Design and Development of a qPCR-Based Mitochondrial Analysis Workflow for Medical Laboratories
Abstract
:1. Introduction
2. State of the Art
2.1. Quantification of mtDNA Parameters Using qPCR
2.2. Legal Requirements for Software Used in Laboratory Diagnostics
2.3. Specific Validation Requirements for Software Used in Laboratory Diagnostics
2.4. Existing qPCR Analysis Software
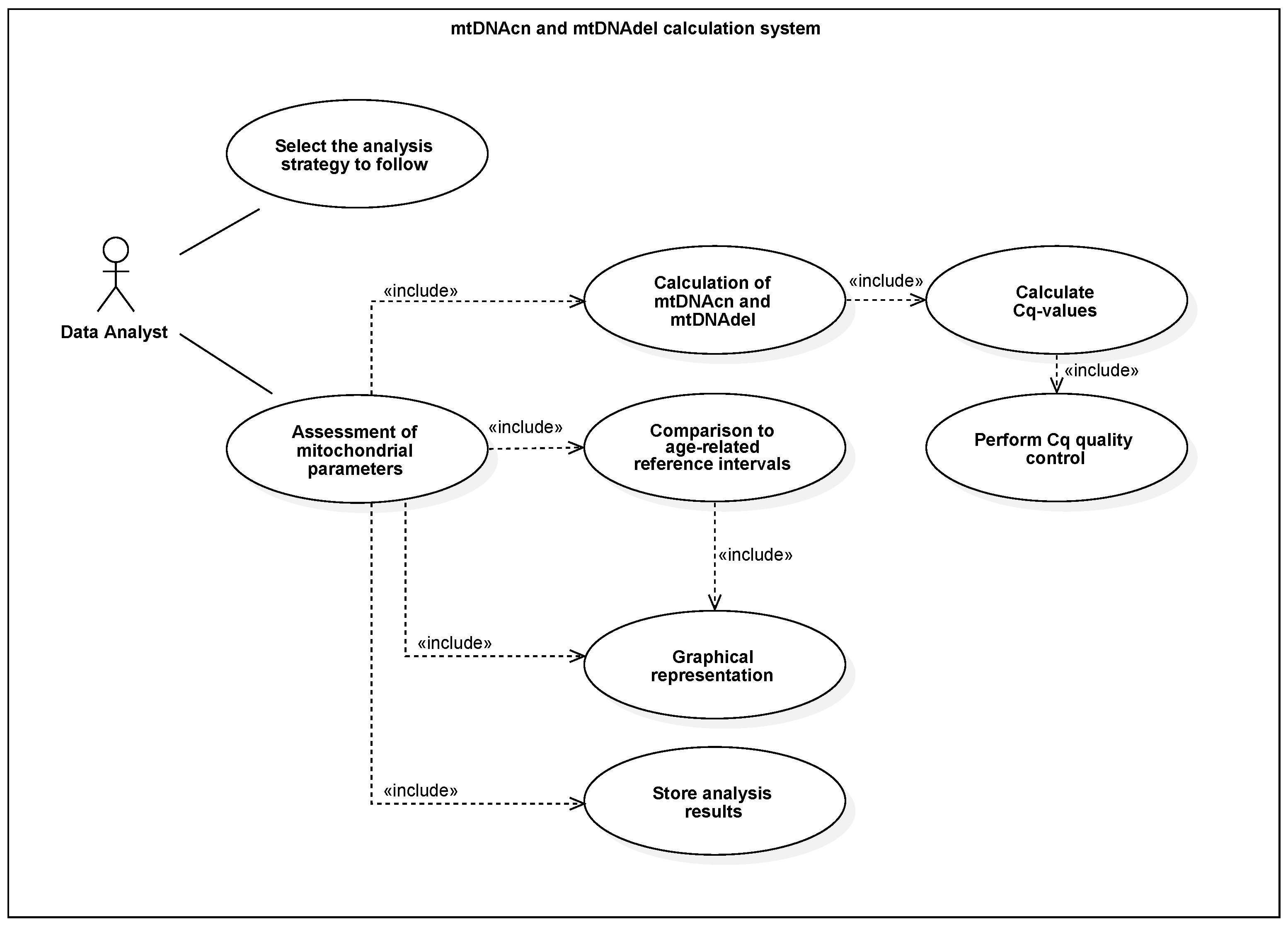
3. Model and Implementation
4. Evaluation and Discussion
5. Conclusions and Future Work
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bock, F.J.; Tait, S.W.G. Mitochondria as multifaceted regulators of cell death. Nat. Rev. Mol. Cell Biol. 2020, 21, 85–100. [Google Scholar] [CrossRef] [PubMed]
- Chocron, E.S.; Munkácsy, E.; Pickering, A.M. Cause or casualty: The role of mitochondrial DNA in aging and age-associated disease. Biochim. Biophys. Acta. Mol. Basis Dis. 2019, 1865, 285–297. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Sumberaz, P. Mitochondrial DNA Damage: Prevalence, Biological Consequence, and Emerging Pathways. Chem. Res. Toxicol. 2020, 33, 2491–2502. [Google Scholar] [CrossRef] [PubMed]
- Park, J.S.; Davis, R.L.; Sue, C.M. Mitochondrial Dysfunction in Parkinson’s Disease: New Mechanistic Insights and Therapeutic Perspectives. Curr. Neurol. Neurosci. Rep. 2018, 18, 21. [Google Scholar] [CrossRef] [Green Version]
- Misrani, A.; Tabassum, S.; Yang, L. Mitochondrial Dysfunction and Oxidative Stress in Alzheimer’s Disease. Front. Aging Neurosci. 2021, 13, 617588. [Google Scholar] [CrossRef]
- Ye, M.; Hu, B.; Shi, W.; Guo, F.; Xu, C.; Li, S. Mitochondrial DNA 4977 bp Deletion in Peripheral Blood Is Associated with Polycystic Ovary Syndrome. Front. Endocrinol. 2021, 12, 675581. [Google Scholar] [CrossRef]
- Wu, H.; Whitcomb, B.W.; Huffman, A.; Brandon, N.; Labrie, S.; Tougias, E.; Lynch, K.; Rahil, T.; Sites, C.K.; Pilsner, J.R. Associations of sperm mitochondrial DNA copy number and deletion rate with fertilization and embryo development in a clinical setting. Hum. Reprod. 2019, 34, 163–170. [Google Scholar] [CrossRef]
