Journal Description
BioMedInformatics
BioMedInformatics
is an international, peer-reviewed, open access journal on all areas of biomedical informatics, as well as computational biology and medicine, published quarterly online by MDPI.
- Open Access— free for readers, with article processing charges (APC) paid by authors or their institutions.
- High Visibility: indexed within Scopus, EBSCO, and other databases.
- Rapid Publication: manuscripts are peer-reviewed and a first decision is provided to authors approximately 22.9 days after submission; acceptance to publication is undertaken in 5.3 days (median values for papers published in this journal in the first half of 2025).
- Journal Rank: CiteScore - Q1 (Health Professions (miscellaneous))
- Recognition of Reviewers: APC discount vouchers, optional signed peer review, and reviewer names published annually in the journal.
Latest Articles
Visualising the Truth: A Composite Evaluation Framework for Score-Based Predictive Model Selection
BioMedInformatics 2025, 5(3), 55; https://doi.org/10.3390/biomedinformatics5030055 - 17 Sep 2025
Abstract
►
Show Figures
Background: The selection of machine learning (ML) models in the biomedical sciences often relies on global performance metrics. When these metrics are closely clustered among candidate models, identifying the most suitable model for real-world deployment becomes challenging. Methods: We developed a novel composite
[...] Read more.
Background: The selection of machine learning (ML) models in the biomedical sciences often relies on global performance metrics. When these metrics are closely clustered among candidate models, identifying the most suitable model for real-world deployment becomes challenging. Methods: We developed a novel composite framework that integrates visual inspection of Model Scoring Distribution Analysis (MSDA) with a new scoring metric (MSDscore). The methodology was implemented within the Digital Phenomics platform as the MSDanalyser tool and tested by generating and evaluating 27 predictive models developed for breast, lung, and renal cancer prognosis. Results: Our approach enabled a detailed inspection of true-positive, false-positive, true-negative, and false-negative distributions across the scoring space, capturing local performance patterns overlooked by conventional metrics. In contrast with the minimal variation between models obtained by global metrics, the MSDA methodology revealed substantial differences in score region behaviour, allowing better discrimination between models. Conclusions: Integrating our composite framework alongside traditional performance metrics provides a complementary and more nuanced approach to model selection in clinical and biomedical settings.
Full article
Open AccessArticle
Correlating Clinical Assessments for Substance Use Disorder Using Unsupervised Machine Learning
by
Kaloso M. Tlotleng, Rodrigo S. Jamisola, Jr. and Jeniffer L. Brown
BioMedInformatics 2025, 5(3), 54; https://doi.org/10.3390/biomedinformatics5030054 - 11 Sep 2025
Abstract
This paper investigates the state of substance use disorder (SUD) and the frequency of substance use by utilizing three unsupervised machine learning techniques, based on the Diagnostic and Statistical Manual 5 (DSM-5) of mental health disorders. We used data obtained from the National
[...] Read more.
This paper investigates the state of substance use disorder (SUD) and the frequency of substance use by utilizing three unsupervised machine learning techniques, based on the Diagnostic and Statistical Manual 5 (DSM-5) of mental health disorders. We used data obtained from the National Survey on Drug Use and Health (NSDUH) 2019 database with random participants who had undergone clinical assessments by mental health professionals and whose clinical diagnoses were not known. This approach classifies SUD status by discovering patterns or correlations from the trained model. Our results were analyzed and validated by a mental health professional. The three unsupervised machine learning techniques that we used comprised k-means clustering, hierarchical clustering, and density-based spatial clustering of applications with noise (DBSCAN), which were applied to alcohol, marijuana, and cocaine datasets. The clustering results were validated using the silhouette score and the 95% confidence interval (CI). The results from this study may be used to supplement psychiatric evaluations.
Full article
(This article belongs to the Topic Machine Learning Empowered Drug Screen)
►▼
Show Figures

Graphical abstract
Open AccessArticle
High-Precision, Automatic, and Fast Segmentation Method of Hepatic Vessels and Liver Tumors from CT Images Using a Fusion Decision-Based Stacking Deep Learning Model
by
Mamoun Qjidaa, Anass Benfares, Mohammed Amine El Azami El Hassani, Amine Benkabbou, Amine Souadka, Anass Majbar, Zakaria El Moatassim, Maroua Oumlaz, Oumayma Lahnaoui, Raouf Mouhcine, Ahmed Lakhssassi and Abdeljabbar Cherkaoui
BioMedInformatics 2025, 5(3), 53; https://doi.org/10.3390/biomedinformatics5030053 - 9 Sep 2025
Abstract
Background: To propose an automatic liver and hepatic vessel segmentation solution based on a stacking model and decision fusion. This model combines the decisions of multiple models to achieve increased accuracy. It exhibits improved robustness due to the reduction of individual errors. Flexibility
[...] Read more.
Background: To propose an automatic liver and hepatic vessel segmentation solution based on a stacking model and decision fusion. This model combines the decisions of multiple models to achieve increased accuracy. It exhibits improved robustness due to the reduction of individual errors. Flexibility is also a key asset, with combination methods such as majority voting or weighted averaging. The model enables managing the uncertainty associated with individual decisions to obtain a more reliable final decision. The combination of decisions improves the overall accuracy of the system. Methods: This research introduces a new deep learning-based architecture for automatically segmenting hepatic vessels and tumors from CT scans, utilizing stacking, decision fusion, and deep transfer learning to achieve high-accuracy and rapid segmentation. This study employed two distinct datasets: the external “Medical Segmentation Decathlon (MSD) task 08” dataset and an internal dataset procured from Ibn Sina University Hospital encompassing a cohort of 112 patients with chronic liver disease who underwent contrast-enhanced abdominal CT scans. Results: The proposed segmentation model reached a DSC of 83.21 and an IoU of 72.76 for hepatic vasculature and tumor segmentation, thereby exceeding the performance benchmarks established by the majority of antecedent studies. Conclusions: This study introduces an automated method for liver vessels and liver tumor segmentation, combining precision and stability to bridge the clinical gap. Furthermore, decision fusion-based stacking models have a significant impact on clinical applications by enhancing diagnostic accuracy, enabling personalized care through the integration of genetic, environmental, and clinical data, optimizing clinical trials, and facilitating the development of personalized medicines and therapies.
Full article
(This article belongs to the Section Methods in Biomedical Informatics)
►▼
Show Figures

