Interpretable Medical Imagery Diagnosis with Self-Attentive Transformers: A Review of Explainable AI for Health Care
Abstract
:1. Introduction
2. Interpretable Model: eXplainable AI (XAI)
3. Primaries on Vision Transformer
4. Explainability Methods in XAI
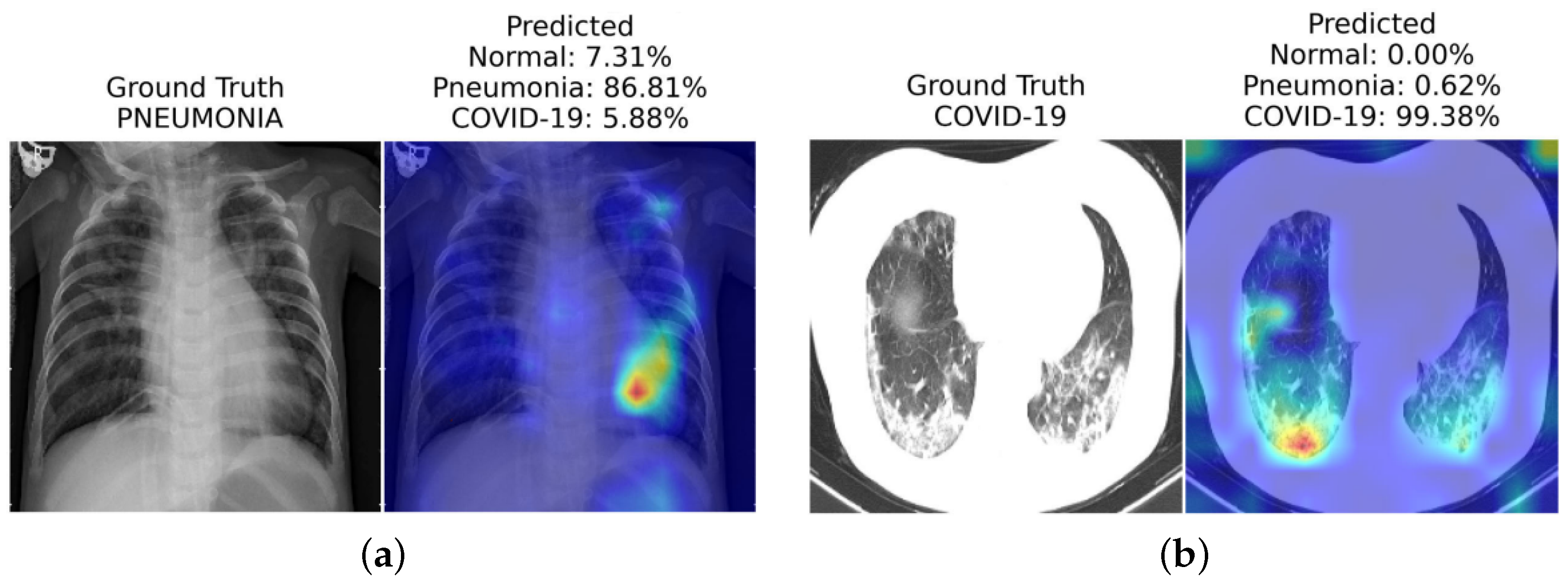
4.1. Gradient-Weighted Class Activation Mapping (Grad-CAM) Method
4.1.1. Saliency Maps
4.1.2. Concept Activation Vectors (CAVs)
4.1.3. Deep Learning Important Features (DeepLift)
4.1.4. Layer-Wise Relevance Propagation (LRP)
4.1.5. Guided Back-Propagation
5. Vision Transformer for Medical Images
5.1. Black Box Methods
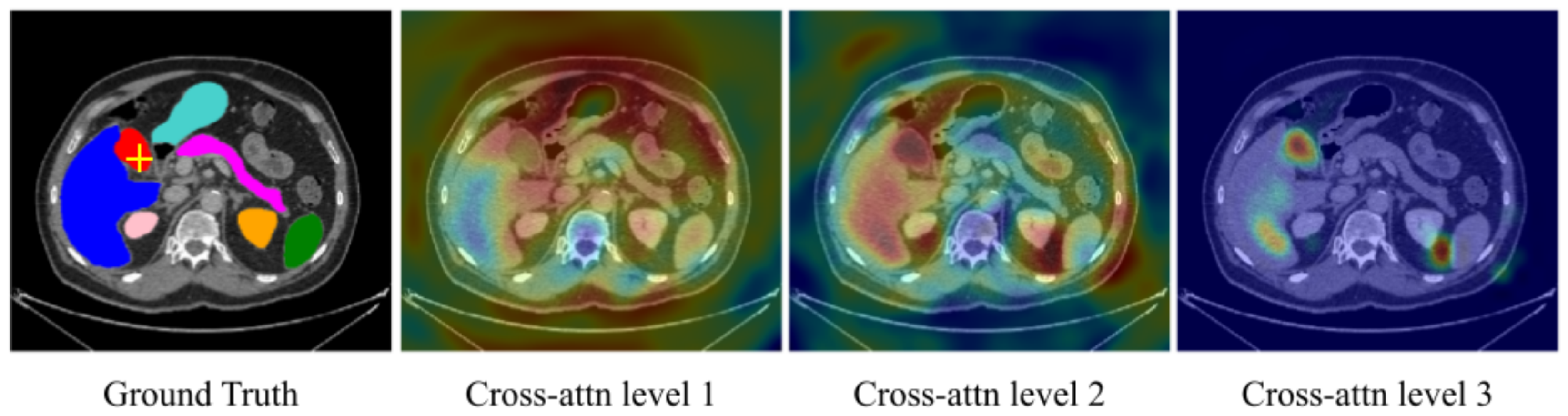
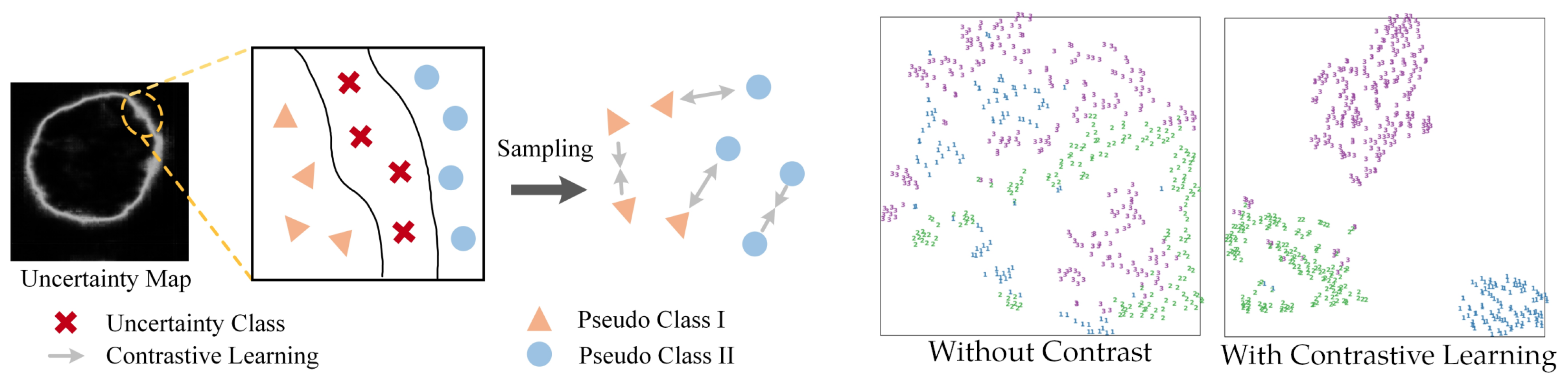
5.2. Interpretable Vision Transformer
| Interoperability Method | Class Specific? | Metrics | Highlights and Summary | |||
|---|---|---|---|---|---|---|
| Pixel Acc. | mAP | mF1 | mIoU | |||
| Raw Attention | 67.87 | 80.24 | 29.44 | 46.37 | Raw attention only consider the attention map of the last block of the transformer architecture | |
| Rollout [60] | 73.54 | 84.76 | 43.68 | 55.42 | Rollout assume a linear Combination of tokens and quantify the influence of skip connections with identity mateix | |
| GradCAM [23] | ✓ | 65.91 | 71.60 | 19.42 | 41.30 | Provides a class-specific explanation by adding weights to gradient based feature map |
| Partial LRP [69] | 76.31 | 84.67 | 38.82 | 57.95 | Considers the information flow within the network by identifying the most important heads in each encoder layer through relevance propagation | |
| Transformer Attribution [61] | ✓ | 76.30 | 85.28 | 41.85 | 58.34 | Combines relevancy and attention-map gradient by regarding the gradient as a weight to the relevance for certain prediction task |
| Generic Attribution [70] | ✓ | 79.68 | 85.99 | 40.10 | 61.92 | Generic attribution extends the usage of Transformer attribution to co-attention and self-attention based models with a generic relevancy update rule |
| Token-wise Approx. [71] | ✓ | 82.15 | 88.04 | 45.72 | 66.32 | Uses head-wise and token-wise approximations to visualize tokens interaction in the pooled vector with noise-decreasing strategy |
6. Conclusions
7. Limitation and Future Directions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Esteva, A.; Chou, K.; Yeung, S.; Naik, N.; Madani, A.; Mottaghi, A.; Liu, Y.; Topol, E.; Dean, J.; Socher, R. Deep learning-enabled medical computer vision. NPJ Digit. Med. 2021, 4, 5. [Google Scholar] [CrossRef] [PubMed]
