of LAMP in plant disease diagnostics.
LAMP has been successfully applied to detect various fungal pathogens. For example, Kaczmarek et al. [80] developed a highly sensitive LAMP assay targeting the cytochrome b gene for the detection of Uromyces betae, the causal agent of sugar beet rust. The test could detect as little as 10 pg of DNA and showed a strong correlation between DNA quantity and spore counts, highlighting its potential for quantifying pathogen loads in environmental samples and improving disease forecasting. LAMP targeting the cytochrome b gene has been successful in rapidly detecting Uromyces betae, with results obtained in less than 30 min [80]. This platform has shown promise in identifying dual infections caused by tomato spotted wilt virus and P. infestans, which can significantly impact tomato crops (Figure 5) [36]. Similarly, both conventional LAMP and quantitative LAMP have been employed to diagnose Fusarium circinatum, the pathogen causing pitch canker in pine and other conifers, with quantitative LAMP demonstrating greater specificity [81]. LAMP also detects Sclerotinia sclerotiorum, a widespread pathogen of rapeseed, and has shown a lower detection threshold than traditional PCR [82]. Other notable LAMP applications include detecting the rice blast fungus Magnaporthe oryzae and the banana Panama wilt fungus Fusarium odoratissimum tropical race 4 (TR4) [36,83]. The method is particularly effective for detecting pathogens in crude samples and is resistant to inhibitors commonly found in plant extracts, making it highly robust for field diagnostics [84].
2.3.2. Sensor-Based Technologies
The development of various sensor-based technologies for detecting fungal diseases in food crops is advancing rapidly, with a range of novel techniques being explored to improve diagnostic accuracy. Sensor-based technologies for detecting fungal diseases in food crops operate by identifying specific physiological or biochemical signals from infected plants, such as volatile organic compounds (VOCs) or pathogen-related enzymes, through portable or embedded electronic systems. Sensor-based diagnostics have the potential to significantly enhance the speed and precision of plant disease identification, particularly in resource-constrained settings. Field practitioners equipped with pathogen-specific sensors can perform on-site diagnostics, enabling rapid responses and minimizing disease spread. Moreover, data generated by these devices can be integrated into regional and national surveillance systems, thereby enhancing our ability to detect and forecast emerging pathogen threats. As electronic and wearable sensor technologies continue to evolve, they will play a crucial role in advancing plant disease monitoring, refining management strategies, and contributing to global efforts to ensure food security. However, initial development and production costs remain a challenge, necessitating government support to scale up deployment in vast agricultural areas.
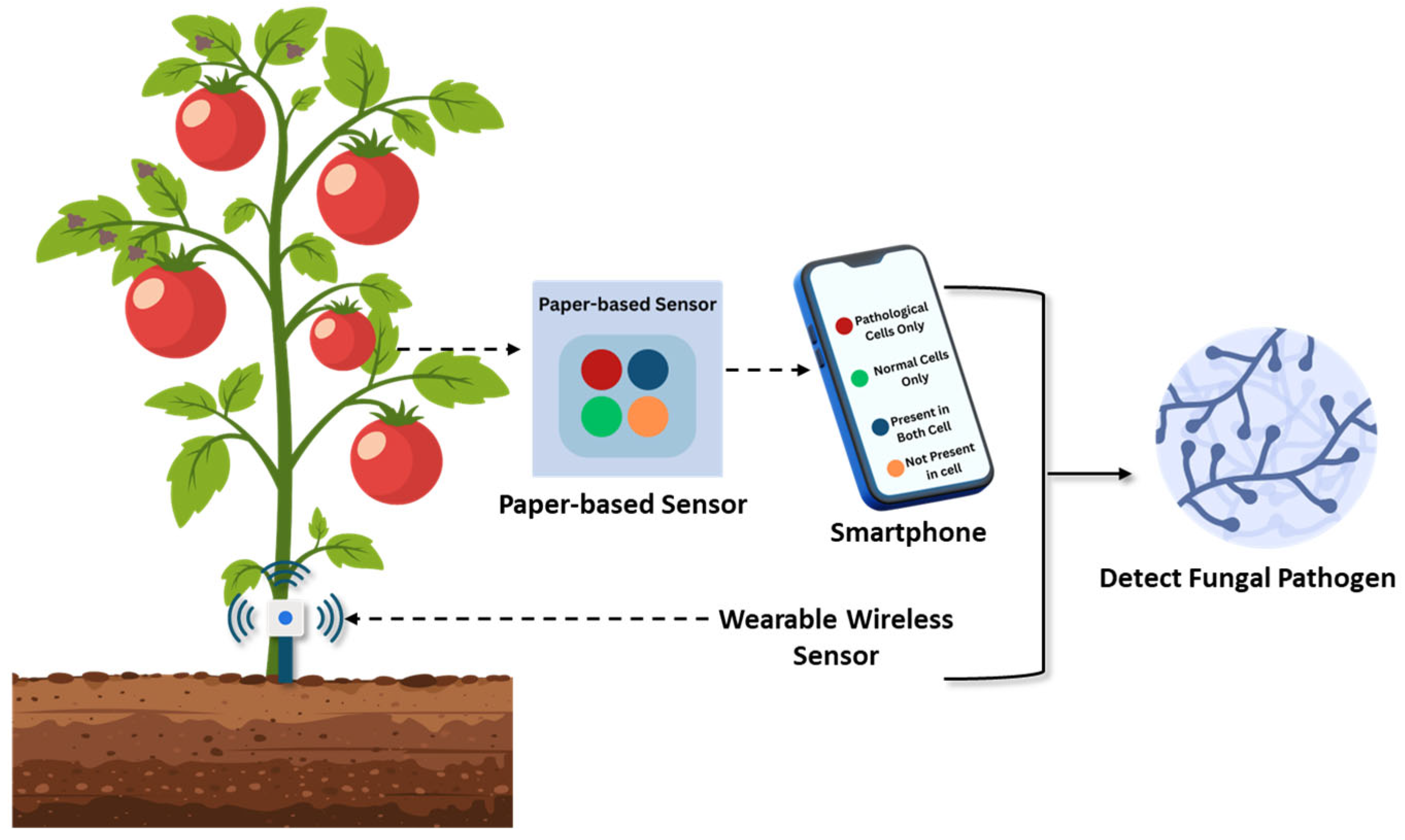
A significant advancement in sensor-based technologies involves smartphone-based VOC sensors that use organic dye arrays on paper substrates and multiplexed nanoplasmonic sensors to detect stress-induced volatiles in plants. The principle here relies on colourimetric or spectral changes in response to specific VOCs, enabling disease identification without visible symptoms (Figure 6). By sensing VOCs emitted by pathogen-stressed plants, these technologies enable early intervention, potentially reducing the spread of disease and crop losses. Key advantages of this approach include portability, low cost, minimal technical expertise required, and compatibility with smartphones, making it accessible to smallholder farmers and those in remote areas. However, limitations include potential interference from environmental volatiles, sensor degradation over time, and variability in VOC emissions across cultivars and environmental conditions. This system has been successfully used to detect diseases such as Septoria blight, late blight, and early blight in tomatoes before visible symptoms appear [36].
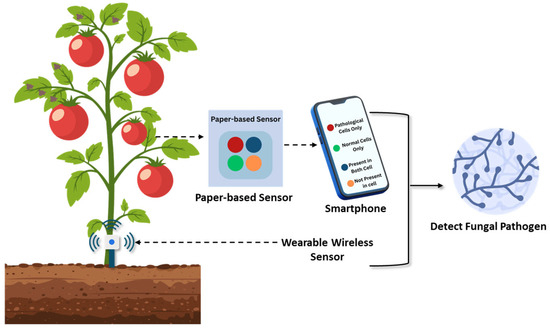
Figure 6.
Emerging sensor-based technologies for the detection of fungal diseases in crops. Paper-based sensors using organic dye arrays detect volatile organic compounds (VOCs) emitted by infected plants, enabling early disease recognition before visual symptoms appear. Smartphone-based diagnostic tools interpret sensor data, facilitating immediate and accurate field diagnosis. These portable, affordable technologies support real-time monitoring of plant health and hold transformative potential for disease surveillance, especially in resource-limited agricultural settings.
In addition to VOC-based platforms, wearable and wireless sensor systems enable continuous, real-time monitoring of plant health metrics, including moisture content, temperature, and biochemical markers, thereby broadening the potential for precision agriculture. These sensors can be deployed directly on plants or in the surrounding environment, enabling the monitoring of plant health and disease diagnostics, particularly in regions lacking access to conventional services such as plant disease clinics or extension support (Figure 6).
Another notable innovation is the development of chitinase-based biosensors, which detect fungal pathogens by recognizing chitin, a key component of fungal cell walls. These biosensors work by measuring changes in electrical or optical signals upon binding of fungal antigens to the enzymatic sensor component. The advantages of chitinase-based biosensors include high sensitivity, potential for early detection, and multifunctionality. However, their disadvantages include limited specificity due to cross-reactivity with non-target fungi and the need for further optimization for in-field deployment. The scope of chitinase-based biosensors in detecting phytopathogenic fungi has been demonstrated by Lucas-Bautista et al. [85]. Their study employed electrochemical, optical, and piezoelectric transduction platforms, utilizing chitinase, to successfully identify pathogens such as Aspergillus niger and Pseudocercospora fijiensis. Beyond detection, the application of chitinase or chitosan enhanced resistance in plants against Sphaerotheca humuli and Phytophthora parasitica, suggesting a dual utility as both a diagnostic and a resistance-inducing agent. These findings highlight the promise of chitinase-based biosensors while also emphasizing the need for improved specificity and real-time accuracy in field diagnostics.