- Wong, L.J.C.; Boles, R.G. Mitochondrial DNA analysis in clinical laboratory diagnostics. Clin. Chim. Acta Int. J. Clin. Chem. 2005, 354, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Rodenburg, R.J.T. Biochemical diagnosis of mitochondrial disorders. J. Inherit. Metab. Dis. 2011, 34, 283–292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phillips, N.R.; Sprouse, M.L.; Roby, R.K. Simultaneous quantification of mitochondrial DNA copy number and deletion ratio: A multiplex real-time PCR assay. Sci. Rep. 2014, 4, 3887. [Google Scholar] [CrossRef]
- de Gonzalo-Calvo, D.; Marchese, M.; Hellemans, J.; Betsou, F.; Skov Frisk, N.L.; Dalgaard, L.T.; Lakkisto, P.; Foy, C.; Scherer, A.; Garcia Bermejo, M.L.; et al. Consensus Guidelines for the Validation of qRT-PCR Assays in Clinical Research by the CardioRNA Consortium. Mol.-Ther.-Methods Clin. Dev. 2022, 24. [Google Scholar] [CrossRef] [PubMed]
- Sanders, R.; Bustin, S.; Huggett, J.; Mason, D. Improving the standardization of mRNA measurement by RT-qPCR. Biomol. Detect. Quantif. 2018, 15, 13–17. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Bodrossy, L.; Frenzel, P.; Hestnes, A.G.; Krause, S.; Lüke, C.; Meima-Franke, M.; Siljanen, H.; Svenning, M.M.; Bodelier, P.L.E. Impacts of inter- and intralaboratory variations on the reproducibility of microbial community analyses. Appl. Environ. Microbiol. 2010, 76, 7451–7458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sepulveda, J.L.; Young, D.S. The ideal laboratory information system. Arch. Pathol. Lab. Med. 2013, 137, 1129–1140. [Google Scholar] [CrossRef] [Green Version]
- Pabinger, S.; Rödiger, S.; Kriegner, A.; Vierlinger, K.; Weinhäusel, A. A survey of tools for the analysis of quantitative PCR (qPCR) data. Biomol. Detect. Quantif. 2014, 1, 23–33. [Google Scholar] [CrossRef] [Green Version]
- Krause, T.; Jolkver, E.; Bruchhaus, S.; Mc Kevitt, P.; Kramer, M.; Hemmje, M. A Preliminary Evaluation of “GenDAI”, an AI-Assisted Laboratory Diagnostics Solution for Genomic Applications. BioMedInformatics 2022, 2, 332–344. [Google Scholar] [CrossRef]
- Krause, T.; Jolkver, E.; Bruchhaus, S.; Kramer, M.; Hemmje, M. An RT-qPCR Data Analysis Platform. In Proceedings of the Collaborative European Research Conference (CERC 2021), Cork, Ireland, 9–10 September 2021. [Google Scholar]