Figure 1
Open AccessArticle
Supervised Machine Learning for PICU Outcome Prediction: A Comparative Analysis Using the TOPICC Study Dataset
by
Amr M. Ali and Orkun Baloglu
BioMedInformatics 2025, 5(3), 52; https://doi.org/10.3390/biomedinformatics5030052 - 5 Sep 2025
Abstract
►▼
Show Figures
Background: Pediatric Intensive Care Unit (PICU) outcome prediction is challenging, and machine learning (ML) can enhance it by leveraging large datasets. Methods: We built an ML model to predict PICU outcomes (“Death vs. Survival”, “Death or Morbidity vs. Survival without morbidity”,
[...] Read more.
Background: Pediatric Intensive Care Unit (PICU) outcome prediction is challenging, and machine learning (ML) can enhance it by leveraging large datasets. Methods: We built an ML model to predict PICU outcomes (“Death vs. Survival”, “Death or Morbidity vs. Survival without morbidity”, and “New Morbidity vs. Survival without new morbidity”) using the Trichotomous Outcome Prediction in Critical Care (TOPICC) study dataset. The model used the Light Gradient-Boosting Machine (LightGBM) algorithm, which was trained on 85% of the dataset and tested on 15% utilizing 10-fold cross validation. Results: The model demonstrated high accuracy across all dichotomies, with 0.98 for “Death vs. Survival”, 0.92 for “Death or New Morbidity vs. Survival without New Morbidity”, and 0.93 for “New Morbidity vs. Survival without New Morbidity.” The AUC-ROC values were also strong, at 0.89, 0.79, and 0.74, respectively. The precision was highest for “Death vs. Survival” (0.92), followed by 0.45 and 0.30 for the other dichotomies. The recalls were low, at 0.26, 0.31, and 0.34, reflecting the model’s difficulty in identifying all positive cases. The AUC-PR values (0.43, 0.37, and 0.20) highlight this trade-off. Conclusions: The LightGBM model demonstrated a predictive performance comparable to previously reported logistic regression models in predicting PICU outcomes. Future work should focus on enhancing the model’s performance and further validation across larger datasets to assess the model’s generalizability and real-world applicability.
Full article

Figure 1
Open AccessArticle
Quantum-Enhanced Dual-Backbone Architecture for Accurate Gastrointestinal Disease Detection Using Endoscopic Imaging
by
Nabil Marzoug, Khidhr Halab, Othmane El Meslouhi, Zouhair Elamrani Abou Elassad and Moulay A. Akhloufi
BioMedInformatics 2025, 5(3), 51; https://doi.org/10.3390/biomedinformatics5030051 - 4 Sep 2025
Abstract
►▼
Show Figures
Background: Quantum machine learning (QML) holds significant promise for advancing medical image classification. However, its practical application to large-scale, high-resolution datasets is constrained by the limited number of qubits and the inherent noise in current quantum hardware. Methods: In this study, we propose
[...] Read more.
Background: Quantum machine learning (QML) holds significant promise for advancing medical image classification. However, its practical application to large-scale, high-resolution datasets is constrained by the limited number of qubits and the inherent noise in current quantum hardware. Methods: In this study, we propose the Fused Quantum Dual-Backbone Network (FQDN), a novel hybrid architecture that integrates classical convolutional neural networks (CNNs) with quantum circuits. This design is optimized for the noisy intermediate-scale quantum (NISQ), enabling efficient computation despite hardware limitations. We evaluate FQDN on the task of gastrointestinal (GI) disease classification using wireless capsule endoscopy (WCE) images. Results: The proposed model achieves a substantial reduction in parameter complexity, with a 29.04% decrease in total parameters and a 94.44% reduction in trainable parameters, while outperforming its classical counterpart. FQDN achieves an accuracy of 95.80% on the validation set and 95.42% on the test set. Conclusions: These results demonstrate the potential of QML to enhance diagnostic accuracy in medical imaging.
Full article