2.3.3. Rolling Circle Amplification (RCA)
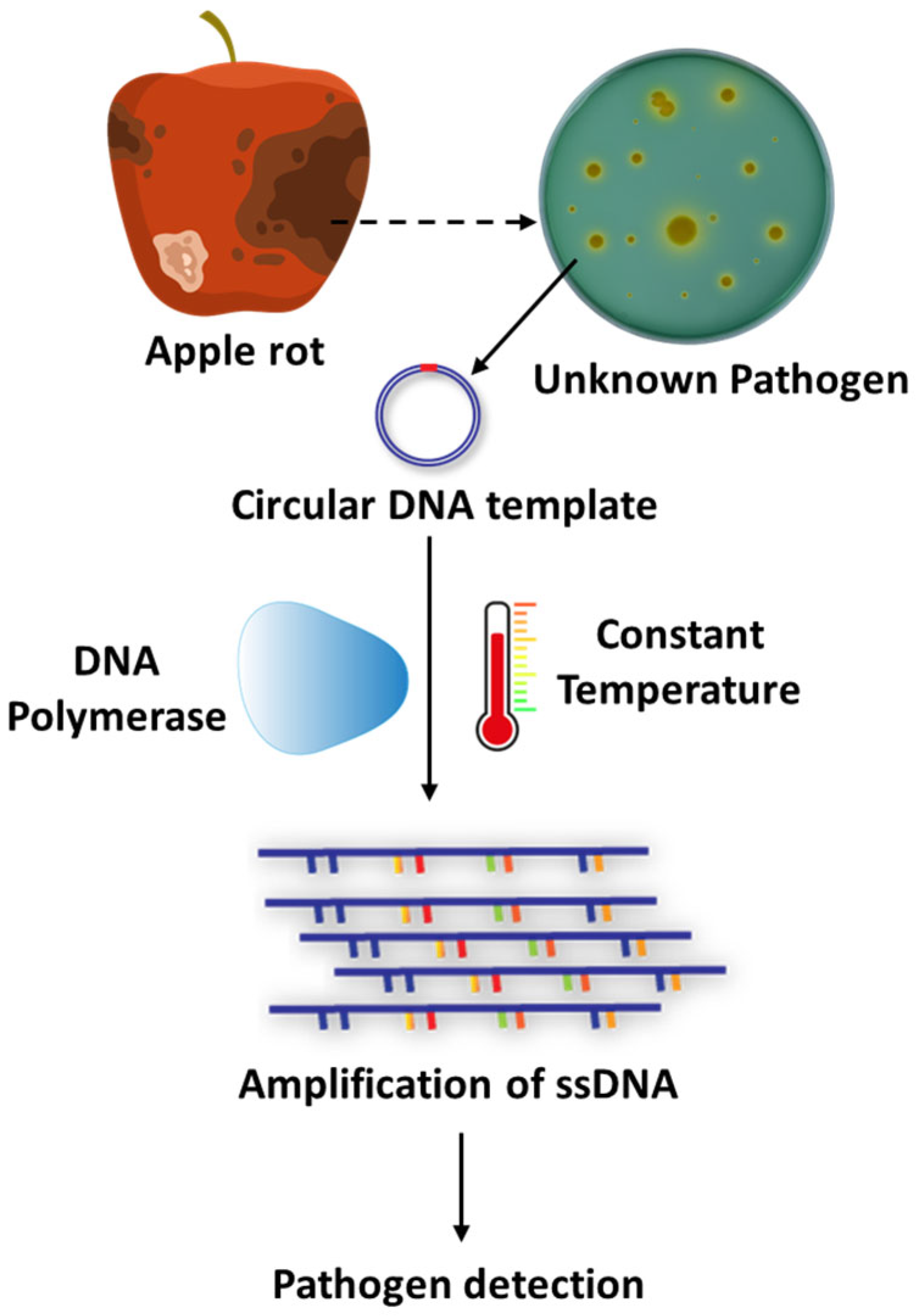
RCA is an isothermal enzymatic amplification technique that offers a simple, efficient, and cost-effective alternative to traditional PCR for detecting pathogens, including those causing fungal diseases. Unlike PCR, which requires thermal cycling, RCA operates at a constant temperature, simplifying the process and making it more accessible in resource-limited settings [86]. RCA employs DNA or RNA polymerases, such as Phi29 DNA polymerase, which possess strand displacement activity that enables the continuous extension of single-stranded DNA (ssDNA) from a circular DNA template, eliminating the need for thermal cycling [87,88] (Figure 7). The advantages of RCA include its simplicity, cost-effectiveness, high sensitivity and specificity, and minimal equipment requirement, making it highly suitable for field-based diagnostics. However, the method has several limitations, including the need for probe circularization, reduced multiplexing capability compared to other molecular tools, and potential technical challenges in probe design and optimization.
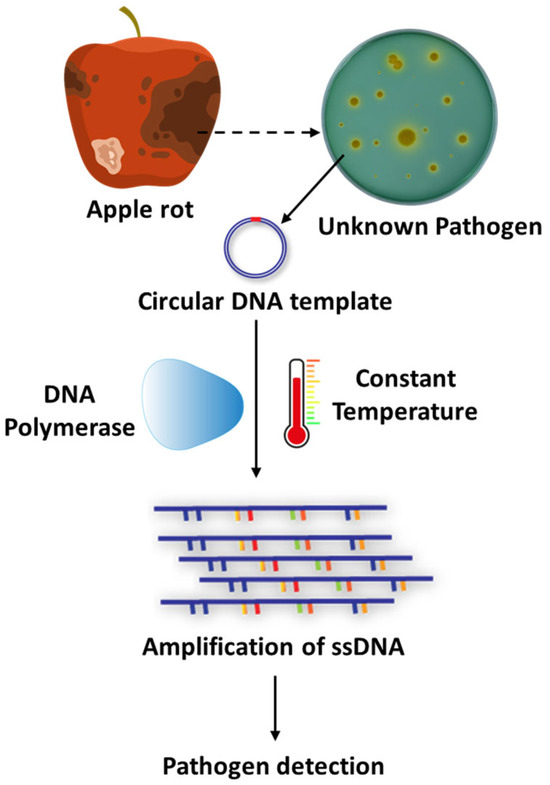
Figure 7.
Rolling Circle Amplification (RCA) for the detection of fungal pathogens. This schematic illustrates the principle of Rolling Circle Amplification (RCA), an isothermal DNA amplification technique employed for detecting fungal pathogens. RCA begins with the recognition of pathogen-specific DNA, which is circularized to serve as a template. A strand-displacing DNA polymerase then synthesizes long single-stranded DNA (ssDNA) concatemers at a constant temperature, eliminating the need for thermal cycling. The amplified products can be visualized or analyzed downstream for accurate pathogen identification.
RCA has been widely exploited for the detection of various plant pathogens, particularly fungi. One notable application is the use of padlock probes (PLPs), which are designed to target specific genetic sequences in pathogens. For instance, Lin et al. [89] developed PLPs targeting the TEF-1α gene to detect Neofabraea species (N. malicorticis, N. perennans, N. kienholzii, and N. vagabunda), which cause bull’s-eye rot in apples. The assay demonstrated high specificity and sensitivity across 28 samples, with no cross-reactivity, offering a reliable tool for pathogen surveillance in port quarantine and orchard epidemiological studies. Similarly, RCA has been applied to detect Fusarium species complex responsible for Fusarium head blight through the use of padlock probes targeting polymorphisms in the TEF-1α gene [90].
2.4. Data Mining/Big Data Analysis/Metagenomics
2.4.1. Data Mining and Big Data Analytics
The application of data mining and big data analysis techniques is a powerful tool for monitoring and managing fungal diseases in food crops. Data mining techniques involve extracting valuable information from vast datasets, which are used to analyze potential disease threats and assess the spread of plant pathogens. These methods can process structured data, such as published research papers, archival documents, and databases like PubMed and CAB Abstracts, as well as unorganized data from sources like Twitter feeds, image posts, and social media. NLP tools are often employed to analyze this diverse data, enabling the tracking and mapping of plant diseases and pathogens in real time [38]. The ability to mine “big data” streams offers decision-makers valuable insights that can inform disease management strategies and enhance the resilience of food supplies, particularly during outbreaks. By periodically surveying the internet and social media for plant disease reports, authorities can more effectively monitor disease dynamics and predict potential disruptions in food production. This data can help identify key control points in agricultural systems, evaluate the robustness of agricultural systems in particular geographic regions, and pinpoint disease outbreak hotspots, thereby facilitating targeted interventions.
The integration of data mining methodologies, including NLP, geoparsing, text mining, and crowdsourced social media data, holds significant promise for enhancing plant disease surveillance. These technologies can be used to monitor historical disease outbreaks and forecast future events, thus improving global food security monitoring, policy development, and the execution of phytosanitary plant health services. By utilizing big data, agriculture, and public health stakeholders can make more informed decisions, ultimately helping safeguard food crops from fungal diseases and other plant pathogens.
One of the key advantages of applying data mining and big data analytics in plant disease management is its cost-effectiveness. Compared to traditional surveillance methods, these approaches allow for rapid and large-scale disease monitoring with minimal labour and operational costs. By leveraging publicly available data from social media, research databases, and sensor networks, authorities can gain real-time insights without significant infrastructure investments. Additionally, predictive modelling using big data can enable preemptive disease control measures, reducing economic losses caused by late interventions. However, the initial setup, including data processing infrastructure and expertise in artificial intelligence, remains a challenge for widespread adoption.
Despite its potential, there have been limited attempts to utilize data mining techniques to examine the dissemination of emerging diseases of plants [38]. In the United States, a notable example is the use of NLP and geoparsing to monitor the dissemination of P. infestans, a pathogen from the 19th century. Tateosian et al. [39] utilized NLP and GIS technologies to analyze historical records from the ports of Philadelphia and New York. They extracted agricultural data on crop yields, disease occurrences, and weather patterns. Using tools like PDFMiner and NLTK, they found a significant rise in the term “potato disease” around 1845, coinciding with the Irish Potato Famine, while “late blight” emerged only around 1900. Reports from 1844 indicated early signs of the epidemic. This approach combined historical text mining with geographic visualization, revealing insights into the epidemic’s origins and management strategies [39].
In another application, more than 12,500 disease reports were analyzed to study the invasiveness of Phytophthora species and the genus-wide distribution. This analysis enabled researchers to predict which countries are most at risk of Phytophthora outbreaks and assess the invasive potential of dominant species, thereby supporting the development of early warning systems for targeted disease management efforts [91].
2.4.2. Genomic Surveillance and Bioinformatics
Genomic surveillance involves the sequencing and analysis of pathogen genomes to monitor the evolution, origin, and spread of plant diseases. This technology relies on high-throughput sequencing platforms and bioinformatic tools to characterize pathogen populations at the molecular level, enabling the identification of outbreak lineages, mutation patterns, and migration routes. The core principle centers on utilizing NGS and comparative genomics to identify genetic variations over time and space. Tools such as “genome skimming,” 12-plex SSR genotyping, Bayesian clustering, and real-time sequencing (e.g., MinION) facilitate both retrospective and in-field diagnostics.
Genomic surveillance systems that focus on the races and genotypes of pathogens, combined with bioinformatic prediction algorithms, hold significant potential for tracing the origins of outbreak strains and discovering novel fungal strains affecting plants. The advent of NGS tools has enabled cost-effective exploration of evolutionary genomics, allowing for more precise tracking of plant disease movements and enhancing our understanding of pathogen evolution. These genomic and bioinformatic tools are crucial for clarifying the functional evolution, ancestry, and origins of significant plant diseases, especially when historical collections are available [92].
Natural history collections, particularly mycological collections, serve as invaluable resources for understanding historical outbreaks and identifying pathogen lineages. They enable researchers to determine whether a new outbreak is caused by a novel lineage or the result of sexual reproduction within existing populations [93]. The digitization of herbarium specimens and the integration of data mining approaches further enhance the utility of these collections, providing essential baseline information on disease outbreaks that impact critical food crops. This data is also increasingly valuable for informing disease spread models. Advanced techniques allow for the deduction of pathogen dispersal patterns based on spatial snapshots over time, and genomic data can help refine these models by more effectively constraining transmission chains [94].