- Shung, K.K.; Smith, M.B.; Tsui, B.M. Principles of Medical Imaging; Academic Press: Cambridge, MA, USA, 2012. [Google Scholar]
- Hu, B.; Vasu, B.; Hoogs, A. X-MIR: EXplainable Medical Image Retrieval. In Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision, Waikoloa, HI, USA, 3–8 January 2022; pp. 440–450. [Google Scholar]
- Lucieri, A.; Bajwa, M.N.; Braun, S.A.; Malik, M.I.; Dengel, A.; Ahmed, S. ExAID: A Multimodal Explanation Framework for Computer-Aided Diagnosis of Skin Lesions. arXiv 2022, arXiv:2201.01249. [Google Scholar] [CrossRef] [PubMed]
- Stieler, F.; Rabe, F.; Bauer, B. Towards Domain-Specific Explainable AI: Model Interpretation of a Skin Image Classifier using a Human Approach. In Proceedings of the 2021 IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops (CVPRW), Nashville, TN, USA, 19–25 June 2021; pp. 1802–1809. [Google Scholar] [CrossRef]
- Lucieri, A.; Bajwa, M.N.; Braun, S.A.; Malik, M.I.; Dengel, A.; Ahmed, S. On interpretability of deep learning based skin lesion classifiers using concept activation vectors. In Proceedings of the 2020 international joint conference on neural networks (IJCNN), Glasgow, UK, 19–24 July 2020; pp. 1–10. [Google Scholar]
- Lenis, D.; Major, D.; Wimmer, M.; Berg, A.; Sluiter, G.; Bühler, K. Domain aware medical image classifier interpretation by counterfactual impact analysis. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Vancouver, BC, Canada, 8–12 October 2020; pp. 315–325. [Google Scholar]
- Brunese, L.; Mercaldo, F.; Reginelli, A.; Santone, A. Explainable deep learning for pulmonary disease and coronavirus COVID-19 detection from X-rays. Comput. Methods Programs Biomed. 2020, 196, 105608. [Google Scholar] [CrossRef]
- Corizzo, R.; Dauphin, Y.; Bellinger, C.; Zdravevski, E.; Japkowicz, N. Explainable image analysis for decision support in medical healthcare. In Proceedings of the 2021 IEEE International Conference on Big Data (Big Data), Orlando, FL, USA, 15–18 December 2021; pp. 4667–4674. [Google Scholar]
- Mondal, A.K.; Bhattacharjee, A.; Singla, P.; Prathosh, A. xViTCOS: Explainable vision transformer based COVID-19 screening using radiography. IEEE J. Transl. Eng. Health Med. 2021, 10, 1–10. [Google Scholar] [CrossRef]