Graphical abstract
Open AccessArticle
Using Large Language Models to Extract Structured Data from Health Coaching Dialogues: A Comparative Study of Code Generation Versus Direct Information Extraction
by
Sai Sangameswara Aadithya Kanduri, Apoorv Prasad and Susan McRoy
BioMedInformatics 2025, 5(3), 50; https://doi.org/10.3390/biomedinformatics5030050 - 4 Sep 2025
Abstract
Background: Virtual coaching can help people adopt new healthful behaviors by encouraging them to set specific goals and helping them review their progress. One challenge in creating such systems is analyzing clients’ statements about their activities. Limiting people to selecting among predefined
[...] Read more.
Background: Virtual coaching can help people adopt new healthful behaviors by encouraging them to set specific goals and helping them review their progress. One challenge in creating such systems is analyzing clients’ statements about their activities. Limiting people to selecting among predefined answers detracts from the naturalness of conversations and user engagement. Large Language Models (LLMs) offer the promise of covering a wide range of expressions. However, using an LLM for simple entity extraction would not necessarily perform better than functions coded in a programming language, while creating higher long-term costs. Methods: This study uses a real data set of annotated human coaching dialogs to develop LLM-based models for two training scenarios: one that generates pattern-matching functions and the other which does direct extraction. We use models of different sizes and complexity, including Meta-Llama, Gemma, and ChatGPT, and calculate their speed and accuracy. Results: LLM-generated pattern-matching functions took an average of 10 milliseconds (ms) per item as compared to 900 ms. (ChatGPT 3.5 Turbo) to 5 s (Llama 2 70B). The accuracy for pattern matching was 99% on real data, while LLM accuracy ranged from 90% (Llama 2 70B) to 100% (ChatGPT 3.5 Turbo), on both real and synthetically generated examples created for fine-tuning. Conclusions: These findings suggest promising directions for future research that combines both methods (reserving the LLM for cases that cannot be matched directly) or that use LLMs to generate synthetic training data with more expressive variety which can be used to improve the coverage of either generated codes or fine-tuned models.
Full article
(This article belongs to the Section Methods in Biomedical Informatics)
►▼
Show Figures

Figure 1
Open AccessArticle
Co-Designing a DSM-5-Based AI-Powered Smart Assistant for Monitoring Dementia and Ongoing Neurocognitive Decline: Development Study
by
Fareed Ud Din, Nabaraj Giri, Namrata Shetty, Tom Hilton, Niusha Shafiabady and Phillip J. Tully
BioMedInformatics 2025, 5(3), 49; https://doi.org/10.3390/biomedinformatics5030049 - 2 Sep 2025
Abstract
Background/Objectives: Dementia is a leading cause of cognitive decline, with significant challenges for early detection and timely intervention. The lack of effective, user-centred technologies further limits clinical response, particularly in underserved areas. This study aimed to develop and describe a co-design process for
[...] Read more.
Background/Objectives: Dementia is a leading cause of cognitive decline, with significant challenges for early detection and timely intervention. The lack of effective, user-centred technologies further limits clinical response, particularly in underserved areas. This study aimed to develop and describe a co-design process for creating a Diagnostic and Statistical Manual of Mental Disorders (DSM-5)-compliant, AI-powered Smart Assistant (SmartApp) to monitor neurocognitive decline, while ensuring accessibility, clinical relevance, and responsible AI integration. Methods: A co-design framework was applied using a novel combination of Agile principles and the Double Diamond Model (DDM). More than twenty iterative Scrum sprints were conducted, involving key stakeholders such as clinicians (psychiatrist, psychologist, physician), designers, students, and academic researchers. Prototype testing and design workshops were organised to gather structured feedback. Feedback was systematically incorporated into subsequent iterations to refine functionality, usability, and clinical applicability. Results: The iterative process resulted in a SmartApp that integrates a DSM-5-based screening tool with 24 items across key cognitive domains. Key features include longitudinal tracking of cognitive performance, comparative visual graphs, predictive analytics using a regression-based machine learning module, and adaptive user interfaces. Workshop participants reported high satisfaction with features such as simplified navigation, notification reminders, and clinician-focused reporting modules. Conclusions: The findings suggest that combining co-design methods with Agile/DDM frameworks provides an effective pathway for developing AI-powered clinical tools as per responsible AI standards. The SmartApp offers a clinically relevant, user-friendly platform for dementia screening and monitoring, with potential to support vulnerable populations through scalable, responsible digital health solutions.
Full article
(This article belongs to the Special Issue Integrating Health Informatics and Artificial Intelligence for Advanced Medicine)
►▼
Show Figures

Figure 1
Open AccessReview
Real-Time Applications of Biophysiological Markers in Virtual-Reality Exposure Therapy: A Systematic Review
by
Marie-Jeanne Fradette, Julie Azrak, Florence Cousineau, Marie Désilets and Alexandre Dumais
BioMedInformatics 2025, 5(3), 48; https://doi.org/10.3390/biomedinformatics5030048 - 28 Aug 2025
Abstract
Virtual-reality exposure therapy (VRET) is an emerging treatment for psychiatric disorders that enables immersive and controlled exposure to anxiety-provoking stimuli. Recent developments integrate real-time physiological monitoring, including heart rate (HR), electrodermal activity (EDA), and electroencephalography (EEG), to dynamically tailor therapeutic interventions. This systematic
[...] Read more.
Virtual-reality exposure therapy (VRET) is an emerging treatment for psychiatric disorders that enables immersive and controlled exposure to anxiety-provoking stimuli. Recent developments integrate real-time physiological monitoring, including heart rate (HR), electrodermal activity (EDA), and electroencephalography (EEG), to dynamically tailor therapeutic interventions. This systematic review examines studies that combine VRET with physiological data to adapt virtual environments in real time. A comprehensive search of major databases identified fifteen studies meeting the inclusion criteria: all employed physiological monitoring and adaptive features, with ten using biofeedback to modulate exposure based on single or multimodal physiological measures. The remaining studies leveraged physiological signals to inform scenario selection or threat modulation using dynamic categorization algorithms and machine learning. Although findings currently show an overrepresentation of anxiety disorders, recent studies are increasingly involving more diverse clinical populations. Results suggest that adaptive VRET is technically feasible and offers promising personalization benefits; however, the limited number of studies, methodological variability, and small sample sizes constrain broader conclusions. Future research should prioritize rigorous experimental designs, standardized outcome measures, and greater diversity in clinical populations. Adaptive VRET represents a frontier in precision psychiatry, where real-time biosensing and immersive technologies converge to enhance individualized mental health care.
Full article
(This article belongs to the Section Applied Biomedical Data Science)
►▼
Show Figures