The genomic analysis of both historical and contemporary outbreak strains plays a crucial role in tracing the timing and origins of plant diseases. It is also vital for identifying the migration routes of previous outbreak strains [95]. Genomic mining can uncover the evolutionary trajectory and geographic origins of pathogen species, helping to pinpoint regions with the highest genetic diversity and identify areas at high risk for crop populations. Additionally, genomic analysis provides insights into the virulence dynamics of historical outbreaks.
One exciting advancement in this field is the use of low-depth sequencing techniques, such as “genome skimming,” which facilitates the tracking of both ancient and contemporary plant diseases. By sequencing museum specimens categorically classified by time and location, researchers can now monitor historical plant disease outbreaks and extend the timeline of initial reports across different countries [96].
Rapid in-field sequencing methods, including those utilizing MinION sequencers, have been deployed to reduce the costs and time associated with identifying fungal strains, such as rust fungi [64]. These accessible and efficient phylogenetic techniques are invaluable for monitoring plant disease outbreak strains and identifying novel pathogen lineages in real-time [97]. However, additional reference genome assemblies for emerging plant diseases are essential to fully leverage the potential of population genomics in outbreak monitoring. Moreover, open sequence reporting systems and diverse panels of sequenced globally diversified plant pathogens are essential tools for advancing the precision and speed of plant disease surveillance.
The integration of genomic surveillance significantly reduces the financial burden of traditional outbreak detection methods. For instance, genomic sequencing technologies such as “genome skimming” and rapid in-field sequencing methods, like MinION sequencers, allow for faster and more affordable pathogen identification compared to conventional diagnostic techniques. These approaches offer substantial cost savings in the long run by minimizing the need for prolonged field testing and manual monitoring while enhancing detection sensitivity. Moreover, the reduced time and labour costs associated with these technologies enable more efficient deployment of resources, ultimately making plant disease surveillance more affordable, scalable, and adaptable in different regions. However, the requirement for bioinformatic expertise, the need for high-quality reference genomes for novel or emerging pathogens, and challenges in building open-access global sequence databases are key limitations.
Application cases demonstrate the effectiveness of this approach. For instance, Saville and Ristaino [96] utilized SSR genotyping and Bayesian clustering to track the global migration of Phytophthora infestans, identifying two dominant genotypes: FAM-1 and US-1. Their research revealed that FAM-1 spread widely during colonial times before being replaced by US-1 in the 1950s. Similarly, Radhakrishnan et al. [64] used rapid sequencing (MinION) to directly identify rust fungi in the field. Additionally, Jombart et al. [94] and Bieker & Martin [95] employed spatial-genomic models to trace dispersal patterns and outbreak timelines. These studies underscore the significance of genomic surveillance in elucidating evolutionary trajectories, pinpointing high-risk regions, and refining the responsiveness of global plant disease monitoring systems.
3. Biovigilance Systems for Detecting Emerging Plant Fungal Diseases
Biovigilance, in the context of plant health, represents a proactive and integrated framework for monitoring, detecting, and responding to the emergence of plant pathogens, including fungal diseases, that threaten agricultural productivity and ecosystem stability. This one decision-support system involves the systematic observation of biotic and abiotic factors influencing pathogen populations, biodiversity, and ecosystem services, with the primary goal of identifying significant temporal and spatial shifts linked to agricultural intensification, climate variability, new cultivar introductions, and emerging pests [98].
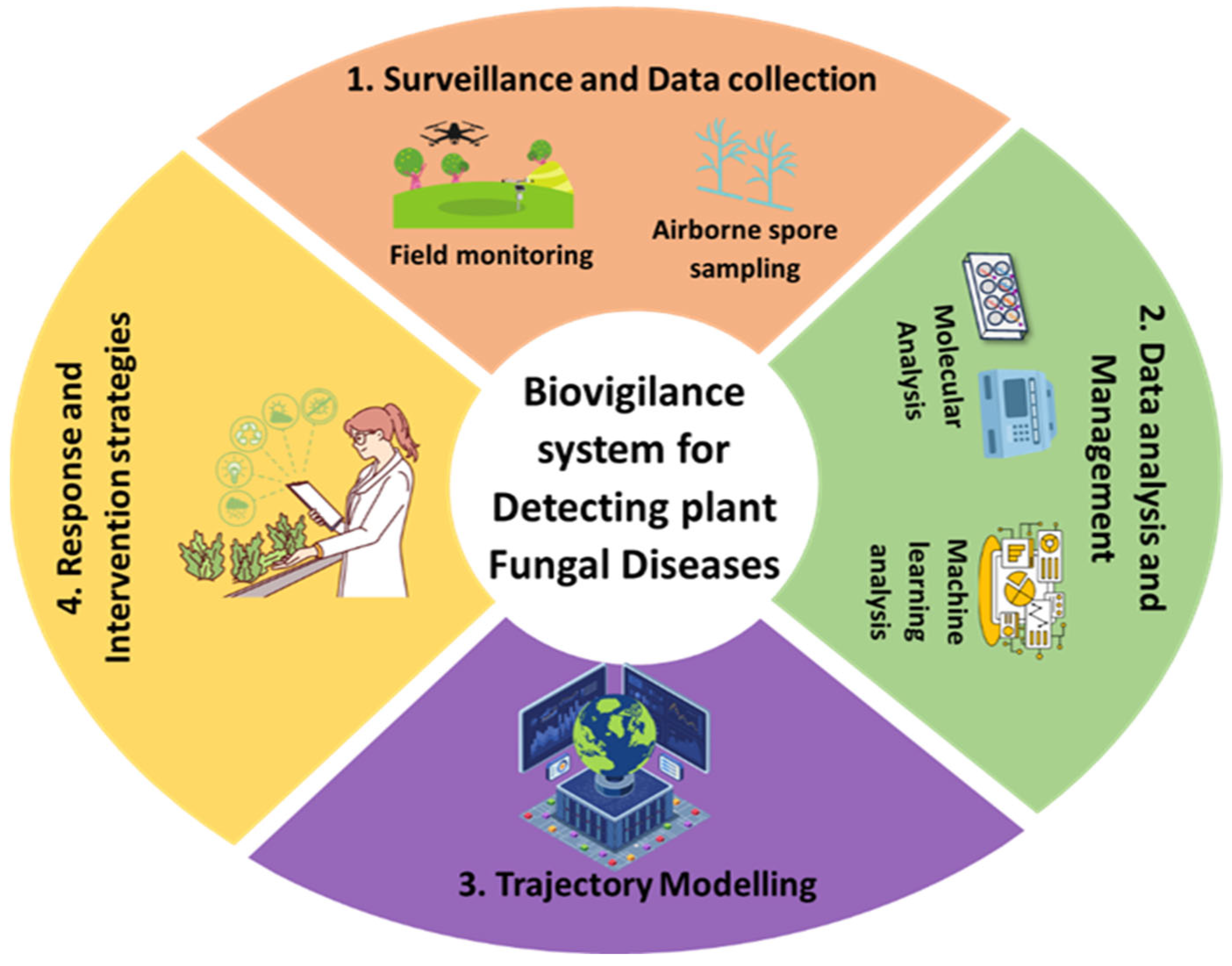
An effective biovigilance system comprises multiple interlinked components, including: (1) surveillance and data collection (e.g., field observations, airborne spore sampling, remote sensing, and meteorological monitoring); (2) data analysis and management (e.g., molecular diagnostics, machine learning algorithms); (3) trajectory and disease spread modeling; and (4) implementation of targeted response strategies [99] (Figure 8). A biovigilance programme should not only focus on developing mitigation strategies, but also include studying their current and potential threats [98]. Therefore, resources should be allocated to activities that focus on surveillance, data collection and data analysis for identifying current and emerging infections. By integrating machine learning algorithms, drones, molecular diagnostics and geospatial analytics, predictive models can uncover hidden transmission patterns and anticipate outbreaks before widespread transmission occurs [100]. Additionally, climate change also affects the diversity and virulence of pathogen populations. Therefore, special emphasis should be given to scaling up predictive models that combine climate change data, environmental variables, and trends in pathogen emergence. Such models can be adopted and optimized across regions using cloud-based platforms, making them scalable and applicable globally, especially in transboundary surveillance systems. Global-scale implementation will require international cooperation, data sharing frameworks, and capacity-building initiatives to ensure equitable access and utility in both high-income and low-resource countries. A biovigilance approach based on this prediction enables the early identification of fungal threats and supports informed decision-making for timely and targeted disease control.
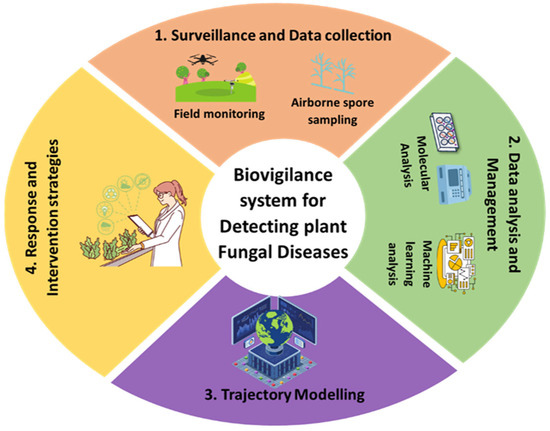
Figure 8.
An integrated biovigilance framework for the detection and management of plant fungal diseases. This circular model outlines four interconnected components of a biovigilance system: (1) Surveillance and Data Collection, involving field monitoring and airborne spore sampling using technologies such as drones and spore traps; (2) Data Analysis and Management, utilizing molecular tools and machine learning to interpret surveillance data; (3) Trajectory Modelling, which combines meteorological and environmental data to forecast the dispersal and spread of fungal pathogens; and (4) Response and Intervention Strategies, supporting informed decision-making for timely and targeted disease control. Together, these components enable early detection, precision management, and sustainable mitigation of fungal threats in dynamic agricultural and climatic contexts.
A vineyard biovigilance study demonstrated the effectiveness of integrating spore trapping networks, field-based surveillance, and decision-support tools informed by meteorological data. This approach enabled early pathogen detection, identification of high-risk infection windows, and optimized fungicide application timing [98]. Similarly, a large-scale biovigilance system was deployed to monitor rust fungi, tracking spatiotemporal patterns of spore abundance and diversity over multiple growing seasons across several provinces, thereby enhancing preparedness and regional disease forecasting [99].
