Identifying the Role of Disulfidptosis in Endometrial Cancer via Machine Learning Methods
Abstract
:1. Introduction
2. Materials And Methods
2.1. Download and Reprocess Data
2.2. Acquisition and Differential Analysis of Disulfidptosis-Related Genes
2.3. Identification of Disulfidptosis-Related Characteristic Genes
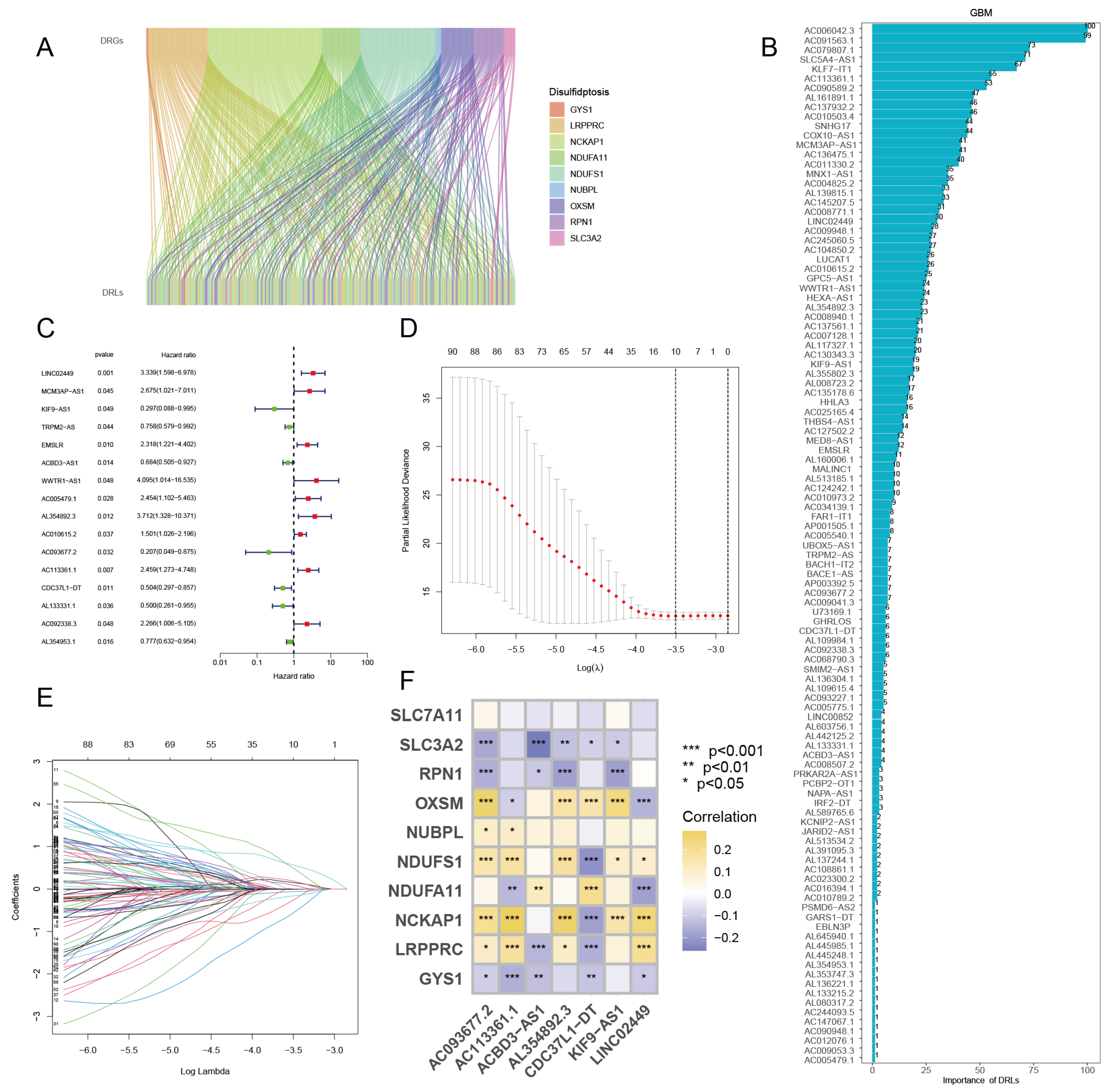
2.4. Construction of Cerna Regulatory Network
2.5. Identification of Disulfidptosis-Related Characteristic lncRNAs
2.6. Construction of Risk Prediction Model
2.7. Performance Evaluation of Risk Prediction Model
2.8. Mining of Drugs for Treating UCEC Patients
2.9. Cluster Analysis of DRGS
2.10. Cluster Analysis of DRCLS
2.11. Statistical Analysis
3. Results
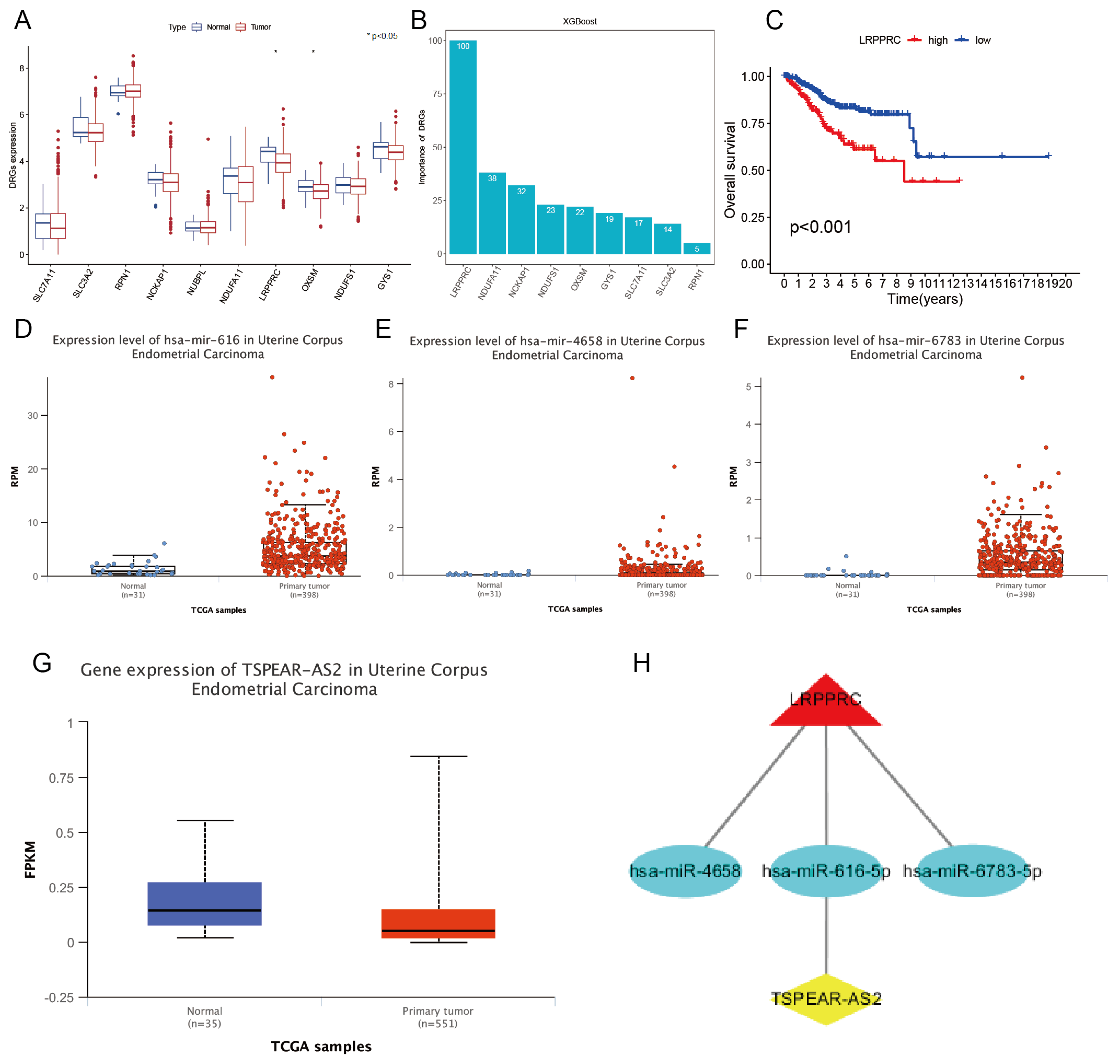
3.1. Identification of Disulfidptosis-Related Characteristic Genes Combined with the XGBoost Algorithm
3.2. Construction of LRPPRC-Related ceRNA Network
3.3. Identification of Disulfidptosis-Related Characteristic lncRNAs via GBM Algorithm
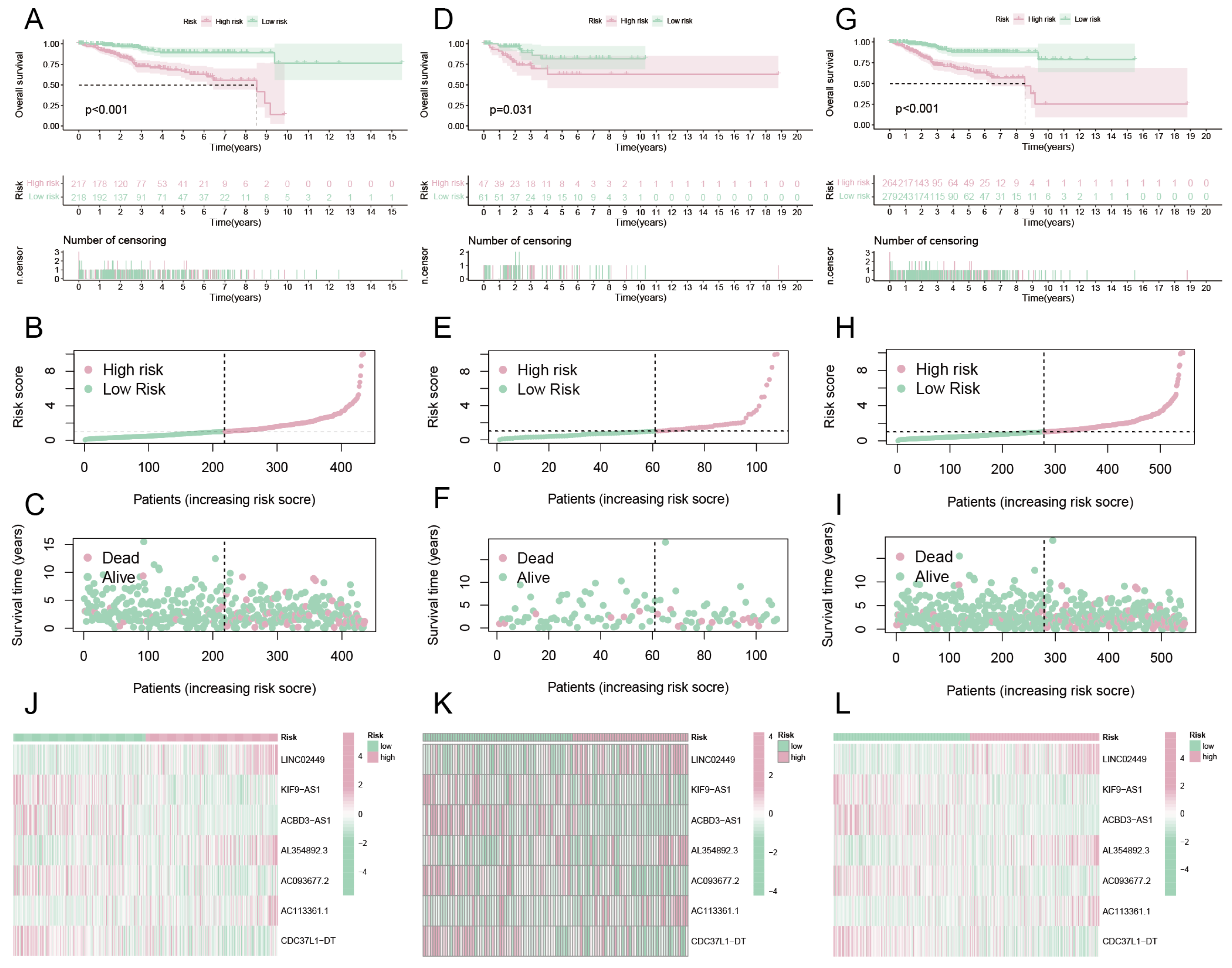
3.4. Constructing a Risk Prediction Model Based on DRCLS
3.5. Survival Analysis
3.6. Evaluate the Performance of the Risk Forecasting Model
3.7. Risk Assessment of DRGS and Survival Analysis of Subgroups of Clinical Traits
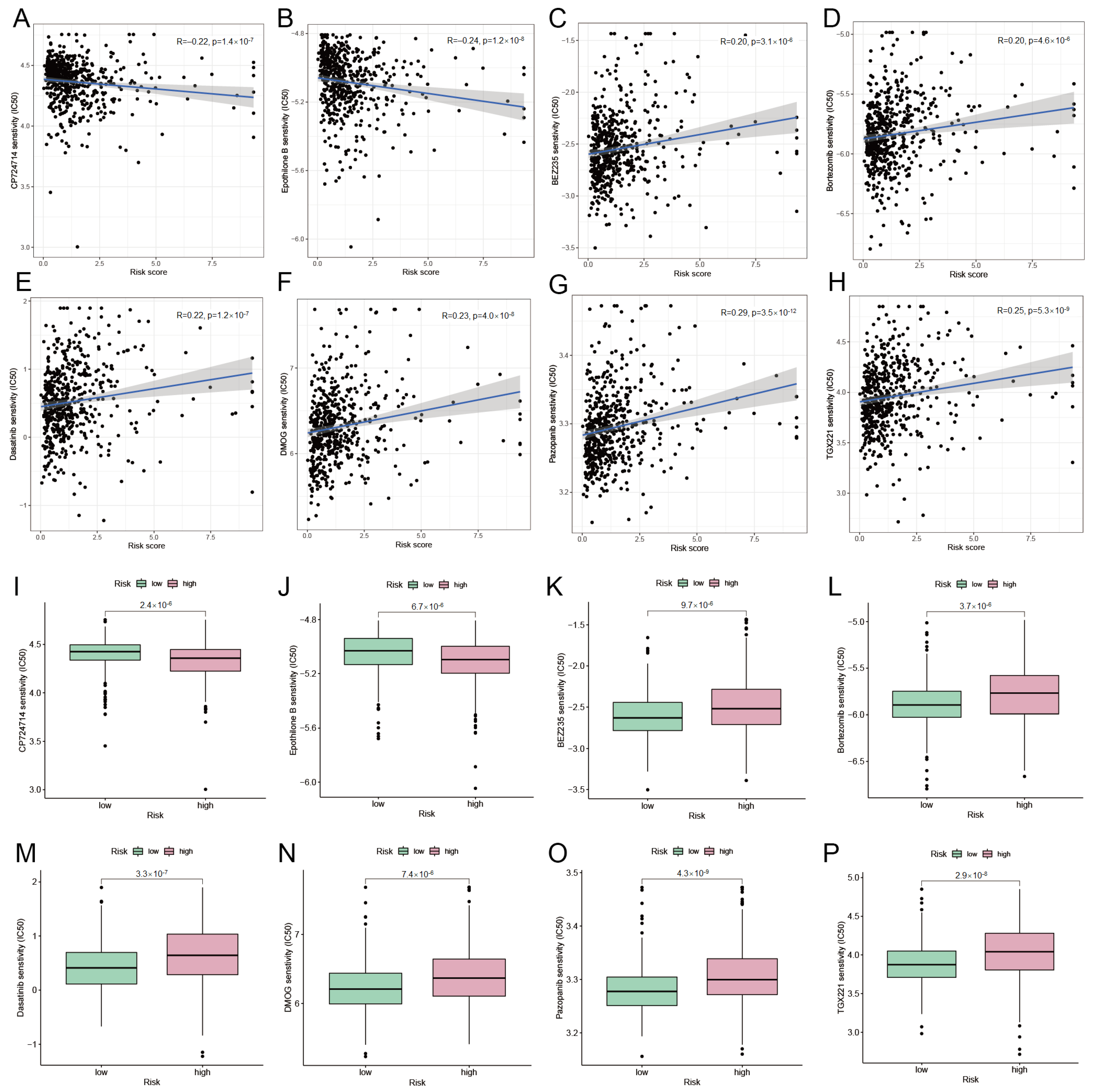
3.8. Mining Potentially Sensitive Drugs for UCEC Treatment
3.9. Biological Characteristics of DRG Cluster
3.10. Biological Characteristics of DRCL Cluster
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| UCEC | Uterine Corpus Endometrial Carcinoma |
| TCGA | The Cancer Genome Atlas |
| FPKM | Fragments per kilobase of transcript per million |
| DRGs | disulfidptosis-related genes |