Figure 1
Open AccessArticle
Stabilizing the Shield: C-Terminal Tail Mutation of HMPV F Protein for Enhanced Vaccine Design
by
Reetesh Kumar, Subhomoi Borkotoky, Rohan Gupta, Jyoti Gupta, Somnath Maji, Savitri Tiwari, Rajeev K. Tyagi and Baldo Oliva
BioMedInformatics 2025, 5(3), 47; https://doi.org/10.3390/biomedinformatics5030047 - 28 Aug 2025
Abstract
Background: Human Metapneumovirus (HMPV) is a respiratory virus in the Pneumoviridae family. HMPV is an enveloped, negative-sense RNA virus encoding three surface proteins: SH, G, and F. The highly immunogenic fusion (F) protein is essential for viral entry and a key target for
[...] Read more.
Background: Human Metapneumovirus (HMPV) is a respiratory virus in the Pneumoviridae family. HMPV is an enveloped, negative-sense RNA virus encoding three surface proteins: SH, G, and F. The highly immunogenic fusion (F) protein is essential for viral entry and a key target for vaccine development. The F protein exists in two conformations: prefusion and postfusion. The prefusion form is highly immunogenic and considered a potent vaccine antigen. However, this conformation needs to be stabilized to improve its immunogenicity for effective vaccine development. Specific mutations are necessary to maintain the prefusion state and prevent it from changing to the postfusion form. Methods: In silico mutagenesis was performed on the C-terminal domain of the pre-F protein, focusing on five amino acids at positions 469 to 473 (LVDQS), using the established pre-F structure (PDB: 8W3Q) as the reference. The amino acid sequence was sequentially mutated based on hydrophobicity, resulting in mutants M1 (IIFLL), M2 (LLIVL), M3 (WWVLL), and M4 (YMWLL). Increasing hydrophobicity was found to enhance protein stability and structural rigidity. Results: Epitope mapping revealed that all mutants displayed significant B and T cell epitopes similar to the reference protein. The structure and stability of all mutants were analyzed using molecular dynamics simulations, free energy calculations, and secondary structure analysis. Based on the lowest RMSD, clash score, MolProbity value, stable radius of gyration, and low RMSF, the M1 mutant demonstrated superior structural stability. Conclusions: Our findings indicate that the M1 mutant of the pre-F protein could be the most stable and structurally accurate candidate for vaccine development against HMPV.
Full article
(This article belongs to the Section Computational Biology and Medicine)
►▼
Show Figures

Figure 1
Open AccessReview
Advancements in Breast Cancer Detection: A Review of Global Trends, Risk Factors, Imaging Modalities, Machine Learning, and Deep Learning Approaches
by
Md. Atiqur Rahman, M. Saddam Hossain Khan, Yutaka Watanobe, Jarin Tasnim Prioty, Tasfia Tahsin Annita, Samura Rahman, Md. Shakil Hossain, Saddit Ahmed Aitijjo, Rafsun Islam Taskin, Victor Dhrubo, Abubokor Hanip and Touhid Bhuiyan
BioMedInformatics 2025, 5(3), 46; https://doi.org/10.3390/biomedinformatics5030046 - 20 Aug 2025
Abstract
Breast cancer remains a critical global health challenge, with over 2.1 million new cases annually. This review systematically evaluates recent advancements (2022–2024) in machine and deep learning approaches for breast cancer detection and risk management. Our analysis demonstrates that deep learning models achieve
[...] Read more.
Breast cancer remains a critical global health challenge, with over 2.1 million new cases annually. This review systematically evaluates recent advancements (2022–2024) in machine and deep learning approaches for breast cancer detection and risk management. Our analysis demonstrates that deep learning models achieve 90–99% accuracy across imaging modalities, with convolutional neural networks showing particular promise in mammography (99.96% accuracy) and ultrasound (100% accuracy) applications. Tabular data models using XGBoost achieve comparable performance (99.12% accuracy) for risk prediction. The study confirms that lifestyle modifications (dietary changes, BMI management, and alcohol reduction) significantly mitigate breast cancer risk. Key findings include the following: (1) hybrid models combining imaging and clinical data enhance early detection, (2) thermal imaging achieves high diagnostic accuracy (97–100% in optimized models) while offering a cost-effective, less hazardous screening option, (3) challenges persist in data variability and model interpretability. These results highlight the need for integrated diagnostic systems combining technological innovations with preventive strategies. The review underscores AI’s transformative potential in breast cancer diagnosis while emphasizing the continued importance of risk factor management. Future research should prioritize multi-modal data integration and clinically interpretable models.
Full article
(This article belongs to the Section Imaging Informatics)
►▼
Show Figures