Molecular technologies have further advanced biovigilance capacity. A clade-specific multiplex qPCR assay was developed for the downy mildew pathogen Pseudoperonospora cubensis, enabling detection of airborne sporangia 2–4 days before symptom onset in cucurbit fields. This assay differentiated between Clade 1 (targeting squash, pumpkin, and watermelon) and Clade 2 (targeting cucumber and cantaloupe), thereby improving precision in disease management [101]. A similar field-validated molecular system integrating spore trapping with qPCR was used to detect cucurbit downy mildew, significantly enhancing early-warning capabilities [102].
Genomic approaches are emerging as powerful tools in biovigilance. For example, Hamelin et al. [103] highlighted the potential of genomic biovigilance to detect cryptic evolutionary events, such as the identification of a novel sexual hybrid of Phytophthora ramorum. These genomic insights not only refine surveillance but also inform regulatory and containment strategies by revealing hidden pathways of pathogen evolution and dispersal.
Despite its promise, the implementation of biovigilance faces several challenges. High operational costs, the complexity of integrating diverse diagnostic platforms, and limited data infrastructure, especially in developing regions, hinder widespread adoption. Moreover, the dynamic evolution of fungal pathogens and increasing climatic unpredictability necessitate continual refinement of biovigilance systems to maintain their efficacy.
4. Cost and Feasibility of Advanced Detection Tools in Low-Resource Settings
The accessibility and affordability of advanced fungal disease detection technologies remain a significant challenge in low-resource settings, particularly in developing countries where smallholder farmers predominate in agricultural production. Many cutting-edge diagnostic tools, such as qPCR, NGS, and drone-based remote sensing, require significant financial investments in equipment, reagents, and technical expertise, making them impractical for widespread use in resource-limited regions. Addressing these constraints necessitates innovative approaches to improve affordability, scalability, and sustainability.
To facilitate the adoption of these technologies in resource-limited settings, public–private partnerships (PPPs) and government support may play a crucial role. Subsidized diagnostic programs, farmer cooperatives investing in shared disease surveillance tools, and collaborations between research institutions and technology companies can help bridge the affordability gap. Additionally, capacity-building initiatives are essential to ensure that farmers and extension workers can effectively use these technologies. Training programs on field-friendly diagnostic kits, mobile-based advisory platforms, and data-driven disease management strategies can enhance adoption and maximize the impact of these innovations.
Incorporating open-source tools and software can further reduce costs and promote transparency, while enabling local adaptation of diagnostic systems. Notably, mobile applications such as PlantVillage Nuru version 14.1 (https://plantvillage.psu.edu/ accessed on 18 December 2024) and Plantix version 4.9.8 represent practical and scalable models for the real-world deployment of plant disease diagnostics. These AI-driven platforms enable farmers to diagnose plant diseases, including those caused by fungal pathogens, using smartphone image recognition. Notably, these platforms are increasingly incorporating farmer-reported observations, indigenous knowledge, and traditional symptom descriptions into their databases and algorithms. By using farmer insights and practical knowledge, these digital tools could enhance diagnostic accuracy and relevance at the community level.
Additionally, supporting the local manufacturing of diagnostic kits and sensor components can reduce dependency on expensive imports, enhance supply chain resilience, and create economic opportunities within agricultural communities. By integrating cost-effective diagnostic tools, AI-powered mobile solutions, and community-driven surveillance networks, it is possible to improve the accessibility of fungal pathogen detection in developing regions. A multi-stakeholder approach—combining technological innovation, policy support, and grassroots implementation—can enhance global plant disease surveillance while making these solutions sustainable for low-income agricultural communities.
5. Advisory Services for Fungal Disease Management: A Strategic Response Framework
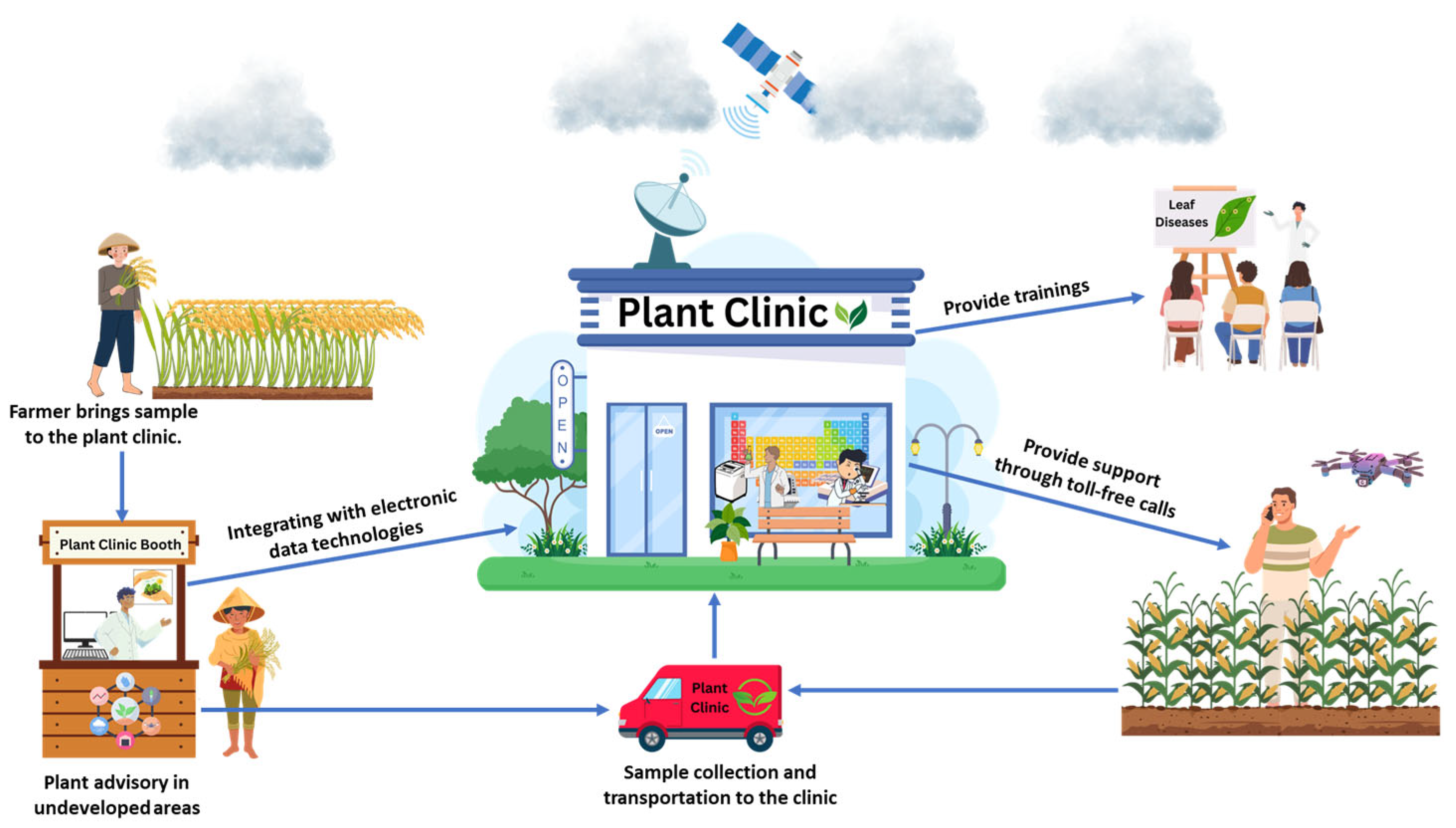
The lack of timely and cost-effective agricultural extension services presents significant challenges for farmers, reducing productivity and increasing crop losses due to extensive biotic stresses from emerging fungal pathogens. Sudden disease outbreaks, which can spread rapidly across borders, are often exacerbated by the absence of early identification systems and practical, cost-effective management solutions across developed, developing, and underdeveloped countries worldwide. Addressing this gap is crucial for safeguarding livelihoods and ensuring food security. Global experiences highlight the need for modern agricultural extension services to provide farmers with sustainable and science-based solutions. One effective initiative is the establishment of local plant clinics staffed by trained plant doctors (Figure 9). These clinics can play a crucial role in diagnosing farmers’ disease problems and providing them with actionable guidance on managing pests and diseases. Strengthening and integrating such clinics with digital disease surveillance systems can significantly enhance plant health management in developing regions. Expanding plant disease diagnostic services in underdeveloped areas empowers regulatory authorities by providing essential plant health information through accessible resources such as fact sheets, distribution maps, diagnostic tools, and pest control guidelines. Additionally, integrating these clinics with electronic data collection tools enables rapid recording and reporting of pathogen information, facilitating early disease warnings and timely interventions [104]. Studies have shown that these networks effectively establish and manage plant clinics, often providing valuable advisory services to farmers at minimal or no cost. Today, plant clinics offer cost-effective solutions and deliver structured services, positioning themselves as essential community resources. From individual clinics to global networks, these initiatives have worked closely with farmers to improve agricultural productivity and quality. Moreover, plant diagnostic networks play a critical role in food security and environmental sustainability by implementing Integrated Pest Management (IPM) and Integrated Nutrient Management (INM) programs, particularly in developed countries. With the advancement of internet-based technologies, some networks and clinics can now share data and information in real-time through toll-free telephony and free social media platforms, offering online advice and expanding farmers’ knowledge through innovative training materials in print and electronic formats. However, plant networks still face challenges in gaining widespread recognition as reliable service centers, particularly among traditional farmers in developing and underdeveloped countries. Governments and national organizations are responsible for introducing these networks to farmers and regulatory bodies, ensuring sustainable agriculture and maximizing income potential worldwide [105].
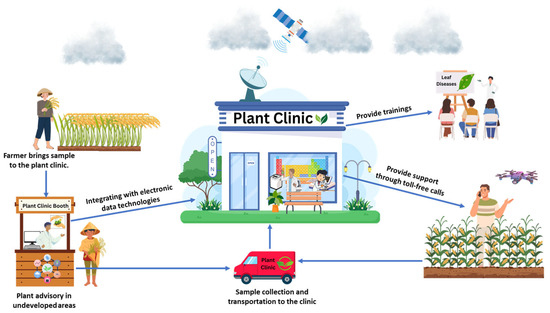
Figure 9.
Crucial roles of a plant disease clinic in supporting modern agricultural extension service to address emerging fungal stress challenges and improve plant health management. This comprehensive approach enables early disease detection, rapid dissemination of information, and accurate recommendations for pest and disease management. Plant clinics provide accessible and cost-effective support systems that integrate field observations with expert diagnoses and technology-driven tools, offering science-based solutions to enhance crop productivity and strengthen food security, particularly in emerging and low-income regions.
PPPs can be highly effective and beneficial when the public and private sectors collaborate. These partnerships harness the strengths of each participant and address the needs of various stakeholders, including extension services, public sector organizations, industry representatives, and crop commodity groups. When feedback from growers identifies a recognized need and consensus is reached on combining complementary skills, expertise, and resources, a solid foundation for successful PPPs is established. However, the development and success of these partnerships rely more on nontechnical skills and strong professional relationships, particularly trust, communication, and a shared pathway to success, than on detailed scientific knowledge [106].