| DRCG | disulfidptosis-related characteristic gene |
| ceRNA | competing endogenous RNA |
| XGBoost | eXtreme Gradient Boosting |
| GBM | Gradient Boosting Machine |
| DRLs | disulfidptosis-related lncRNAs |
| DRCLs | disulfidptosis-related characteristic lncRNAs |
| LASSO | Least absolute shrinkage and selection operator |
| OS | Overall survival |
| ROC | Receiver operating characteristic curve |
| AUC | Area under curve |
| CI | Concordance index |
| IC50 | the half maximal inhibitory concentration |
| NMF | non-negative matrix factorization |
| PCA | Principal Component Analysis |
| GSVA | Gene Set Variation Analysis |
| ssGSEA | single-sample Gene Set Enrichment Analysis |
References
- Crosbie, E.J.; Kitson, S.J.; McAlpine, J.N.; Mukhopadhyay, A.; Powell, M.E.; Singh, N. Endometrial cancer. Lancet 2022, 399, 1412–1428. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer statistics, 2023. CA A Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef] [PubMed]
- Brooks, R.A.; Fleming, G.F.; Lastra, R.R.; Lee, N.K.; Moroney, J.W.; Son, C.H.; Tatebe, K.; Veneris, J.L. Current recommendations and recent progress in endometrial cancer. CA A Cancer J. Clin. 2019, 69, 258–279. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.Q.; Xiang, Y.; Meng, F.; Ji, C.H.; Xiao, R.; Li, J.P.; Dai, Z.T.; Liao, X.H. ALDH2 promotes uterine corpus endometrial carcinoma proliferation and construction of clinical survival prognostic model. Aging 2021, 13, 23588. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Yin, X.; Zhang, L.; Ye, Z.; Liang, G. Identification of drug targets and prognosis projection for uterine carcinosarcoma based on alternative splicing events. Comput. Biol. Med. 2023, 152, 106346. [Google Scholar] [CrossRef] [PubMed]