- The European Parliament and the Council of the European Union. In Vitro Diagnostic Regulation; European Union: Maastricht, The Netherlands, 2017. [Google Scholar]
- Krause, T.; Jolkver, E.; Bruchhaus, S.; Kramer, M.; Hemmje, M. GenDAI—AI-Assisted Laboratory Diagnostics for Genomic Applications. In Proceedings of the 2021 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Houston, TX, USA, 9–12 December 2021. [Google Scholar] [CrossRef]
- Krause, T.; Jolkver, E.; Mc Kevitt, P.; Kramer, M.; Hemmje, M. A Systematic Approach to Diagnostic Laboratory Software Requirements Analysis. Bioengineering 2022, 9, 144. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Standard ISO 13485:2016; Medical Devices—Quality Management Systems—Requirements for Regulatory Purposes. ISO International Organization for Standardization: Geneva, Switzerland, 2016.
- Standard ISO 15189:2012; Medical Laboratories—Requirements for Quality and Competence. ISO International Organization for Standardization: Geneva, Switzerland, 2012.
- Standard ISO 14971:2019; Medical Devices—Application of Risk Management to Medical Devices. ISO International Organization for Standardization: Geneva, Switzerland, 2019.
- Standard ISO 22367:2020; Medical Laboratories—Application of Risk Management to Medical Laboratories. ISO International Organization for Standardization: Geneva, Switzerland, 2020.
- Standard IEC 62304:2006; Medical Device Software—Software Life Cycle Processes. IEC International Electrotechnical Commission: Geneva, Switzerland, 2006.
- Johner, C. IEC 62304 2. Ausgabe: Alle Anwendungsbereiche und Änderungen. Available online: https://www.johner-institut.de/blog/iec-62304-medizinische-software/iec-62304-zweite-ausgabe/ (accessed on 13 October 2022).
- Standard IEC 82304-1:2016; Health Software—Part 1: General Requirements for Product Safety. IEC International Electrotechnical Commission: Geneva, Switzerland, 2016.
- Standard ISO/IEC 27001:2013; Information Technology—Security Techniques—Information Security Management Systems—Requirements. ISO International Organization for Standardization: Geneva, Switzerland, 2013.
- Standard IEC 62366-1:2015; Medical Devices—Part 1: Application of Usability Engineering to Medical Devices. IEC International Electrotechnical Commission: Geneva, Switzerland, 2015.