For example, in November 2004, Asian soybean rust (ASR), caused by Phakopsora pachyrhizi, was first identified in the United States [107,108,109]. This pathogen had already led to significant yield losses in South America, prompting the USDA to estimate potential economic losses in the U.S. ranging from $240 million to $2 billion [110]. Scientists recognized ASR as a serious threat to North American soybeans. In response, several public–private partnerships were formed. Federal agencies, agrochemical companies, and Extension plant pathologists worked together to ensure that fungicides were available for the 2005 growing season, resulting in Section 18 exemptions for eight fungicides [111]. Over the following eight years, various extension events and hands-on workshops trained more than 700 participants in identifying and managing ASR. These partnerships successfully detected and provided synergistic benefits to all parties involved, ultimately creating a win-win situation for growers.
Another example of a successful PPP is the development of the wheat blast disease management strategy in Bangladesh. Wheat blast, caused by the fungus Magnaporthe oryzae pathotype triticum, posed a significant threat to wheat production in South Asia [10]. In response, a collaboration was established among international research organizations, such as the International Maize and Wheat Improvement Center (CIMMYT), national agricultural research institutions, and private seed companies. This partnership led to the rapid development and dissemination of blast-resistant wheat varieties, along with targeted recommendations for fungicides [112,113]. The joint effort between public and private entities ensured the successful transfer of technology, farmer training, and deployment of effective disease management strategies. Additionally, the partnership between the Bill & Melinda Gates Foundation, research institutions, and private agricultural companies has played a critical role in combating late blight in potatoes. Through initiatives such as the Feed the Future Biotechnology Potato Partnership, genetically engineered late blight-resistant potato varieties were developed and tested in collaboration with local farmers [114]. This initiative demonstrated how PPPs could accelerate the adoption of innovative agricultural solutions while ensuring that smallholder farmers benefit from new technologies. These examples highlight how PPPs can drive rapid responses to emerging plant disease threats, enhance resource mobilization, and facilitate knowledge exchange between public research institutions and private industry. Strengthening these partnerships with long-term commitments and clear frameworks can further improve their impact on plant disease management.
6. Key Issues and Challenges in the Adoption of Tech-Driven Approaches
Over the past decade, significant progress has been made in diagnosing emerging fungal infections in plants, largely due to the development of innovative technologies. Although the integration of these technologies into standard agricultural diagnostics has begun, challenges still remain in effectively detecting fungal pathogens. PCR, which relies on a thermal cycler and DNA polymerases, cannot be performed at room temperature in a solution, on a solid base, or in a mixed biological environment (e.g., on the cell surface or inside a cell) [115]. RCA, though useful, involves multiple discrete steps, making it less suitable for rapid diagnostics. Additionally, sample matrix interference may require extensive preparation, while digital signal readout increases complexity and cost. High multiplexing also complicates data processing and liquid handling, limiting its use in routine diagnostics [116]. LAMP appears to be a promising method for point-of-care (POC) detection, offering high sensitivity and specificity without significantly increasing analytical costs. However, it requires careful optimization and skilled handling, necessitating expertise in genetics and diagnostics for proper implementation [37]. Although software exists for LAMP primer design, many programs are not freely accessible, limiting their usability in developing countries [117]. Given the high cost of antifungal fungicides, the expense of next-generation sequencing (NGS) for detecting fungal pathogens is relatively justifiable. However, a structured NGS framework is needed, including protocols for sample collection, nucleic acid extraction, quality control, and standardized result interpretation [118]. The application of drones for disease detection also presents challenges, primarily in dataset quality and model development. Dataset-related issues include image deformations, limited expert-labeled data, strong randomness, and insufficient class diversity. Model-building challenges involve small training sample sizes, long training times, and extended processing durations [43]. Considering these technological limitations, clinicians should actively incorporate these diagnostic tools when fungal infections are suspected. Additionally, a clinical consensus should be established to ensure that technological advancements align with practical applications.
The primary challenge for developing countries in utilizing advanced technologies for plant disease detection is the high cost. Commercial tools from developed nations are frequently too expensive for smallholder farmers in poorer countries. Many cutting-edge diagnostic tools, such as qPCR, NGS, and drone-based remote sensing, require significant financial investments. Additionally, a considerable knowledge gap exists in how these farmers can effectively use these tools and techniques. Therefore, a multifaceted approach is necessary to make modern diagnostic and surveillance innovations accessible and affordable. Research organizations and governments should invest in budget-friendly diagnostic kits, like portable PCR tools and paper-based assays. Farmer cooperatives investing in shared disease surveillance tools can help bridge the affordability gap. Open-source AI and remote sensing can also provide cost-effective monitoring solutions. Public–private partnerships can subsidize costs and enhance access to new technologies. Additionally, skill-building training is essential to ensure that farmers and extension workers can effectively use these technologies. A multi-stakeholder approach—combining technological innovation, policy support, and grassroots implementation—can enhance plant disease surveillance and monitoring, making these solutions sustainable for low-income agricultural communities.
7. Conclusions and Future Perspective
Emerging fungal plant pathogens represent one of the most serious biotic threats to crop plants. Advances in the diagnosis and surveillance of these pathogens have highlighted the importance of integrating modern technologies with traditional farming practices to mitigate the local and global socioeconomic impacts of fungal diseases. This review has highlighted the rapid evolution of detection methods, including remote sensing, sensor-based technologies, PCR, and NGS, which are revolutionizing our ability to monitor, identify, and respond to fungal plant pathogens at unprecedented speeds and scales.
While these innovations represent remarkable progress, the challenges of global implementation, resource accessibility, and technical capacity remain significant, particularly for farmers in developing regions. The urgency of developing scalable, affordable, and easy-to-use diagnostic tools cannot be overstated. Strengthening extension services and digital advisory platforms is vital to ensure that even the most vulnerable farming communities receive timely and actionable insights.
Looking forward, a multidisciplinary approach that combines advanced diagnostics, big data analytics, and global surveillance systems is essential. Equally critical are national and international efforts to promote data sharing, policy alignment, and equitable access to new technologies. Integrating climate adaptation strategies into plant health policies will also be crucial, as changing climate conditions increasingly influence pathogen dynamics, geographic spread, and outbreak intensity. Investment in research to enhance detection sensitivity, reduce costs, and develop portable, user-friendly tools will empower local communities to address these challenges directly.
Ultimately, combating the rising threats of emerging fungal plant diseases requires more than just technological advancements. It demands a global commitment to fostering resilient agricultural systems. By prioritizing innovation, accessibility, and global cooperation, disease surveillance can be transformed into a cornerstone of food security and ensure sustainable agriculture for future generations.
Author Contributions
Conceptualization, M.M.H. and F.S.; methodology, software M.M.; validation, H.F. and N.A.; formal analysis, investigation, resources, data curation, M.M.H., F.S., M.M., H.F. and N.A.; writing—original draft preparation, M.M.H., F.S., N.A. and M.M.; writing—review and editing, M.M.H., F.S., M.M., N.A. and H.F.; visualization, F.S. and M.M.; supervision, M.M.H.; project administration, M.M.H.; funding acquisition, M.M.H. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
Not applicable.
Acknowledgments
We are grateful to the Ministry of Science and Technology, Bangladesh, for supporting this research.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- FAO; IFAD; UNICEF; WFP; WHO. The State of Food Security and Nutrition in the World 2018; Building climate resilience for food security and nutrition; FAO: Rome, Italy, 2018; Licence: CC BY-NC-SA 3.0 IGO; Available online: https://creativecommons.org/licenses/by-nc-sa/3.0/igo (accessed on 22 December 2024).
- Shuping, D.S.S.; Eloff, J.N. The Use of Plants to Protect Plants and Food Against Fungal Pathogens: A Review. Afr. J. Tradit. Complement. Altern. Med. 2017, 14, 120–127. [Google Scholar] [CrossRef] [PubMed]
- Doehlemann, G.; Ökmen, B.; Zhu, W.; Sharon, A. Plant Pathogenic Fungi. Fungal Kingd. 2017, 5, 701–726. [Google Scholar] [CrossRef]
- Malcolm, G.M.; Kuldau, G.A.; Gugino, B.K.; Jiménez-Gasco, M.D.M. Hidden Host Plant Associations of Soilborne Fungal Pathogens: An Ecological Perspective. Phytopathology 2013, 103, 538–544. [Google Scholar] [CrossRef]
- Fisher, M.C.; Gurr, S.J.; Cuomo, C.A.; Blehert, D.S.; Jin, H.; Stukenbrock, E.H.; Cowen, L.E. Threats posed by the fungal kingdom to humans, wildlife, and agriculture. MBio 2020, 11, 10–1128. [Google Scholar] [CrossRef]
- Narayanasamy, P.; Narayanasamy, P. Diagnosis of fungal diseases of plants. In Microbial Plant Pathogens-Detection and Disease Diagnosis: Fungal Pathogens; Springer: Dordrecht, The Netherlands, 2011; Volume 1, pp. 273–284. [Google Scholar] [CrossRef]
- Iqbal, Z.; Khan, M.A.; Sharif, M.; Shah, J.H.; ur Rehman, M.H.; Javed, K. An automated detection and classification of citrus plant diseases using image processing techniques: A review. Comput. Electron. Agric. 2018, 153, 12–32. [Google Scholar] [CrossRef]
- Rodriguez-Moreno, L.; Ebert, M.K.; Bolton, M.D.; Thomma, B.P. Tools of the crook-infection strategies of fungal plant pathogens. Plant J. 2018, 93, 664–674. [Google Scholar] [CrossRef]
- Hossain, M.M.; Sultana, F.; Mostafa, M.; Ferdus, H.; Rahman, M.; Rana, J.A.; Al Sabbir, M.A. Plant disease dynamics in a changing climate: Impacts, molecular mechanisms, and climate-informed strategies for sustainable management. Discov. Agric. 2024, 2, 132. [Google Scholar] [CrossRef]
- Hossain, M.M. Wheat blast: A review from a genetic and genomic perspective. Front. Microbiol. 2022, 13, 983243. [Google Scholar] [CrossRef]
- Mutiga, S.K.; Hoffmann, V.; Harvey, J.W.; Milgroom, M.G.; Nelson, R.J. Assessment of aflatoxin and fumonisin contamination of maize in western Kenya. Phytopathology 2015, 105, 1250–1261. [Google Scholar] [CrossRef] [PubMed]
- Prova, A.; Akanda, A.M.; Islam, S.; Hossain, M.M. Characterization of Sclerotinia sclerotiorum, an Emerging Fungal Pathogen Causing Blight in Hyacinth Bean (Lablab purpureus). Plant Pathol. J. 2018, 34, 367–380. [Google Scholar] [CrossRef]
- Jahan, R.; Siddique, S.S.; Jannat, R.; Hossain, M.M. Cosmos white rot: First characterization, physiology, host range, disease resistance, and chemical control. J Basic Microbiol 2022, 62, 911–929. [Google Scholar] [CrossRef]
- Hossain, M.M.; Sultana, F.; Li, W.; Tran, L.-S.P.; Mostofa, M.G. Sclerotinia sclerotiorum (Lib.) de Bary: Insights into the Pathogenomic Features of a Global Pathogen. Cells 2023, 12, 1063. [Google Scholar] [CrossRef]
- Islam, M.R.; Akanda, A.M.; Hossain, M.M.; Hossain, M.M. First Characterization of a Newly Emerging Phytopathogen, Sclerotinia sclerotiorum Causing White Mold in Pea. J. Basic Microbiol. 2021, 61, 923–939. [Google Scholar] [CrossRef]
- Avelino, J.; Cristancho, M.; Georgiou, S.; Imbach, P.; Aguilar, L.; Bornemann, G.; Morales, C. The Coffee Rust Crises in Colombia and Central America (2008–2013): Impacts, Plausible Causes and Proposed Solutions. Food Secur. 2015, 7, 303–321. [Google Scholar] [CrossRef]
- Sieff, K. The Migration Problem Is a Coffee Problem. The Washington Post, 11 June 2019. Available online: https://www.washingtonpost.com/world/2019/06/11/falling-coffee-prices-drive-guatemalan-migration-united-states/ (accessed on 7 October 2019).