- Budczies, J.; Seidel, A.; Christopoulos, P.; Endris, V.; Kloor, M.; Gyorffy, B.; Seliger, B.; Schirmacher, P.; Stenzinger, A.; Denkert, C. Integrated analysis of the immunological and genetic status in and across cancer types: Impact of mutational signatures beyond tumor mutational burden. Oncoimmunology 2018, 7, e1526613. [Google Scholar] [CrossRef] [PubMed]
- El Khoury, L.Y.; Lin, W.H.; Smadbeck, J.B.; Barrett, M.T.; Sadeghian, D.; McCune, A.F.; Karagouga, G.; Cheville, J.C.; Harris, F.R.; Kinsella, L.M.; et al. Epigenomics may begin to explain in vitro differential response to hypomethylating agents in MMR-D hypermethylated endometrial cancer. Epigenomics 2023, 15, 283–292. [Google Scholar] [CrossRef]
- Riedinger, C.J.; Brown, M.; Haight, P.J.; Backes, F.J.; Cohn, D.E.; Goodfellow, P.J.; Cosgrove, C.M. Epigenetic MMR defect identifies a risk group not accounted for through traditional risk stratification algorithms in endometrial cancer. Front. Oncol. 2023, 13, 1147657. [Google Scholar] [CrossRef]
- Cosgrove, C.M.; Cohn, D.E.; Hampel, H.; Frankel, W.L.; Jones, D.; McElroy, J.P.; Suarez, A.A.; Zhao, W.; Chen, W.; Salani, R.; et al. Epigenetic silencing of MLH1 in endometrial cancers is associated with larger tumor volume, increased rate of lymph node positivity and reduced recurrence-free survival. Gynecol. Oncol. 2017, 146, 588–595. [Google Scholar] [CrossRef]
- Liu, X.; Nie, L.; Zhang, Y.; Yan, Y.; Wang, C.; Colic, M.; Olszewski, K.; Horbath, A.; Chen, X.; Lei, G.; et al. Actin cytoskeleton vulnerability to disulfide stress mediates disulfidptosis. Nat. Cell Biol. 2023, 25, 404–414. [Google Scholar] [CrossRef]