- Kraft, F.; Begemann, M.; Bietenbeck, A.; Ungelenk, M.; Kuhle, M.; Krawitz, P.; von Neuhoff, N.; Streichert, T. Hinweise der AWMF Subgruppe Software zur Umsetzung der Verordnung (EU) 2017/746 (IVDR) bei Software aus Eigenherstellung. Available online: https://www.awmf.org/fileadmin/user%5fupload/dateien/arbeitshilfen%5fund%5fmusterformblaetter/validierung%5fsoftware.docx (accessed on 21 November 2022).
- German Medical Association. Neufassung der Richtlinie der Bundesärztekammer zur Qualitätssicherung laboratoriumsmedizinischer Untersuchungen – Rili-BÄK. Dtsch. ÄRzteblatt Online 2019, 111. [Google Scholar] [CrossRef]
- Nielsen, J. Enhancing the explanatory power of usability heuristics. In Proceedings of the SIGCHI Conference on Human Factors in Computing Systems Celebrating Interdependence-CHI ’94, Toronto, ON, Canada, 26 April–1 May 2014; Adelson, B., Dumais, S., Olson, J., Eds.; ACM Press: New York, NY, USA, 1994; pp. 152–158. [Google Scholar] [CrossRef]
- What Makes Qbase+ Unique? Available online: https://www.qbaseplus.com/features (accessed on 14 October 2022).
- qbase+ | qPCR Analysis Software. Available online: https://www.qbaseplus.com/ (accessed on 14 October 2022).
- MultiD Analyses AB. GenEx. Available online: https://multid.se/genex/ (accessed on 23 October 2022).
- Hellemans, J.; Mortier, G.; de Paepe, A.; Speleman, F.; Vandesompele, J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007, 8, R19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vandesompele, J.; de Preter, K.; Pattyn, F.; Poppe, B.; van Roy, N.; de Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dopp, A.R.; Parisi, K.E.; Munson, S.A.; Lyon, A.R. A glossary of user-centered design strategies for implementation experts. Transl. Behav. Med. 2019, 9, 1057–1064. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kushniruk, A.W.; Patel, V.L. Cognitive and usability engineering methods for the evaluation of clinical information systems. J. Biomed. Inform. 2004, 37, 56–76. [Google Scholar] [CrossRef] [PubMed]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Krause, T.; Glau, L.; Jolkver, E.; Leonardi-Essmann, F.; Kevitt, P.M.; Kramer, M.; Hemmje, M. Design and Development of a qPCR-Based Mitochondrial Analysis Workflow for Medical Laboratories. BioMedInformatics 2022, 2, 643-653. https://doi.org/10.3390/biomedinformatics2040042
Krause T, Glau L, Jolkver E, Leonardi-Essmann F, Kevitt PM, Kramer M, Hemmje M. Design and Development of a qPCR-Based Mitochondrial Analysis Workflow for Medical Laboratories. BioMedInformatics. 2022; 2(4):643-653. https://doi.org/10.3390/biomedinformatics2040042
Chicago/Turabian StyleKrause, Thomas, Laura Glau, Elena Jolkver, Fernando Leonardi-Essmann, Paul Mc Kevitt, Michael Kramer, and Matthias Hemmje. 2022. "Design and Development of a qPCR-Based Mitochondrial Analysis Workflow for Medical Laboratories" BioMedInformatics 2, no. 4: 643-653. https://doi.org/10.3390/biomedinformatics2040042
APA StyleKrause, T., Glau, L., Jolkver, E., Leonardi-Essmann, F., Kevitt, P. M., Kramer, M., & Hemmje, M. (2022). Design and Development of a qPCR-Based Mitochondrial Analysis Workflow for Medical Laboratories. BioMedInformatics, 2(4), 643-653. https://doi.org/10.3390/biomedinformatics2040042