- Bang, J.S.; Lee, M.H.; Fazli, S.; Guan, C.; Lee, S.W. Spatio-Spectral Feature Representation for Motor Imagery Classification Using Convolutional Neural Networks. IEEE Trans. Neural Netw. Learn. Syst. 2021, 33, 3038–3049. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Shi, H.; Hwang, K.S. An explainable ensemble feedforward method with Gaussian convolutional filter. Knowl.-Based Syst. 2021, 225, 107103. [Google Scholar] [CrossRef]
- Mohagheghi, S.; Foruzan, A.H. Developing an explainable deep learning boundary correction method by incorporating cascaded x-Dim models to improve segmentation defects in liver CT images. Comput. Biol. Med. 2022, 140, 105106. [Google Scholar] [CrossRef]
- Hu, H.; Lai, T.; Farid, F. Feasibility Study of Constructing a Screening Tool for Adolescent Diabetes Detection Applying Machine Learning Methods. Sensors 2022, 22, 6155. [Google Scholar] [CrossRef]
- Yang, S.; Zhu, F.; Ling, X.; Liu, Q.; Zhao, P. Intelligent Health Care: Applications of Deep Learning in Computational Medicine. Front. Genet. 2021, 12, 607471. [Google Scholar] [CrossRef]
- Lai, T.; Shi, Y.; Du, Z.; Wu, J.; Fu, K.; Dou, Y.; Wang, Z. Psy-LLM: Scaling up Global Mental Health Psychological Services with AI-based Large Language Models. arXiv 2023, arXiv:2307.11991. [Google Scholar]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An image is worth 16x16 words: Transformers for image recognition at scale. arXiv 2020, arXiv:2010.11929. [Google Scholar]
- Ker, J.; Wang, L.; Rao, J.; Lim, T. Deep learning applications in medical image analysis. IEEE Access 2017, 6, 9375–9389. [Google Scholar] [CrossRef]
- MacDonald, S.; Steven, K.; Trzaskowski, M. Interpretable AI in healthcare: Enhancing fairness, safety, and trust. In Artificial Intelligence in Medicine: Applications, Limitations and Future Directions; Springer: Berlin/Heidelberg, Germany, 2022; pp. 241–258. [Google Scholar]
- Ghosh, A.; Kandasamy, D. Interpretable artificial intelligence: Why and when. Am. J. Roentgenol. 2020, 214, 1137–1138. [Google Scholar] [CrossRef] [PubMed]
- Arrieta, A.B.; Díaz-Rodríguez, N.; Del Ser, J.; Bennetot, A.; Tabik, S.; Barbado, A.; García, S.; Gil-López, S.; Molina, D.; Benjamins, R.; et al. Explainable Artificial Intelligence (XAI): Concepts, taxonomies, opportunities and challenges toward responsible AI. Inf. Fusion 2020, 58, 82–115. [Google Scholar] [CrossRef]