- Zheng, P.; Zhou, C.; Ding, Y.; Duan, S. Disulfidptosis: A new target for metabolic cancer therapy. J. Exp. Clin. Cancer Res. 2023, 42, 103. [Google Scholar] [CrossRef] [PubMed]
- Sezgin, B.; Pirinççi, F.; Camuzcuoğlu, A.; Erel, Ö.; Neşelioğlu, S.; Camuzcuoğlu, H. Assessment of thiol disulfide balance in early-stage endometrial cancer. J. Obstet. Gynaecol. Res. 2020, 46, 1140–1147. [Google Scholar] [CrossRef] [PubMed]
- Fernando, S.R.; Kottawatta, K.S.A.; Jiang, L.; Chen, X.; Cheng, K.W.; Wong, B.P.C.; Ng, E.H.Y.; Yeung, W.S.B.; Lee, K.F. Differential expression of protein disulfide isomerase (PDI) in regulating endometrial receptivity in humans. Reprod. Biol. 2021, 21, 100498. [Google Scholar] [CrossRef] [PubMed]
- Cheng, D.L.; Xiang, Y.Y.; Ji, L.j.; Lu, X.J. Competing endogenous RNA interplay in cancer: Mechanism, methodology, and perspectives. Tumor Biol. 2015, 36, 479–488. [Google Scholar] [CrossRef] [PubMed]
- Shi, R.; Zhang, W.; Zhang, J.; Yu, Z.; An, L.; Zhao, R.; Zhou, X.; Wang, Z.; Wei, S.; Wang, H. CircESRP1 enhances metastasis and epithelial–mesenchymal transition in endometrial cancer via the miR-874-3p/CPEB4 axis. J. Transl. Med. 2022, 20, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Xin, W.; Gao, X.; Zhao, S.; Zhao, P.; Yu, H.; Wu, Q.; Hua, K. LncRNA RP11-395G23. 3 suppresses the endometrial cancer progression via regulating microRNA-205-5p/PTEN axis. Am. J. Transl. Res. 2020, 12, 4422. [Google Scholar]
- Vallone, C.; Rigon, G.; Gulia, C.; Baffa, A.; Votino, R.; Morosetti, G.; Zaami, S.; Briganti, V.; Catania, F.; Gaffi, M.; et al. Non-coding RNAs and endometrial cancer. Genes 2018, 9, 187. [Google Scholar] [CrossRef] [PubMed]