Figure 1
Open AccessArticle
Using Geometric Approaches to the Common Transcriptomics in Acute Lymphoblastic Leukemia and Rhabdomyosarcoma: Expanding and Integrating Pathway Simulations
by
Christos Tselios, Ioannis Vezakis, Apostolos Zaravinos and George I. Lambrou
BioMedInformatics 2025, 5(3), 45; https://doi.org/10.3390/biomedinformatics5030045 - 15 Aug 2025
Abstract
Background: The amount of data produced from biological experiments has increased geometrically, posing a challenge for the development of new methodologies that could enable their interpretation. We propose a novel approach for the analysis of transcriptomic data derived from acute lymphoblastic leukemia
[...] Read more.
Background: The amount of data produced from biological experiments has increased geometrically, posing a challenge for the development of new methodologies that could enable their interpretation. We propose a novel approach for the analysis of transcriptomic data derived from acute lymphoblastic leukemia (ALL) and rhabdomyosarcoma (RMS) cell lines, using bioinformatics, systems biology and geometrical approaches. Methods: The expression profile of each cell line was investigated using microarrays, and identified genes were used to create a systems pathway model, which was then simulated using differential equations. The transcriptomic profile used involved genes with similar expression levels. The simulated results were further analyzed using geometrical approaches to identify common expressional dynamics. Results: We simulated and analyzed the system network using time series, regression analysis and helical functions, detecting predictable structures after iterating the modelled biological network, focusing on TIE1, STAT1, MAPK14 and ADAM17. Our results show that such common attributes in gene expression patterns can lead to more effective treatment options and help in the discovery of universal tumor biomarkers. Discussion: Our approach was able to identify complex structures in gene expression patterns, indicating that such approaches could prove useful towards the understanding of the complex tumor dynamics.
Full article
(This article belongs to the Section Methods in Biomedical Informatics)
►▼
Show Figures

Figure 1
Open AccessArticle
Medical Segmentation of Kidney Whole Slide Images Using Slicing Aided Hyper Inference and Enhanced Syncretic Mask Merging Optimized by Particle Swarm Metaheuristics
by
Marko Mihajlovic and Marina Marjanovic
BioMedInformatics 2025, 5(3), 44; https://doi.org/10.3390/biomedinformatics5030044 - 11 Aug 2025
Abstract
►▼
Show Figures
Accurate segmentation of kidney microstructures in whole slide images (WSIs) is essential for the diagnosis and monitoring of renal diseases. In this study, an end-to-end instance segmentation pipeline was developed for the detection of glomeruli and blood vessels in hematoxylin and eosin (H&E)
[...] Read more.
Accurate segmentation of kidney microstructures in whole slide images (WSIs) is essential for the diagnosis and monitoring of renal diseases. In this study, an end-to-end instance segmentation pipeline was developed for the detection of glomeruli and blood vessels in hematoxylin and eosin (H&E) stained kidney tissue. A tiling-based strategy was employed using Slicing Aided Hyper Inference (SAHI) to manage the resolution and scale of WSIs and the performance of two segmentation models, YOLOv11 and YOLOv12, was comparatively evaluated. The influence of tile overlap ratios on segmentation quality and inference efficiency was assessed, with configurations identified that balance object continuity and computational cost. To address object fragmentation at tile boundaries, an Enhanced Syncretic Mask Merging algorithm was introduced, incorporating morphological and spatial constraints. The algorithm’s hyperparameters were optimized using Particle Swarm Optimization (PSO), with vessel and glomerulus-specific performance targets. The optimization process revealed key parameters affecting segmentation quality, particularly for vessel structures with fine, elongated morphology. When compared with a baseline without postprocessing, improvements in segmentation precision were observed, notably a 48% average increase for glomeruli and up to 17% for blood vessels. The proposed framework demonstrates a balance between accuracy and efficiency, supporting scalable histopathology analysis and contributing to the Vasculature Common Coordinate Framework (VCCF) and Human Reference Atlas (HRA).
Full article

Figure 1
Open AccessArticle
SCCM: An Interpretable Enhanced Transfer Learning Model for Improved Skin Cancer Classification
by
Md. Rifat Aknda, Fahmid Al Farid, Jia Uddin, Sarina Mansor and Muhammad Golam Kibria
BioMedInformatics 2025, 5(3), 43; https://doi.org/10.3390/biomedinformatics5030043 - 5 Aug 2025
Abstract
Skin cancer is the most common cancer worldwide, for which early detection is crucial to improve survival rates. Visual inspection and biopsies have limitations, including being error-prone, costly, and time-consuming. Although several deep learning models have been developed, they demonstrate significant limitations. An
[...] Read more.
Skin cancer is the most common cancer worldwide, for which early detection is crucial to improve survival rates. Visual inspection and biopsies have limitations, including being error-prone, costly, and time-consuming. Although several deep learning models have been developed, they demonstrate significant limitations. An interpretable and improved transfer learning model for binary skin cancer classification is proposed in this research, which uses the last convolutional block of VGG16 as the feature extractor. The methodology focuses on addressing the existing limitations in skin cancer classification, to support dermatologists and potentially saving lives through advanced, reliable, and accessible AI-driven diagnostic tools. Explainable AI is incorporated for the visualization and explanation of classifications. Multiple optimization techniques are applied to avoid overfitting, ensure stable training, and enhance the classification accuracy of dermoscopic images into benign and malignant classes. The proposed model shows 90.91% classification accuracy, which is better than state-of-the-art models and established approaches in skin cancer classification. An interactive desktop application integrating the model is developed, enabling real-time preliminary screening with offline access.
Full article
(This article belongs to the Section Imaging Informatics)
►▼
Show Figures