- Stokstad, E. Devastating Banana Disease May Have Reached Latin America, Could Drive Up Global Prices. Science 2019, 365, 207–208. [Google Scholar] [CrossRef]
- Vurro, M.; Bonciani, B.; Vannacci, G. Emerging Infectious Diseases of Crop Plants in Developing Countries: Impact on Agriculture and Socio-Economic Consequences. Food Secur. 2010, 2, 113–132. [Google Scholar] [CrossRef]
- Gewin, V. Bioterrorism: Agriculture shock. Nature 2003, 421, 106–109. [Google Scholar] [CrossRef]
- Dutech, C.; Barrès, B.; Bridier, J.; Robin, C.; Milgroom, M.G.; Ravigné, V. The chestnut blight fungus world tour: Successive introduction events from diverse origins in an invasive plant fungal pathogen. Mol. Ecol. 2012, 21, 3931–3946. [Google Scholar] [CrossRef] [PubMed]
- Hovmøller, M.S.; Rodriguez-Algaba, J.; Thach, T.; Sørensen, C.K. Race typing of Puccinia striiformis on wheat. In Wheat Rust Diseases: Methods and Protocols; Springer: New York, NY, USA, 2017; pp. 29–40. [Google Scholar] [CrossRef]
- Abdissa, T.; Fininsa, C.; Abeyo, B.; Woldeab, G. Virulence analysis of wheat stem rust (Puccinia graminis f. sp. tritici) in Horro Guduru Wollega, Western Ethiopia. J. Phytopathol. 2022, 170, 315–325. [Google Scholar] [CrossRef]
- Fones, H.N.; Fisher, M.C.; Gurr, S.J. Emerging Fungal Threats to Plants and Animals Challenge Agriculture and Ecosystem Resilience. Microbiol. Spectr. 2017, 5, 787–809. [Google Scholar] [CrossRef] [PubMed]
- Friesen, T.L.; Stukenbrock, E.H.; Liu, Z.; Meinhardt, S.; Ling, H.; Faris, J.D.; Rasmussen, J.B.; Solomon, P.S.; McDonald, B.A.; Oliver, R.P. Emergence of a new disease as a result of interspecific virulence gene transfer. Nat. Genet. 2006, 38, 953–956. [Google Scholar] [CrossRef] [PubMed]
- Gladieux, P.; Feurtey, A.; Hood, M.E.; Snirc, A.; Clavel, J.; Dutech, C.; Roy, M.; Giraud, T. The Population Biology of Fungal Invasions. In Invasion Genetics; Barrett, S.C.H., Colautti, R.I., Dlugosch, K.M., Rieseberg, L.H., Eds.; Wiley: Hoboken, NJ, USA, 2016; pp. 81–100. [Google Scholar] [CrossRef]
- Ceresini, P.C.; Castroagudín, V.L.; Rodrigues, F.Á.; Rios, J.A.; Aucique-Pérez, C.E.; Moreira, S.I.; Alves, E.; Croll, D.; Maciel, J.L.N. Wheat blast: Past, present, and future. Annu. Rev. Phytopathol. 2018, 56, 427–456. [Google Scholar] [CrossRef] [PubMed]
- Capote, N.; Pastrana, A.M.; Aguado, A.; Sánchez-Torres, P. Molecular Tools for Detection of Plant Pathogenic Fungi and Fungicide Resistance. Plant Pathol. 2012, 12, 151–202. [Google Scholar] [CrossRef]
- Rollins, L.; Coats, K.; Elliott, M.; Chastagner, G. Comparison of five detection and quantification methods for Phytophthora ramorum in stream and irrigation water. Plant Dis. 2016, 100, 1202–1211. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Sharma, S. Paradigm Shift in Plant Disease Diagnostics: A Journey from Conventional Diagnostics to Nano-Diagnostics. In Current Trends in Plant Disease Diagnostics and Management Practices; Springer: Cham, Switzerland, 2016; pp. 237–264. [Google Scholar] [CrossRef]
- Luchi, N.; Ioos, R.; Santini, A. Fast and Reliable Molecular Methods to Detect Fungal Pathogens in Woody Plants. Appl. Microbiol. Biotechnol. 2020, 104, 2453–2468. [Google Scholar] [CrossRef]
- Schmale III, D.G.; Ross, S.D. Highways in the Sky: Scales of Atmospheric Transport of Plant Pathogens. Annu. Rev. Phytopathol. 2015, 53, 591–611. [Google Scholar] [CrossRef]
- Lievens, B.; Claes, L.; Vanachter, A.C.; Cammue, B.P.; Thomma, B.P. Detecting single nucleotide polymorphisms using DNA arrays for plant pathogen diagnosis. FEMS Microbiol. Lett. 2006, 255, 129–139. [Google Scholar] [CrossRef]
- Adams, I.P.; Fox, A.; Boonham, N.; Massart, S.; De Jonghe, K. The impact of high throughput sequencing on plant health diagnostics. Eur. J. Plant Pathol. 2018, 152, 909–919. [Google Scholar] [CrossRef]
- Le, D.T.; Vu, N.T. Progress of Loop-Mediated Isothermal Amplification Technique in Molecular Diagnosis of Plant Diseases. Appl. Biol. Chem. 2017, 60, 169–180. [Google Scholar] [CrossRef]
- Li, Z.; Paul, R.; Ba Tis, T.; Saville, A.C.; Hansel, J.C.; Yu, T.; Ristaino, J.B.; Wei, Q. Non-Invasive Plant Disease Diagnostics Enabled by Smartphone-Based Fingerprinting of Leaf Volatiles. Nat. Plants 2019, 5, 856–866. [Google Scholar] [CrossRef]
- Notomi, T.; Mori, Y.; Tomita, N.; Kanda, H. Loop-mediated isothermal amplification (LAMP): Principle, features, and future prospects. J. Microbiol. 2015, 53, 1–5. [Google Scholar] [CrossRef]
- O’Shea, J. Digital disease detection: A systematic review of event-based internet biosurveillance systems. Int. J. Med. Inform. 2017, 101, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Tateosian, L.; Guenter, R.; Yang, Y.P.; Ristaino, J. Tracking 19th Century Late Blight from Archival Documents Using Text Analytics and Geoparsing. In Free and Open Source Software for Geospatial (FOSS4G) Conference Proceedings; the Open Source Geospatial Foundation: Beaverton, OR, USA, 2017; Volume 17, p. 17. [Google Scholar] [CrossRef]
- Agrawal, S.; Azar, P.D.; Lo, A.W.; Singh, T. Momentum, mean-reversion, and social media: Evidence from StockTwits and Twitter. J. Portf. Manag. 2018, 44, 85–95. [Google Scholar] [CrossRef]
- Mahlein, A.K. Plant Disease Detection by Imaging Sensors–Parallels and Specific Demands for Precision Agriculture and Plant Phenotyping. Plant Dis. 2016, 100, 241–251. [Google Scholar] [CrossRef] [PubMed]
- Franke, J.; Menz, G. Multi-temporal wheat disease detection by multi-spectral remote sensing. Precis. Agric. 2007, 8, 161–172. [Google Scholar] [CrossRef]
- Chin, R.; Catal, C.; Kassahun, A. Plant Disease Detection Using Drones in Precision Agriculture. Precis. Agric. 2023, 24, 1663–1682. [Google Scholar] [CrossRef]
- Abdulridha, J.; Min, A.; Rouse, M.N.; Kianian, S.; Isler, V.; Yang, C. Evaluation of Stem Rust Disease in Wheat Fields by Drone Hyperspectral Imaging. Sensors 2023, 23, 4154. [Google Scholar] [CrossRef] [PubMed]
- Parnell, S.; van den Bosch, F.; Gottwald, T.; Gilligan, C.A. Surveillance to inform control of emerging plant diseases: An epidemiological perspective. Annu. Rev. Phytopathol. 2017, 55, 591–610. [Google Scholar] [CrossRef]
- Ma, Z.; Michailides, T.J. Approaches for Eliminating PCR Inhibitors and Designing PCR Primers for the Detection of Phytopathogenic Fungi. Crop. Prot. 2007, 26, 145–161. [Google Scholar] [CrossRef]
- Sikdar, P.; Okubara, P.A.; Mazzola, M.; Xiao, C.L. Development of PCR Assays for Diagnosis and Detection of the Pathogens Phacidiopycnis washingtonensis and Sphaeropsis pyriputrescens in Apple Fruit. Plant Dis. 2014, 98, 241–246. [Google Scholar] [CrossRef]
- Yang, X.; Hameed, U.; Zhang, A.F.; Zang, H.Y.; Gu, C.Y.; Chen, Y.; Xu, Y.L. Development of a Nested-PCR Assay for the Rapid Detection of Pilidiella granati in Pomegranate Fruit. Sci. Rep. 2017, 7, 40954. [Google Scholar] [CrossRef] [PubMed]
- Conti, M.; Cinget, B.; Vivancos, J.; Oudemans, P.; Bélanger, R.R. A Molecular Assay Allows the Simultaneous Detection of 12 Fungi Causing Fruit Rot in Cranberry. Plant Dis. 2019, 103, 2843–2850. [Google Scholar] [CrossRef]
- Ji Cho, H.; Hong, S.W.; Kim, H.J.; Kwak, Y.S. Development of a multiplex PCR method to detect fungal pathogens for quarantine on exported cacti. Plant Pathol. J. 2016, 32, 53. [Google Scholar] [CrossRef][Green Version]
- Chandelier, A.; Massot, M.; Fabreguettes, O.; Gischer, F.; Teng, F.; Robin, C. Early Detection of Cryphonectria parasitica by Real-Time PCR. Eur. J. Plant Pathol. 2019, 153, 29–46. [Google Scholar] [CrossRef]
- Havis, N.D.; Gorniak, K.; Carmona, M.A.; Formento, A.N.; Luque, A.G.; Scandiani, M.M. First molecular detection of Ramularia leaf spot (Ramularia collo-cygni) in seeds and leaves of barley in Argentina. Plant Dis. 2014, 98, 277. [Google Scholar] [CrossRef]
- Pecchia, S.; Caggiano, B.; Da Lio, D.; Cafà, G.; Le Floch, G.; Baroncelli, R. Molecular detection of the seed-borne pathogen Colletotrichum lupini targeting the hyper-variable IGS region of the ribosomal cluster. Plants 2019, 8, 222. [Google Scholar] [CrossRef]