- Ribeiro, M.T.; Singh, S.; Guestrin, C. “Why should i trust you?” Explaining the predictions of any classifier. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 1135–1144. [Google Scholar]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-cam: Visual explanations from deep networks via gradient-based localization. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 618–626. [Google Scholar]
- Shin, D. User perceptions of algorithmic decisions in the personalized AI system: Perceptual evaluation of fairness, accountability, transparency, and explainability. J. Broadcast. Electron. Media 2020, 64, 541–565. [Google Scholar] [CrossRef]
- Balasubramaniam, N.; Kauppinen, M.; Hiekkanen, K.; Kujala, S. Transparency and explainability of AI systems: Ethical guidelines in practice. In Proceedings of the International Working Conference on Requirements Engineering: Foundation for Software Quality, Birmingham, UK, 21–24 March 2022; pp. 3–18. [Google Scholar]
- Lai, T.; Farid, F.; Bello, A.; Sabrina, F. Ensemble Learning based Anomaly Detection for IoT Cybersecurity via Bayesian Hyperparameters Sensitivity Analysis. arXiv 2023, arXiv:2307.10596. [Google Scholar]
- Imai, T. Legal regulation of autonomous driving technology: Current conditions and issues in Japan. IATSS Res. 2019, 43, 263–267. [Google Scholar] [CrossRef]
- Gilpin, L.H.; Bau, D.; Yuan, B.Z.; Bajwa, A.; Specter, M.; Kagal, L. Explaining explanations: An overview of interpretability of machine learning. In Proceedings of the 2018 IEEE 5th International Conference on data science and advanced analytics (DSAA), Turin, Italy, 1–3 October 2018; pp. 80–89. [Google Scholar]
- Banerjee, A.; Chakraborty, C.; Rathi Sr, M. Medical imaging, artificial intelligence, internet of things, wearable devices in terahertz healthcare technologies. In Terahertz Biomedical and Healthcare Technologies; Elsevier: Amsterdam, The Netherlands, 2020; pp. 145–165. [Google Scholar]
- Loh, H.W.; Ooi, C.P.; Seoni, S.; Barua, P.D.; Molinari, F.; Acharya, U.R. Application of explainable artificial intelligence for healthcare: A systematic review of the last decade (2011–2022). Comput. Methods Programs Biomed. 2022, 226, 107161. [Google Scholar] [CrossRef]
- He, Z.; Tang, X.; Yang, X.; Guo, Y.; George, T.J.; Charness, N.; Quan Hem, K.B.; Hogan, W.; Bian, J. Clinical trial generalizability assessment in the big data era: A review. Clin. Transl. Sci. 2020, 13, 675–684. [Google Scholar] [CrossRef]