Figure 1
Open AccessArticle
Deep Learning Treatment Recommendations for Patients Diagnosed with Non-Metastatic Castration-Resistant Prostate Cancer Receiving Androgen Deprivation Treatment
by
Chunyang Li, Julia Bohman, Vikas Patil, Richard Mcshinsky, Christina Yong, Zach Burningham, Matthew Samore and Ahmad S. Halwani
BioMedInformatics 2025, 5(3), 42; https://doi.org/10.3390/biomedinformatics5030042 - 4 Aug 2025
Abstract
Background: Prostate cancer (PC) is the second leading cause of cancer-related death in men in the United States. A subset of patients develops non-metastatic, castration-resistant PC (nmCRPC), for which management requires a personalized consideration for appropriate treatment. However, there is no consensus regarding
[...] Read more.
Background: Prostate cancer (PC) is the second leading cause of cancer-related death in men in the United States. A subset of patients develops non-metastatic, castration-resistant PC (nmCRPC), for which management requires a personalized consideration for appropriate treatment. However, there is no consensus regarding when to switch from androgen deprivation therapy (ADT) to more aggressive treatments like abiraterone or enzalutamide. Methods: We analyzed 5037 nmCRPC patients and employed a Weibull Time to Event Recurrent Neural Network to identify patients who would benefit from switching from ADT to abiraterone/enzalutamide. We evaluated this model using differential treatment benefits measured by the Kaplan–Meier estimation and milestone probabilities. Results: The model achieved an area under the curve of 0.738 (standard deviation (SD): 0.057) for patients treated with abiraterone/enzalutamide and 0.693 (SD: 0.02) for patients exclusively treated with ADT at the 2-year milestone. The model recommended 14% of ADT patients switch to abiraterone/enzalutamide. Analysis showed a statistically significant absolute improvement using model-recommended treatments in progression-free survival (PFS) of 0.24 (95% confidence interval (CI): 0.23–0.24) at the 2-year milestone (PFS rate increasing from 0.50 to 0.74) with a hazard ratio of 0.44 (95% CI: 0.39–0.50). Conclusions: Our model successfully identified nmCRPC patients who would benefit from switching to abiraterone/enzalutamide, demonstrating potential outcome improvements.
Full article
(This article belongs to the Special Issue Integrating Health Informatics and Artificial Intelligence for Advanced Medicine)
►▼
Show Figures

Figure 1
Open AccessArticle
Skin Lesion Classification Using Hybrid Feature Extraction Based on Classical and Deep Learning Methods
by
Maryem Zahid, Mohammed Rziza and Rachid Alaoui
BioMedInformatics 2025, 5(3), 41; https://doi.org/10.3390/biomedinformatics5030041 - 16 Jul 2025
Abstract
►▼
Show Figures
This paper proposes a hybrid method for skin lesion classification combining deep learning features with conventional descriptors such as HOG, Gabor, SIFT, and LBP. Feature extraction was performed by extracting features of interest within the tumor area using suggested fusion methods. We tested
[...] Read more.
This paper proposes a hybrid method for skin lesion classification combining deep learning features with conventional descriptors such as HOG, Gabor, SIFT, and LBP. Feature extraction was performed by extracting features of interest within the tumor area using suggested fusion methods. We tested and compared features obtained from different deep learning models coupled to HOG-based features. Dimensionality reduction and performance improvement were achieved by Principal Component Analysis, after which SVM was used for classification. The compared methods were tested on the reference database skin cancer-malignant-vs-benign. The results show a significant improvement in terms of accuracy due to complementarity between the conventional and deep learning-based methods. Specifically, the addition of HOG descriptors led to an accuracy increase of 5% for EfficientNetB0, 7% for ResNet50, 5% for ResNet101, 1% for NASNetMobile, 1% for DenseNet201, and 1% for MobileNetV2. These findings confirm that feature fusion significantly enhances performance compared to the individual application of each method.
Full article

Figure 1
Open AccessArticle
Identifying Themes in Social Media Discussions of Eating Disorders: A Quantitative Analysis of How Meaningful Guidance and Examples Improve LLM Classification
by
Apoorv Prasad, Setayesh Abiazi Shalmani, Lu He, Yang Wang and Susan McRoy
BioMedInformatics 2025, 5(3), 40; https://doi.org/10.3390/biomedinformatics5030040 - 11 Jul 2025
Abstract
►▼
Show Figures
Background: Social media represents a unique opportunity to investigate the perspectives of people with eating disorders at scale. One forum alone, r/EatingDisorders, now has 113,000 members worldwide. In less than a day, where a manual analysis might sample a few dozen items, automatic
[...] Read more.
Background: Social media represents a unique opportunity to investigate the perspectives of people with eating disorders at scale. One forum alone, r/EatingDisorders, now has 113,000 members worldwide. In less than a day, where a manual analysis might sample a few dozen items, automatic classification using large language models (LLMs) can analyze thousands of posts. Methods: Here, we compare multiple strategies for invoking an LLM, including ones that include examples (few-shot) and annotation guidelines, to classify eating disorder content across 14 predefined themes using Llama3.1:8b on 6850 social media posts. In addition to standard metrics, we calculate four novel dimensions of classification quality: a Category Divergence Index, confidence scores (overall model certainty), focus scores (a measure of decisiveness for selected subsets of themes), and dominance scores (primary theme identification strength). Results: By every measure, invoking an LLM without extensive guidance and examples (zero-shot) is insufficient. Zero-shot had worse mean category divergence (7.17 versus 3.17). Whereas, few-shot yielded higher mean confidence, 0.42 versus 0.27, and higher mean dominance, 0.81 versus 0.46. Overall, a few-shot approach improved quality measures across nearly 90% of predictions. Conclusions: These findings suggest that LLMs, if invoked with expert instructions and helpful examples, can provide instantaneous high-quality annotation, enabling automated mental health content moderation systems or future clinical research.
Full article