- Konstantinova, P.; Bonants, P.J.; Van Gent-Pelzer, M.P.; Van Der Zouwen, P. Development of Specific Primers for Detection and Identification of Alternaria spp. in Carrot Material by PCR and Comparison with Blotter and Plating Assays. Mycol. Res. 2002, 106, 23–33. [Google Scholar] [CrossRef]
- Ha, Y.; Fessehaie, A.; Ling, K.S.; Wechter, W.P.; Keinath, A.P.; Walcott, R.R. Simultaneous detection of Acidovorax avenae subsp. citrulli and Didymella bryoniae in cucurbit seedlots using magnetic capture hybridization and real-time polymerase chain reaction. Phytopathology 2009, 99, 666–678. [Google Scholar] [CrossRef]
- Depotter, J.R.; Rodriguez-Moreno, L.; Thomma, B.P.; Wood, T.A. The Emerging British Verticillium longisporum Population Consists of Aggressive Brassica Pathogens. Phytopathology 2017, 107, 1399–1405. [Google Scholar] [CrossRef] [PubMed]
- Nikitin, D.A.; Ivanova, E.A.; Semenov, M.V.; Zhelezova, A.D.; Ksenofontova, N.A.; Tkhakakhova, A.K.; Kholodov, V.A. Diversity, Ecological Characteristics and Identification of Some Problematic Phytopathogenic Fusarium in Soil: A Review. Diversity 2023, 15, 49. [Google Scholar] [CrossRef]
- Liebe, S.; Christ, D.S.; Ehricht, R.; Varrelmann, M. Development of a DNA Microarray-Based Assay for the Detection of Sugar Beet Root Rot Pathogens. Phytopathology 2016, 106, 76–86. [Google Scholar] [CrossRef]
- Wöhrle, J.; Krämer, S.D.; Meyer, P.A.; Rath, C.; Hügle, M.; Urban, G.A.; Roth, G. Digital DNA Microarray Generation on Glass Substrates. Sci. Rep. 2020, 10, 5770. [Google Scholar] [CrossRef] [PubMed]
- Chalupowicz, L.; Dombrovsky, A.; Gaba, V.; Luria, N.; Reuven, M.; Beerman, A.; Manulis-Sasson, S. Diagnosis of Plant Diseases Using the Nanopore Sequencing Platform. Plant Pathol. 2019, 68, 229–238. [Google Scholar] [CrossRef]
- Potgieter, L.; Feurtey, A.; Dutheil, J.Y.; Stukenbrock, E.H. On variant discovery in genomes of fungal plant pathogens. Front. Microbiol. 2020, 11, 626. [Google Scholar] [CrossRef]
- Huang, J.; Jiang, E.; Yang, D.; Wei, J.; Zhao, M.; Feng, J.; Cao, J. Metagenomic next-generation sequencing versus traditional pathogen detection in the diagnosis of peripheral pulmonary infectious lesions. Infect. Drug Resist. 2020, 13, 567–576. [Google Scholar] [CrossRef]
- Boykin, L.M.; Sseruwagi, P.; Alicai, T.; Ateka, E.; Mohammed, I.U.; Stanton, J.A.L.; Ndunguru, J. Tree lab: Portable genomics for early detection of plant viruses and pests in sub-Saharan Africa. Genes 2019, 10, 632. [Google Scholar] [CrossRef]
- Radhakrishnan, G.V.; Cook, N.M.; Bueno-Sancho, V.; Lewis, C.M.; Persoons, A.; Mitiku, A.D.; Saunders, D.G. MARPLE, a point-of-care, strain-level disease diagnostics and surveillance tool for complex fungal pathogens. BMC Biol. 2019, 17, 65. [Google Scholar] [CrossRef]
- Goodwin, S.; McPherson, J.D.; McCombie, W.R. Coming of age: Ten years of next-generation sequencing technologies. Nat. Rev. Genet. 2016, 17, 333–351. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, G.L.; MacCannell, D.R.; Taylor, J.; Carleton, H.A.; Neuhaus, E.B.; Bradbury, R.S.; Posey, J.E.; Gwinn, M. Pathogen genomics in public health. N. Engl. J. Med. 2019, 381, 2569–2580. [Google Scholar] [CrossRef] [PubMed]
- Malapi-Wight, M.; Salgado-Salazar, C.; Demers, J.E.; Clement, D.L.; Rane, K.K.; Crouch, J.A. Sarcococca Blight: Use of Whole-Genome Sequencing for Fungal Plant Disease Diagnosis. Plant Dis. 2016, 100, 1093–1100. [Google Scholar] [CrossRef]
- Hubbard, A.; Lewis, C.M.; Yoshida, K.; Ramirez-Gonzalez, R.H.; de Vallavieille-Pope, C.; Thomas, J.; Saunders, D.G. Field pathogenomics reveals the emergence of a diverse wheat yellow rust population. Genome Biol. 2015, 16, 23. [Google Scholar] [CrossRef]
- Withers, S.; Gongora-Castillo, E.; Gent, D.; Thomas, A.; Ojiambo, P.S.; Quesada-Ocampo, L.M. Using Next-Generation Sequencing to Develop Molecular Diagnostics for Pseudoperonospora cubensis, the Cucurbit Downy Mildew Pathogen. Phytopathology 2016, 106, 1105–1116. [Google Scholar] [CrossRef]
- Chang, C.C.; Chen, C.C.; Wei, S.C.; Lu, H.H.; Liang, Y.H.; Lin, C.W. Diagnostic Devices for Isothermal Nucleic Acid Amplification. Sensors 2012, 12, 8319–8337. [Google Scholar] [CrossRef]
- Van Beckhoven, J.R.C.M.; Stead, D.E.; Van der Wolf, J.M. Detection of Clavibacter michiganensis subsp. sepedonicus by AmpliDet RNA, a New Technology Based on Real-Time Monitoring of NASBA Amplicons with a Molecular Beacon. J. Appl. Microbiol. 2002, 93, 840–849. [Google Scholar] [CrossRef]
- Heo, S.; Kim, H.R.; Lee, H.J. Development of a quantitative real-time nucleic acid sequence-based amplification (NASBA) assay for early detection of apple scar skin viroid. Plant Pathol. J. 2019, 35, 164. [Google Scholar] [CrossRef]
- Corthell, J.T. In Situ Hybridization. In Basic Molecular Protocols in Neuroscience: Tips, Tricks, and Pitfalls; Academic Press: San Diego, CA, USA, 2014; pp. 105–111. [Google Scholar] [CrossRef]
- Ellison, M.A.; McMahon, M.B.; Bonde, M.R.; Palmer, C.L.; Luster, D.G. In Situ Hybridization for the Detection of Rust Fungi in Paraffin Embedded Plant Tissue Sections. Plant Methods 2016, 12, 37. [Google Scholar] [CrossRef] [PubMed]
- Shakoori, A.R. Fluorescence In Situ Hybridization (FISH) and Its Applications. In Chromosome Structure and Aberrations; Springer: Berlin/Heidelberg, Germany, 2017; pp. 343–367. [Google Scholar] [CrossRef]
- Milner, H.; Ji, P.; Sabula, M.; Wu, T. Quantitative Polymerase Chain Reaction (Q-PCR) and Fluorescent In Situ Hybridization (FISH) Detection of Soilborne Pathogen Sclerotium rolfsii. Appl. Soil Ecol. 2019, 136, 86–92. [Google Scholar] [CrossRef]
- Panno, S.; Matić, S.; Tiberini, A.; Caruso, A.G.; Bella, P.; Torta, L.; Davino, S. Loop mediated isothermal amplification: Principles and applications in plant virology. Plants 2020, 9, 461. [Google Scholar] [CrossRef]
- Dong, J.; Feng, W.; Lin, M.; Chen, S.; Liu, X.; Wang, X.; Chen, Q. Comparative evaluation of PCR-based, LAMP and RPA-CRISPR/Cas12a assays for the rapid detection of Diaporthe aspalathi. Int. J. Mol. Sci. 2024, 25, 5773. [Google Scholar] [CrossRef] [PubMed]
- Tomlinson, J.A.; Boonham, N.; Dickinson, M. Development and evaluation of a one-hour DNA extraction and loop-mediated isothermal amplification assay for rapid detection of phytoplasmas. Plant Pathol. 2010, 59, 465–471. [Google Scholar] [CrossRef]
- Kaczmarek, A.M.; King, K.M.; West, J.S.; Stevens, M.; Sparkes, D.; Dickinson, M.J. A Loop-Mediated Isothermal Amplification (LAMP) Assay for Rapid and Specific Detection of Airborne Inoculum of Uromyces betae (Sugar Beet Rust). Plant Dis. 2019, 103, 417–421. [Google Scholar] [CrossRef]
- Stehlíková, D.; Luchi, N.; Aglietti, C.; Pepori, A.L.; Diez, J.J.; Santini, A. Real-Time Loop-Mediated Isothermal Amplification Assay for Rapid Detection of Fusarium circinatum. Biotechniques 2020, 69, 11–17. [Google Scholar] [CrossRef]
- Hossain, M.M.; Sultana, F.; Rubayet, M.T.; Khan, S.; Mostafa, M.; Mishu, N.J.; Mostofa, M.G. White Mold: A Global Threat to Crops and Key Strategies for Its Sustainable Management. Microorganisms 2024, 13, 4. [Google Scholar] [CrossRef]
- Ordóñez, N.; Salacinas, M.; Mendes, O.; Seidl, M.F.; Meijer, H.J.G.; Schoen, C.D.; Kema, G.H.J. A loop-mediated isothermal amplification (LAMP) assay based on unique markers derived from genotyping by sequencing data for rapid in planta diagnosis of Panama disease caused by Tropical Race 4 in banana. Plant Pathol. 2019, 68, 1682–1693. [Google Scholar] [CrossRef]
- Panno, S.; Ruiz-Ruiz, S.; Caruso, A.G.; Alfaro-Fernandez, A.; San Ambrosio, M.I.F.; Davino, S. Real-time reverse transcription polymerase chain reaction development for rapid detection of Tomato brown rugose fruit virus and comparison with other techniques. PeerJ 2019, 7, e7928. [Google Scholar] [CrossRef] [PubMed]
- Lucas-Bautista, J.A.; Bautista-Baños, S.; Ventura-Aguilar, R.I.; Hernández-López, M. Chitinases in Plants and Possible Use as Biomarkers for the Design of Biosensors in the Detection of Phytopathogenic Fungi. Rev. Mex. Cienc. Agríc. 2022, 13, 701–713. [Google Scholar]