- Autio, L.; Juhola, M.; Laurikkala, J. On the neural network classification of medical data and an endeavour to balance non-uniform data sets with artificial data extension. Comput. Biol. Med. 2007, 37, 388–397. [Google Scholar] [CrossRef]
- Chen, C.; Zhang, P.; Zhang, H.; Dai, J.; Yi, Y.; Zhang, H.; Zhang, Y. Deep learning on computational-resource-limited platforms: A survey. Mob. Inf. Syst. 2020, 2020, 8454327. [Google Scholar] [CrossRef]
- Abirami, S.; Chitra, P. Energy-efficient edge based real-time healthcare support system. In Advances in Computers; Elsevier: Amsterdam, The Netherlands, 2020; Volume 117, pp. 339–368. [Google Scholar]
- Zhang, Z.; Genc, Y.; Wang, D.; Ahsen, M.E.; Fan, X. Effect of ai explanations on human perceptions of patient-facing ai-powered healthcare systems. J. Med. Syst. 2021, 45, 64. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.R.; Hullman, J.; Bertini, E. Human factors in model interpretability: Industry practices, challenges, and needs. Proc. Acm -Hum.-Comput. Interact. 2020, 4, 1–26. [Google Scholar] [CrossRef]
- Felzmann, H.; Fosch-Villaronga, E.; Lutz, C.; Tamò-Larrieux, A. Towards transparency by design for artificial intelligence. Sci. Eng. Ethics 2020, 26, 3333–3361. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Ethics and Governance of Artificial Intelligence for Health: WHO Guidance; World Health Organization: Geneva, Switzerland, 2021. [Google Scholar]
- Ahmad, M.A.; Teredesai, A.; Eckert, C. Interpretable Machine Learning in Healthcare. In Proceedings of the 2018 IEEE International Conference on Healthcare Informatics (ICHI), New York, NY, USA, 4–7 June 2018; p. 447. [Google Scholar] [CrossRef]
- Burrell, J. How the machine ‘thinks’: Understanding opacity in machine learning algorithms. Big Data Soc. 2016, 3, 2053951715622512. [Google Scholar] [CrossRef]
- Arnold, M.H. Teasing out artificial intelligence in medicine: An ethical critique of artificial intelligence and machine learning in medicine. J. Bioethical Inq. 2021, 18, 121–139. [Google Scholar] [CrossRef] [PubMed]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention is all you need. In Proceedings of the Advances in Neural Information Processing Systems, Long Beach, CA, USA, 4–9 December 2017; Volume 30. [Google Scholar]
- Hendrycks, D.; Gimpel, K. Gaussian error linear units (gelus). arXiv 2016, arXiv:1606.08415. [Google Scholar]