- He, W.P.; Chen, Y.Y.; Wu, L.X.; Guo, Y.; You, Z.S.; Yang, G. A novel necroptosis-related lncRNA signature for predicting prognosis and anti-cancer treatment response in endometrial cancer. Front. Immunol. 2022, 13, 1018544. [Google Scholar] [CrossRef]
- Liu, J.; Cui, G.; Ye, J.; Wang, Y.; Wang, C.; Bai, J. Comprehensive analysis of the prognostic signature of mutation-derived genome instability-related lncRNAs for patients with endometrial cancer. Front. Cell Dev. Biol. 2022, 10, 753957. [Google Scholar] [CrossRef]
- Winterhoff, B.; Hamidi, H.; Wang, C.; Kalli, K.R.; Fridley, B.L.; Dering, J.; Chen, H.W.; Cliby, W.A.; Wang, H.J.; Dowdy, S.; et al. Molecular classification of high grade endometrioid and clear cell ovarian cancer using TCGA gene expression signatures. Gynecol. Oncol. 2016, 141, 95–100. [Google Scholar] [CrossRef]
- Urick, M.E.; Bell, D.W. Clinical actionability of molecular targets in endometrial cancer. Nat. Rev. Cancer 2019, 19, 510–521. [Google Scholar] [CrossRef]
- Miller, K.D.; Nogueira, L.; Devasia, T.; Mariotto, A.B.; Yabroff, K.R.; Jemal, A.; Kramer, J.; Siegel, R.L. Cancer treatment and survivorship statistics, 2022. CA A Cancer J. Clin. 2022, 72, 409–436. [Google Scholar] [CrossRef] [PubMed]
- Miao, C.; Fang, X.; Chen, Y.; Zhao, Y.; Guo, Q. Identification of an eight-m6A RNA methylation regulator prognostic signature of uterine corpus endometrial carcinoma based on bioinformatics analysis. Medicine 2021, 100, e27689. [Google Scholar] [CrossRef] [PubMed]