Figure 1
Open AccessArticle
AI-Driven Bayesian Deep Learning for Lung Cancer Prediction: Precision Decision Support in Big Data Health Informatics
by
Natalia Amasiadi, Maria Aslani-Gkotzamanidou, Leonidas Theodorakopoulos, Alexandra Theodoropoulou, George A. Krimpas, Christos Merkouris and Aristeidis Karras
BioMedInformatics 2025, 5(3), 39; https://doi.org/10.3390/biomedinformatics5030039 - 9 Jul 2025
Abstract
►▼
Show Figures
Lung-cancer incidence is projected to rise by 50% by 2035, underscoring the need for accurate yet accessible risk-stratification tools. We trained a Bayesian neural network on 300 annotated chest-CT scans from the public LIDC–IDRI cohort, integrating clinical metadata. Hamiltonian Monte-Carlo sampling (10 000
[...] Read more.
Lung-cancer incidence is projected to rise by 50% by 2035, underscoring the need for accurate yet accessible risk-stratification tools. We trained a Bayesian neural network on 300 annotated chest-CT scans from the public LIDC–IDRI cohort, integrating clinical metadata. Hamiltonian Monte-Carlo sampling (10 000 posterior draws) captured parameter uncertainty; performance was assessed with stratified five-fold cross-validation and on three independent multi-centre cohorts. On the locked internal test set, the model achieved 99.0% accuracy, AUC = 0.990 and macro-F1 = 0.987. External validation across 824 scans yielded a mean AUC of 0.933 and an expected calibration error
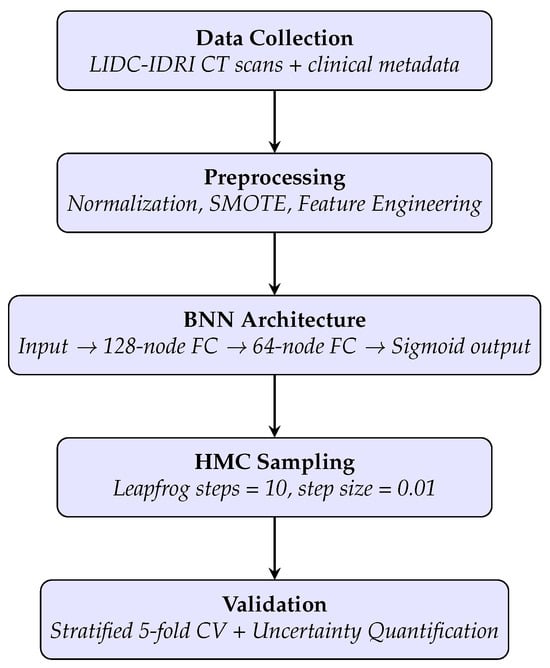
Figure 1
Open AccessArticle
An Effective Approach for Wearable Sensor-Based Human Activity Recognition in Elderly Monitoring
by
Youssef Errafik, Younes Dhassi, Mohamed Baghrous and Adil Kenzi
BioMedInformatics 2025, 5(3), 38; https://doi.org/10.3390/biomedinformatics5030038 - 9 Jul 2025
Abstract
Technological advancements and AI-based research have significantly influenced our daily lives. Human activity recognition (HAR) is a key area at the intersection of various AI technologies and application domains. In this study, we present our novel time series classification approach for monitoring the
[...] Read more.
Technological advancements and AI-based research have significantly influenced our daily lives. Human activity recognition (HAR) is a key area at the intersection of various AI technologies and application domains. In this study, we present our novel time series classification approach for monitoring the physical behaviors of the elderly and patients. This approach, which integrates supervised and unsupervised methods with generative models, has been validated for HAR, showing promising results. Our method was specifically adapted for healthcare and surveillance applications, enhancing the classification of physical behaviors in the elderly. The hybrid approach proved its effectiveness on the HAR70+ dataset, surpassing traditional recurrent convolutional network-based approaches. We further evaluated the surveillance system for the elderly (Surv-Sys-Elderly) model on the HARTH and HAR70+ datasets, achieving an accuracy of 94,3% on the HAR70+ dataset for recognizing elderly behaviors, highlighting its robustness and suitability for both clinical and domestic environments.
Full article
(This article belongs to the Topic Computational Intelligence and Bioinformatics (CIB))
►▼
Show Figures
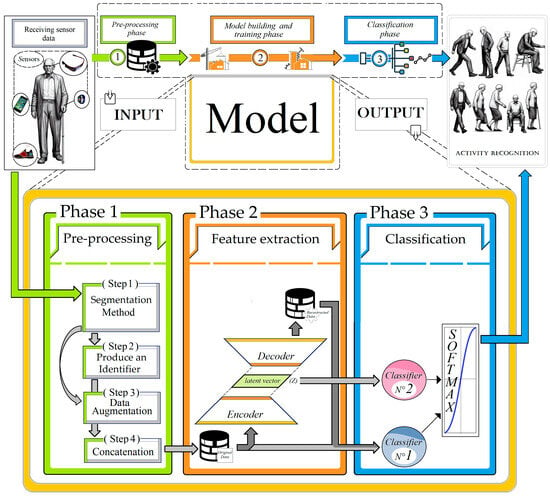
Figure 1
Open AccessReview
Generative Artificial Intelligence in Healthcare: Applications, Implementation Challenges, and Future Directions
by
Syed Arman Rabbani, Mohamed El-Tanani, Shrestha Sharma, Syed Salman Rabbani, Yahia El-Tanani, Rakesh Kumar and Manita Saini
BioMedInformatics 2025, 5(3), 37; https://doi.org/10.3390/biomedinformatics5030037 - 7 Jul 2025
Cited by 1
Abstract
Generative artificial intelligence (AI) is rapidly transforming healthcare systems since the advent of OpenAI in 2022. It encompasses a class of machine learning techniques designed to create new content and is classified into large language models (LLMs) for text generation and image-generating models
[...] Read more.
Generative artificial intelligence (AI) is rapidly transforming healthcare systems since the advent of OpenAI in 2022. It encompasses a class of machine learning techniques designed to create new content and is classified into large language models (LLMs) for text generation and image-generating models for creating or enhancing visual data. These generative AI models have shown widespread applications in clinical practice and research. Such applications range from medical documentation and diagnostics to patient communication and drug discovery. These models are capable of generating text messages, answering clinical questions, interpreting CT scan and MRI images, assisting in rare diagnoses, discovering new molecules, and providing medical education and training. Early studies have indicated that generative AI models can improve efficiency, reduce administrative burdens, and enhance patient engagement, although most findings are preliminary and require rigorous validation. However, the technology also raises serious concerns around accuracy, bias, privacy, ethical use, and clinical safety. Regulatory bodies, including the FDA and EMA, are beginning to define governance frameworks, while academic institutions and healthcare organizations emphasize the need for transparency, supervision, and evidence-based implementation. Generative AI is not a replacement for medical professionals but a potential partner—augmenting decision-making, streamlining communication, and supporting personalized care. Its responsible integration into healthcare could mark a paradigm shift toward more proactive, precise, and patient-centered systems.
Full article
(This article belongs to the Special Issue Integrating Health Informatics and Artificial Intelligence for Advanced Medicine)
►▼
Show Figures
Figure 1
Open AccessArticle
Self-Explaining Neural Networks for Food Recognition and Dietary Analysis
by
Zvinodashe Revesai and Okuthe P. Kogeda
BioMedInformatics 2025, 5(3), 36; https://doi.org/10.3390/biomedinformatics5030036 - 2 Jul 2025
Cited by 1
Abstract
►▼
Show Figures
Food pattern recognition plays a crucial role in modern healthcare by enabling automated dietary monitoring and personalised nutritional interventions, particularly for vulnerable populations with complex dietary needs. Current food recognition systems struggle to balance high accuracy with interpretability and computational efficiency when analysing
[...] Read more.
Food pattern recognition plays a crucial role in modern healthcare by enabling automated dietary monitoring and personalised nutritional interventions, particularly for vulnerable populations with complex dietary needs. Current food recognition systems struggle to balance high accuracy with interpretability and computational efficiency when analysing complex meal compositions in real-world settings. We developed a novel self-explaining neural architecture that integrates specialised attention mechanisms with temporal modules within a streamlined framework. Our methodology employs hierarchical feature extraction through successive convolution operations, multi-head attention mechanisms for pattern classification, and bidirectional LSTM networks for temporal analysis. Architecture incorporates self-explaining components utilising attention-based mechanisms and interpretable concept encoders to maintain transparency. We evaluated our model on the FOOD101 dataset using 5-fold cross-validation, ablation studies, and comprehensive computational efficiency assessments. Training employed multi-objective optimisation with adaptive learning rates and specialised loss functions designed for dietary pattern recognition. Experiments demonstrate our model’s superior performance, achieving 94.1% accuracy with only 29.3 ms inference latency and 3.8 GB memory usage, representing a 63.3% parameter reduction compared to baseline transformers. The system maintains detection rates above 84% in complex multi-item recognition scenarios, whilst feature attribution analysis achieved scores of 0.89 for primary components. Cross-validation confirmed consistent performance with accuracy ranging from 92.8% to 93.5% across all folds. This research advances automated dietary analysis by providing an efficient, interpretable solution for food recognition with direct applications in nutritional monitoring and personalised healthcare, particularly benefiting vulnerable populations who require transparent and trustworthy dietary guidance.
Full article