- Lipton, Z.C. The Mythos of Model Interpretability. arXiv 2016, arXiv:1606.03490. [Google Scholar] [CrossRef]
- Freitas, A.A. Comprehensible Classification Models: A Position Paper. SIGKDD Explor. Newsl. 2014, 15, 1–10. [Google Scholar] [CrossRef]
- Miller, T. Explanation in Artificial Intelligence: Insights from the Social Sciences. arXiv 2017, arXiv:1706.07269. [Google Scholar] [CrossRef]
- Simonyan, K.; Vedaldi, A.; Zisserman, A. Deep Inside Convolutional Networks: Visualizing Image Classification Models and Saliency Maps. arXiv 2013, arXiv:1312.6034. [Google Scholar] [CrossRef]
- Molnar, C. Interpretable Machine Learning; Lulu Press: Morrisville, NC, USA, 2020. [Google Scholar]
- Kim, B.; Wattenberg, M.; Gilmer, J.; Cai, C.; Wexler, J.; Viegas, F.; Sayres, R. Interpretability Beyond Feature Attribution: Quantitative Testing with Concept Activation Vectors (TCAV). arXiv 2017, arXiv:1711.11279. [Google Scholar] [CrossRef]
- Shrikumar, A.; Greenside, P.; Kundaje, A. Learning important features through propagating activation differences. In Proceedings of the International Conference on Machine Learning, PMLR, Sydney, Australia, 6–11 August 2017; pp. 3145–3153. [Google Scholar]
- Bach, S.; Binder, A.; Montavon, G.; Klauschen, F.; Müller, K.R.; Samek, W. On Pixel-Wise Explanations for Non-Linear Classifier Decisions by Layer-Wise Relevance Propagation. PLoS ONE 2015, 10, e0130140. [Google Scholar] [CrossRef] [PubMed]
- Springenberg, J.; Dosovitskiy, A.; Brox, T.; Riedmiller, M. Striving for Simplicity: The All Convolutional Net. arXiv 2014, arXiv:1412.6806. [Google Scholar]
- Dai, Y.; Gao, Y.; Liu, F. Transmed: Transformers advance multi-modal medical image classification. Diagnostics 2021, 11, 1384. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.; Pan, Y.; Nie, D.; Liu, F.; Shi, F.; Xia, Y.; Shen, D. SMILE: Sparse-Attention based Multiple Instance Contrastive Learning for Glioma Sub-Type Classification Using Pathological Images. In Proceedings of the MICCAI Workshop on Computational Pathology, PMLR, Virtual Event, 27 September 2021; pp. 159–169. [Google Scholar]
- Napel, S.; Plevritis, S.K. NSCLC Radiogenomics: Initial Stanford Study of 26 Cases. The Cancer Imaging Archive2014. Available online: https://wiki.cancerimagingarchive.net/pages/viewpage.action?pageId=6883610 (accessed on 29 July 2023).