- Shan, J.; Geng, R.; Zhang, Y.; Wei, J.; Liu, J.; Bai, J. Identification of cuproptosis-related subtypes, establishment of a prognostic model and tumor immune landscape in endometrial carcinoma. Comput. Biol. Med. 2022, 149, 105988. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Ye, Z.; Xiao, G.; He, T. Prognostic signature construction and immunotherapy response analysis for Uterine Corpus Endometrial Carcinoma based on cuproptosis-related lncRNAs. Comput. Biol. Med. 2023, 159, 106905. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Xu, Y.; Liu, B.; Zheng, L.; Cao, C.; Wu, P.; Ding, W.; Ren, F. A novel cuproptosis-related gene signature for overall survival prediction in uterine corpus endometrial carcinoma (UCEC). Heliyon 2023, 9, e14613. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y. Identification and validation of cuproptosis-related prognostic signature and associated regulatory axis in uterine corpus endometrial carcinoma. Front. Genet. 2022, 13, 912037. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Tian, R.; Tan, H.; Liu, J.; Ou, C.; Li, Y.; Fu, X. A comprehensive analysis focusing on cuproptosis to investigate its clinical and biological relevance in uterine corpus endometrial carcinoma and its potential in indicating prognosis. Front. Mol. Biosci. 2022, 9, 1048356. [Google Scholar] [CrossRef] [PubMed]
- Mahmoodi, N.; Ai, J.; Ebrahimi-Barough, S.; Hassannejad, Z.; Hasanzadeh, E.; Basiri, A.; Vaccaro, A.R.; Rahimi-Movaghar, V. Microtubule stabilizer epothilone B as a motor neuron differentiation agent for human endometrial stem cells. Cell Biol. Int. 2020, 44, 1168–1183. [Google Scholar] [CrossRef]
- Moxley, K.M.; McMeekin, D.S. Endometrial carcinoma: A review of chemotherapy, drug resistance, and the search for new agents. Oncologist 2010, 15, 1026–1033. [Google Scholar] [CrossRef]