Figure 1
Highly Accessed Articles
Latest Books
E-Mail Alert
News
Topics
Topic in
AI, Algorithms, Applied Sciences, BioMedInformatics, Computers, Electronics, BDCC
Theoretical Foundations and Applications of Deep Learning Techniques
Topic Editors: Juan Ernesto Solanes Galbis, Adolfo Muñoz GarcíaDeadline: 31 March 2026
Topic in
Electronics, AgriEngineering, AI Sensors, Healthcare, AI, BioMedInformatics, BDCC, Information
Generative AI and Interdisciplinary Applications
Topic Editors: Jisheng Dang, Wenjie Wang, Yongqi Li, Juncheng LiDeadline: 31 August 2026

Special Issues
Special Issue in
BioMedInformatics
Feature Papers on Methods in Biomedical Informatics
Guest Editor: Rosalba GiugnoDeadline: 31 October 2025
Special Issue in
BioMedInformatics
Editor's Choice Series for Medical Statistics and Data Science Section
Guest Editor: Pentti NieminenDeadline: 31 October 2025
Special Issue in
BioMedInformatics
Editor's Choice Series for the Computational Biology and Medicine Section
Guest Editor: Hans BinderDeadline: 31 October 2025
Special Issue in
BioMedInformatics
Advances in Structural Bioinformatics and Next-Generation Sequence Analysis for Drug Design
Guest Editor: Alexandre G. De BrevernDeadline: 31 December 2025