- Gheflati, B.; Rivaz, H. Vision Transformers for Classification of Breast Ultrasound Images. arXiv 2021, arXiv:2110.14731. [Google Scholar]
- Khan, A.; Lee, B. Gene Transformer: Transformers for the Gene Expression-based Classification of Lung Cancer Subtypes. arXiv 2021, arXiv:2108.11833. [Google Scholar]
- Chen, H.; Li, C.; Li, X.; Wang, G.; Hu, W.; Li, Y.; Liu, W.; Sun, C.; Yao, Y.; Teng, Y.; et al. GasHis-Transformer: A Multi-scale Visual Transformer Approach for Gastric Histopathology Image Classification. arXiv 2021, arXiv:2104.14528. [Google Scholar]
- Jiang, Z.; Dong, Z.; Wang, L.; Jiang, W. Method for Diagnosis of Acute Lymphoblastic Leukemia Based on ViT-CNN Ensemble Model. Comput. Intell. Neurosci. 2021, 2021, 7529893. [Google Scholar] [CrossRef]
- Abnar, S.; Zuidema, W. Quantifying attention flow in transformers. arXiv 2020, arXiv:2005.00928. [Google Scholar]
- Chefer, H.; Gur, S.; Wolf, L. Transformer interpretability beyond attention visualization. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Virtual Conference, 19–25 June 2021; pp. 782–791. [Google Scholar]
- Park, S.; Kim, G.; Oh, Y.; Seo, J.B.; Lee, S.M.; Kim, J.H.; Moon, S.; Lim, J.K.; Ye, J.C. Vision Transformer for COVID-19 CXR Diagnosis using Chest X-ray Feature Corpus. arXiv 2021, arXiv:2103.07055. [Google Scholar]
- Chen, T.; Kornblith, S.; Norouzi, M.; Hinton, G. A simple framework for contrastive learning of visual representations. In Proceedings of the International Conference on Machine Learning, PMLR, Virtual Event, 13–18 July 2020; pp. 1597–1607. [Google Scholar]
- Irvin, J.; Rajpurkar, P.; Ko, M.; Yu, Y.; Ciurea-Ilcus, S.; Chute, C.; Marklund, H.; Haghgoo, B.; Ball, R.; Shpanskaya, K.; et al. Chexpert: A large chest radiograph dataset with uncertainty labels and expert comparison. In Proceedings of the AAAI Conference on Artificial Intelligence, Honolulu, HI, USA, 27 January–1 February 2019; Volume 33, pp. 590–597. [Google Scholar]
- Gao, X.; Qian, Y.; Gao, A. COVID-VIT: Classification of COVID-19 from CT chest images based on vision transformer models. arXiv 2021, arXiv:2107.01682. [Google Scholar]
- Kollias, D.; Arsenos, A.; Soukissian, L.; Kollias, S. MIA-COV19D: COVID-19 Detection through 3-D Chest CT Image Analysis. arXiv 2021, arXiv:2106.07524. [Google Scholar]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708. [Google Scholar]
- Gunraj, H.; Sabri, A.; Koff, D.; Wong, A. COVID-Net CT-2: Enhanced Deep Neural Networks for Detection of COVID-19 from Chest CT Images Through Bigger, More Diverse Learning. arXiv 2021, arXiv:2101.07433. [Google Scholar] [CrossRef] [PubMed]
- Voita, E.; Talbot, D.; Moiseev, F.; Sennrich, R.; Titov, I. Analyzing Multi-Head Self-Attention: Specialized Heads Do the Heavy Lifting, the Rest Can Be Pruned. In Proceedings of the 57th Annual Meeting of the Association for Computational Linguistics. Association for Computational Linguistics, Florence, Italy, 28 July–2 August 2019. [Google Scholar]
- Chefer, H.; Gur, S.; Wolf, L. Generic attention-model explainability for interpreting bi-modal and encoder-decoder transformers. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Virtual Conference, 11–17 October 2021; pp. 397–406. [Google Scholar]
- Chen, J.; Li, X.; Yu, L.; Dou, D.; Xiong, H. Beyond Intuition: Rethinking Token Attributions inside Transformers. Transactions on Machine Learning Research. 2023. Available online: https://openreview.net/pdf?id=rm0zIzlhcX (accessed on 29 July 2023).