- Sorolla, A.; Yeramian, A.; Valls, J.; Dolcet, X.; Bergadà, L.; Llombart-Cussac, A.; Martí, R.M.; Matias-Guiu, X. Blockade of NFκB activity by Sunitinib increases cell death in Bortezomib-treated endometrial carcinoma cells. Mol. Oncol. 2012, 6, 530–541. [Google Scholar] [CrossRef] [PubMed]
- Coleman, R.L.; Hu, W.; Soliman, P.; Nick, A.; Ramirez, P.T.; Westin, S.N.; Garcia, M.E.; Zhu, Z.; Palancia, J.; Fellman, B.M.; et al. Dasatinib, paclitaxel, and carboplatin in women with advanced-stage or recurrent endometrial cancer: A pilot clinical and translational study. Gynecol. Oncol. 2021, 161, 104–112. [Google Scholar] [CrossRef] [PubMed]
- Pazarentzos, E.; Giannikopoulos, P.; Hrustanovic, G.; St John, J.; Olivas, V.; Gubens, M.; Balassanian, R.; Weissman, J.; Polkinghorn, W.; Bivona, T. Oncogenic activation of the PI3-kinase p110β isoform via the tumor-derived PIK3CβD1067V kinase domain mutation. Oncogene 2016, 35, 1198–1205. [Google Scholar] [CrossRef] [PubMed]
- Troisi, J.; Sarno, L.; Landolfi, A.; Scala, G.; Martinelli, P.; Venturella, R.; Di Cello, A.; Zullo, F.; Guida, M. Metabolomic signature of endometrial cancer. J. Proteome Res. 2018, 17, 804–812. [Google Scholar] [CrossRef] [PubMed]
- Gu, M.; Chen, X.; Sun, Y.; Wang, L.; Shu, H.; Qian, C. A metabolomic signature of FIGO stage I and II endometrial cancer. Neoplasma 2021, 68, 1283–1291. [Google Scholar] [CrossRef] [PubMed]
- Shi, K.; Wang, Q.; Su, Y.; Xuan, X.; Liu, Y.; Chen, W.; Qian, Y.; Lash, G.E. Identification and functional analyses of differentially expressed metabolites in early stage endometrial carcinoma. Cancer Sci. 2018, 109, 1032–1043. [Google Scholar] [CrossRef] [PubMed]