- Goo, N.I.; Kim, D.E. Rolling circle amplification as isothermal gene amplification in molecular diagnostics. Biochip J. 2016, 10, 262–271. [Google Scholar] [CrossRef]
- Gu, L.; Yan, W.; Liu, L.; Wang, S.; Zhang, X.; Lyu, M. Research progress on rolling circle amplification (RCA)-based biomedical sensing. Pharmaceuticals 2018, 11, 35. [Google Scholar] [CrossRef]
- van Emmerik, C.L.; Gachulincova, I.; Lobbia, V.R.; Daniëls, M.A.; Heus, H.A.; Soufi, A.; van Ingen, H. Ramified Rolling Circle Amplification for Synthesis of Nucleosomal DNA Sequences. Anal. Biochem. 2020, 588, 113469. [Google Scholar] [CrossRef]
- Lin, H.; Jiang, X.; Yi, J.; Wang, X.; Zuo, R.; Jiang, Z.; Wang, W.; Zhou, E. Molecular Identification of Neofabraea Species Associated with Bull’s-Eye Rot on Apple Using Rolling-Circle Amplification of Partial EF-1α Sequence. Can. J. Microbiol. 2018, 64, 57–68. [Google Scholar] [CrossRef] [PubMed]
- Davari, M.; van Diepeningen, A.D.; Babai-Ahari, A.; Arzanlou, M.; Najafzadeh, M.J.; van der Lee, T.A.; de Hoog, G.S. Rapid Identification of Fusarium graminearum Species Complex Using Rolling Circle Amplification (RCA). J. Microbiol. Methods 2012, 89, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Scott, P.; Bader, M.K.F.; Burgess, T.; Hardy, G.; Williams, N. Global Biogeography and Invasion Risk of the Plant Pathogen Genus Phytophthora. Environ. Sci. Policy 2019, 101, 175–182. [Google Scholar] [CrossRef]
- Ristaino, J.B.; Cooke, D.E.; Acuña, I.; Muñoz, M. The threat of late blight to global food security. In Emerging Plant Diseases and Global Food Security; The American Phytopathological Society: Saint Paul, MN, USA, 2020; pp. 101–132. [Google Scholar] [CrossRef]
- Ristaino, J.B. The importance of mycological and plant herbaria in tracking plant killers. Front. Ecol. Evol. 2020, 7, 521. [Google Scholar] [CrossRef]
- Jombart, T.; Cori, A.; Didelot, X.; Cauchemez, S.; Fraser, C.; Ferguson, N. Bayesian Reconstruction of Disease Outbreaks by Combining Epidemiologic and Genomic Data. PLoS Comput. Biol. 2014, 10, e1003457. [Google Scholar] [CrossRef]
- Bieker, V.C.; Martin, M.D. Implications and Future Prospects for Evolutionary Analyses of DNA in Historical Herbarium Collections. Bot. Lett. 2018, 165, 409–418. [Google Scholar] [CrossRef]
- Saville, A.C.; Ristaino, J.B. Global historic pandemics caused by the FAM-1 genotype of the Irish potato pathogen Phytophthora infestans. Sci. Rep. 2021, 11, 12335. [Google Scholar] [CrossRef] [PubMed]
- Gardy, J.L.; Loman, N.J. Towards a genomics-informed, real-time, global pathogen surveillance system. Nat. Rev. Genet. 2018, 19, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Carisse, O.; Fall, M.L.; Vincent, C. Using a biovigilance approach for pest and disease management in Quebec vineyards. Can. J. Plant Pathol. 2017, 39, 393–404. [Google Scholar] [CrossRef]
- Chen, W.; Newlands, N.; Hambleton, S.; Laroche, A.; Davoodi, S.M.; Bakkeren, G. Optimizing an integrated biovigilance toolbox to study the spatial distribution and dynamic changes of airborne mycobiota, with a focus on cereal rust fungi in western Canada. Mol. Ecol. Resour. 2024, 24, e13983. [Google Scholar] [CrossRef]
- Folasole, A. Data analytics and predictive modelling approaches for identifying emerging zoonotic infectious diseases: Surveillance techniques, prediction accuracy, and public health implications. Int. J. Eng. Technol. Res. Manag. 2023, 7, 292. [Google Scholar] [CrossRef]
- Rahman, A.; Standish, J.R.; D’arcangelo, K.N.; Quesada-Ocampo, L.M. Clade-specific biosurveillance of Pseudoperonospora cubensis using spore traps for precision disease management of cucurbit downy mildew. Phytopathology 2021, 111, 312–320. [Google Scholar] [CrossRef]
- Rahman, A.; Miles, T.D.; Martin, F.N.; Quesada-Ocampo, L.M. Molecular approaches for biosurveillance of the cucurbit downy mildew pathogen, Pseudoperonospora cubensis. Can. J. Plant Pathol. 2017, 39, 282–296. [Google Scholar] [CrossRef]
- Hamelin, R.C.; Bilodeau, G.J.; Heinzelmann, R.; Hrywkiw, K.; Capron, A.; Dort, E.; Feau, N. Genomic bio-surveillance detects a sexual hybrid in the sudden oak death pathogen. Commun. Biol. 2022, 5, 477. [Google Scholar] [CrossRef]
- Carvajal-Yepes, M.; Cardwell, K.; Nelson, A.; Garrett, K.A.; Giovani, B.; Saunders, D.G.; Tohme, J. A Global Surveillance System for Crop Diseases. Science 2019, 364, 1237–1239. [Google Scholar] [CrossRef]
- Vakilian, K.A. Using Networks in Plant Disease Diagnosis. CABI Rev. 2017, 1–12. [Google Scholar] [CrossRef]
- Markell, S.G.; Tylka, G.L.; Anderson, E.J.; van Esse, H.P. Developing Public–Private Partnerships in Plant Pathology Extension: Case Studies and Opportunities in the United States. Annu. Rev. Phytopathol. 2020, 58, 161–180. [Google Scholar] [CrossRef] [PubMed]
- Schneider, R.W.; Hollier, C.A.; Whitam, H.K.; Palm, M.E.; McKemy, J.M.; Hernandez, J.R.; DeVries-Paterson, R. First Report of Soybean Rust Caused by Phakopsora pachyrhizi in the Continental United States. Plant Dis. 2005, 89, 774. [Google Scholar] [CrossRef]
- Hossain, M.M. Pathogenesis and Virulence of Phakopsora pachyrhizi. In CRC Press eBooks; CRC Press: Boca Raton, FL, USA, 2024; pp. 220–242. [Google Scholar] [CrossRef]
- Hossain, M.M.; Sultana, F.; Yesmin, L.; Rubayet, M.T.; Abdullah, H.M.; Siddique, S.S.; Bhuiyan, M.A.B.; Yamanaka, N. Understanding Phakopsora pachyrhizi in Soybean: Comprehensive Insights, Threats, and Interventions from the Asian Perspective. Front. Microbiol. 2024, 14, 1304205. [Google Scholar] [CrossRef] [PubMed]
- Livingston, M.; Johansson, R.; Daberkow, S.; Roberts, M.; Ash, M.; Breneman, V. Economic and Policy Implications of Wind-Borne Entry of Asian Soybean Rust in the United States. In Oil Crop Outlook Report; OCS04D-02; USA Department of Agriculture Economic Research Service: Washington, DC, USA, 2004. Available online: https://ers.usda.gov/sites/default/files/_laserfiche/outlooks/37823/50153_ocs04d02.pdf?v=54405 (accessed on 18 December 2024).
- Sikora, E.J.; Allen, T.W.; Wise, K.W.; Baniecki, J.; Bergstrom, G.; Bradley, C.A.; Bond, J.; Brown-Rytlewski, D.; Chilvers, M.; Damicone, J.; et al. A Coordinated Effort to Manage Soybean Rust in North America: A Success Story in Soybean Disease Monitoring. Plant Dis. 2014, 98, 864–875. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.T.; Gupta, D.R.; Hossain, A.; Roy, K.K.; He, X.; Kabir, M.R.; Singh, P.K.; Khan, M.A.R.; Rahman, M.; Wang, G.L. Wheat blast: A new threat to food security. Phytopathol. Res. 2020, 2, 28. [Google Scholar] [CrossRef]
- Yesmin, N.; Jenny, F.; Abdullah, H.M.; Hossain, M.M.; Kader, M.A.; Solomon, P.S.; Bhuiyan, M.A. A review on South Asian wheat blast: The present status and future perspective. Plant Pathol. 2020, 69, 1618–1629. [Google Scholar] [CrossRef]
- Haverkort, A.J.; Boonekamp, P.M.; Hutten, R.; Jacobsen, E.; Lotz, L.A.P.; Kessel, G.J.T.; Vossen, J.H.; Visser, R.G.F. Durable late blight resistance in potato through dynamic varieties obtained by cisgenesis: Scientific and societal advances in the DuRPh project. Potato Res. 2016, 59, 35–66. [Google Scholar] [CrossRef]
- Reem, A.; Almezgagi, M.; Al-Shaebi, F.; Al-Shehari, W.A.; Kumal, J.P.P. Application of rolling circle amplification (RCA) to detect the pathogens of infectious diseases. Infect. Dis. Diagn. Treat. 2020, 4, 158. [Google Scholar]
- Silva Zatti, M.; Domingos Arantes, T.; Cordeiro Theodoro, R. Isothermal Nucleic Acid Amplification Techniques for Detection and Identification of Pathogenic Fungi: A Review. Mycoses 2020, 63, 1006–1020. [Google Scholar] [CrossRef]
- Lau, H.Y.; Botella, J.R. Advanced DNA-Based Point-of-Care Diagnostic Methods for Plant Disease Detection. Front. Plant Sci. 2017, 8, 2016. [Google Scholar] [CrossRef]
- Jiang, S.; Chen, Y.; Han, S.; Lv, L.; Li, L. Next-Generation Sequencing Applications for the Study of Fungal Pathogens. Microorganisms 2022, 10, 1882. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).