- Shome, D.; Kar, T.; Mohanty, S.N.; Tiwari, P.; Muhammad, K.; AlTameem, A.; Zhang, Y.; Saudagar, A.K.J. COVID-Transformer: Interpretable COVID-19 Detection Using Vision Transformer for Healthcare. Int. J. Environ. Res. Public Health 2021, 18, 11086. [Google Scholar] [CrossRef]
- Shao, Z.; Bian, H.; Chen, Y.; Wang, Y.; Zhang, J.; Ji, X.; Zhang, Y. TransMIL: Transformer based Correlated Multiple Instance Learning for Whole Slide Image Classication. arXiv 2021, arXiv:2106.00908. [Google Scholar]
- Huang, J.; Xing, X.; Gao, Z.; Yang, G. Swin deformable attention u-net transformer (sdaut) for explainable fast mri. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Singapore, 18–22 September 2022; pp. 538–548. [Google Scholar]
- Petit, O.; Thome, N.; Rambour, C.; Themyr, L.; Collins, T.; Soler, L. U-net transformer: Self and cross attention for medical image segmentation. In Proceedings of the Machine Learning in Medical Imaging: 12th International Workshop, MLMI 2021, Held in Conjunction with MICCAI 2021, Strasbourg, France, 27 September 2021; Proceedings 12. Springer: Berlin/Heidelberg, Germany, 2021; pp. 267–276. [Google Scholar]
- Fung, G.; Dundar, M.; Krishnapuram, B.; Rao, R.B. Multiple instance learning for computer aided diagnosis. Adv. Neural Inf. Process. Syst. 2007, 19, 425. [Google Scholar]
- Bejnordi, B.E.; Veta, M.; Van Diest, P.J.; Van Ginneken, B.; Karssemeijer, N.; Litjens, G.; Van Der Laak, J.A.; Hermsen, M.; Manson, Q.F.; Balkenhol, M.; et al. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. Jama 2017, 318, 2199–2210. [Google Scholar] [CrossRef]
- Zhang, H.; Meng, Y.; Zhao, Y.; Qiao, Y.; Yang, X.; Coupland, S.E.; Zheng, Y. DTFD-MIL: Double-tier feature distillation multiple instance learning for histopathology whole slide image classification. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, New Orleans, LA, USA, 18–24 June 2022; pp. 18802–18812. [Google Scholar]
- Playout, C.; Duval, R.; Boucher, M.C.; Cheriet, F. Focused attention in transformers for interpretable classification of retinal images. Med. Image Anal. 2022, 82, 102608. [Google Scholar] [CrossRef]
- Zheng, Y.; Gindra, R.; Betke, M.; Beane, J.; Kolachalama, V.B. A deep learning based graph-transformer for whole slide image classification. medRxiv 2021. [Google Scholar] [CrossRef]
- Kipf, T.N.; Welling, M. Semi-supervised classification with graph convolutional networks. arXiv 2016, arXiv:1609.02907. [Google Scholar]
- Xie, Y.; Zhang, J.; Shen, C.; Xia, Y. Cotr: Efficiently bridging cnn and transformer for 3d medical image segmentation. In Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2021: 24th International Conference, Strasbourg, France, 27 September–1 October 2021; Proceedings, Part III 24. Springer: Abingdon, UK, 2021; pp. 171–180. [Google Scholar]
- Wang, T.; Lu, J.; Lai, Z.; Wen, J.; Kong, H. Uncertainty-guided pixel contrastive learning for semi-supervised medical image segmentation. In Proceedings of the Thirty-First International Joint Conference on Artificial Intelligence, IJCAI, Vienna, Austria, 23–29 July; pp. 1444–1450.


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lai, T. Interpretable Medical Imagery Diagnosis with Self-Attentive Transformers: A Review of Explainable AI for Health Care. BioMedInformatics 2024, 4, 113-126. https://doi.org/10.3390/biomedinformatics4010008
Lai T. Interpretable Medical Imagery Diagnosis with Self-Attentive Transformers: A Review of Explainable AI for Health Care. BioMedInformatics. 2024; 4(1):113-126. https://doi.org/10.3390/biomedinformatics4010008
Chicago/Turabian StyleLai, Tin. 2024. "Interpretable Medical Imagery Diagnosis with Self-Attentive Transformers: A Review of Explainable AI for Health Care" BioMedInformatics 4, no. 1: 113-126. https://doi.org/10.3390/biomedinformatics4010008
APA StyleLai, T. (2024). Interpretable Medical Imagery Diagnosis with Self-Attentive Transformers: A Review of Explainable AI for Health Care. BioMedInformatics, 4(1), 113-126. https://doi.org/10.3390/biomedinformatics4010008