- Si, L.; Liu, L.; Yang, R.; Li, W.; Xu, X. High expression of TARS is associated with poor prognosis of endometrial cancer. Aging 2023, 15, 1524. [Google Scholar] [CrossRef]
- Kolben, T.; Mannewitz, M.; Perleberg, C.; Schnell, K.; Anz, D.; Hahn, L.; Meister, S.; Schmoeckel, E.; Burges, A.; Czogalla, B.; et al. Presence of regulatory T-cells in endometrial cancer predicts poorer overall survival and promotes progression of tumor cells. Cell. Oncol. 2022, 45, 1171–1185. [Google Scholar] [CrossRef]
- Liu, X.; Cheng, C.; Cai, Y.; Gu, Y.; Wu, Y.; Chen, K.; Wu, Z. Pan-cancer analyses reveal the regulation and clinical outcome association of PCLAF in human tumors. Int. J. Oncol. 2022, 60, 66. [Google Scholar] [CrossRef]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fu, F.; Lu, X.; Zhang, Z.; Li, Z.; Xie, Q. Identifying the Role of Disulfidptosis in Endometrial Cancer via Machine Learning Methods. BioMedInformatics 2023, 3, 908-925. https://doi.org/10.3390/biomedinformatics3040056
Fu F, Lu X, Zhang Z, Li Z, Xie Q. Identifying the Role of Disulfidptosis in Endometrial Cancer via Machine Learning Methods. BioMedInformatics. 2023; 3(4):908-925. https://doi.org/10.3390/biomedinformatics3040056
Chicago/Turabian StyleFu, Fei, Xuesong Lu, Zhushanying Zhang, Zhi Li, and Qinlan Xie. 2023. "Identifying the Role of Disulfidptosis in Endometrial Cancer via Machine Learning Methods" BioMedInformatics 3, no. 4: 908-925. https://doi.org/10.3390/biomedinformatics3040056
APA StyleFu, F., Lu, X., Zhang, Z., Li, Z., & Xie, Q. (2023). Identifying the Role of Disulfidptosis in Endometrial Cancer via Machine Learning Methods. BioMedInformatics, 3(4), 908-925. https://doi.org/10.3390/biomedinformatics3040056








