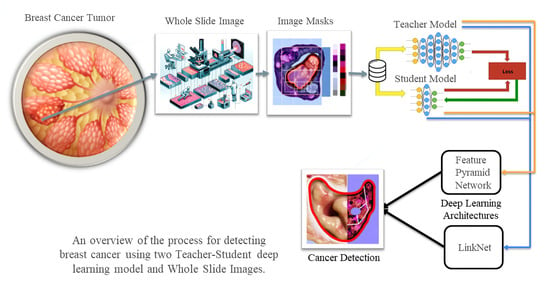
Deep Segmentation Techniques for Breast Cancer Diagnosis
Abstract
1. Introduction
2. Literature Review
2.1. Global Disparities in Breast Cancer
2.2. Deep Learning Models: Convolutional Neurol Network (CNN)
2.3. Deep Learning Models, Backpropagation, and Gradient Descent
2.4. Algorithm and Equations
2.5. Combining Backpropagation and Gradient Descent
2.6. Whole Slide Images (WSIs)
2.7. Segmentation
2.8. Common Deep Learning Models Used for Cancer Detection
3. Methodology
3.1. Segmentation Model Training Strategy
3.1.1. Dice Coefficient (Sørensen–Dice Index)
3.1.2. Jaccard Index (Intersection over Union)
3.2. About the Dataset
3.3. Deep Learning Models Being Compared
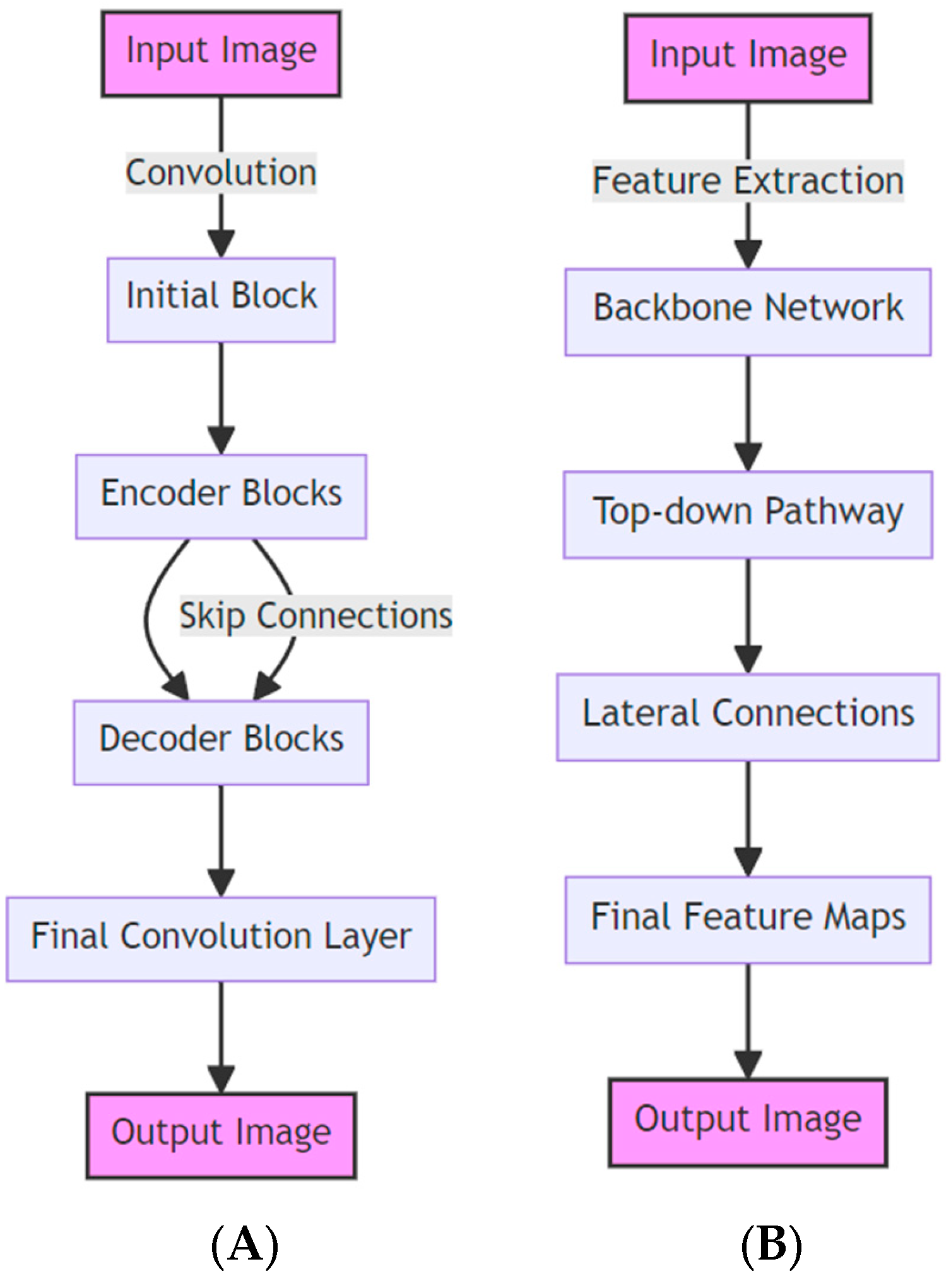
3.3.1. LinkNet
3.3.2. Feature Pyramid Network
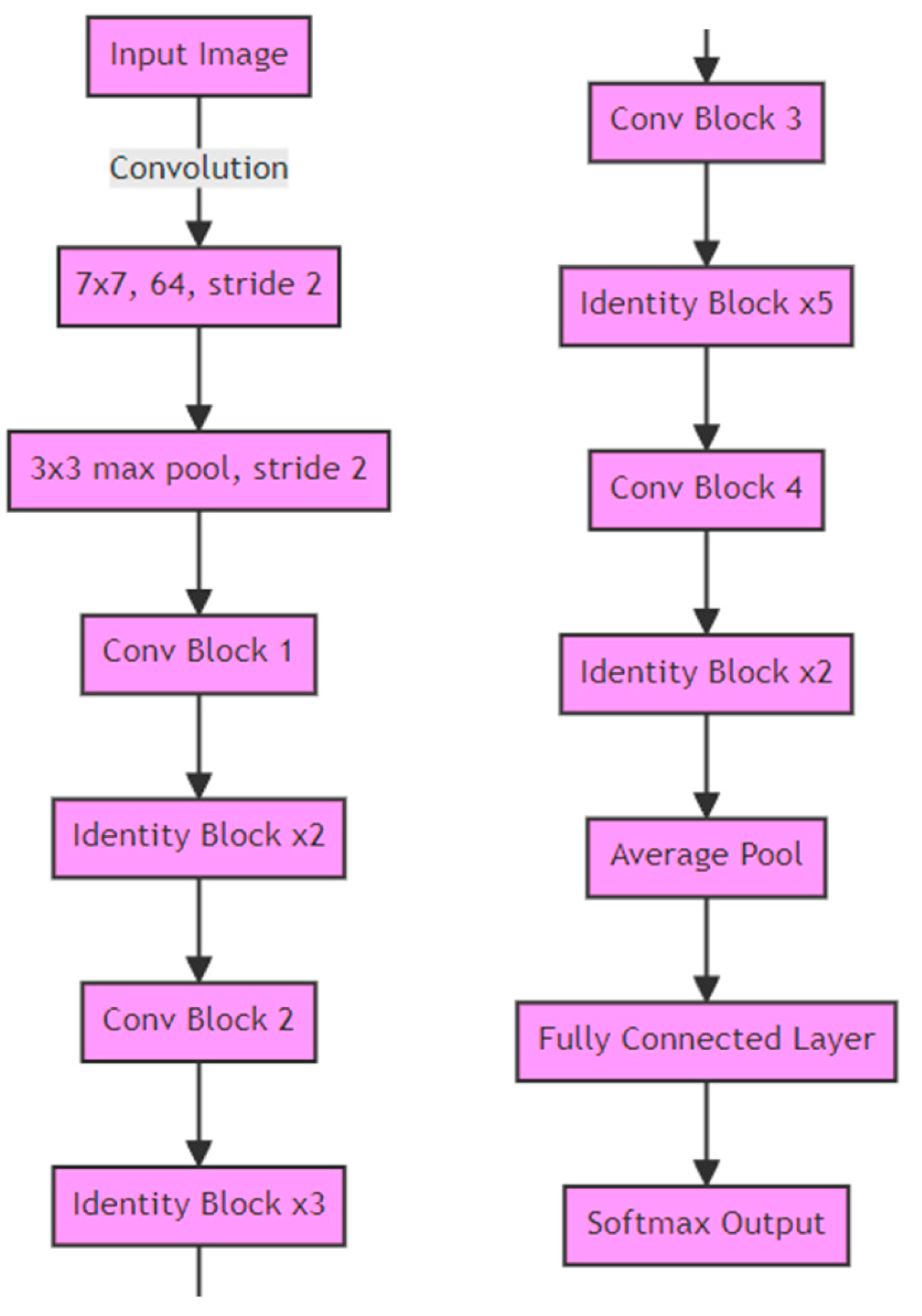
3.3.3. ResNet34 Layer Encoder Used in Both Models
3.4. Summary
4. Results Analysis
4.1. Experiment with Linknet Architecture and Resent34 Base
4.1.1. Teacher Model Findings
4.1.2. Student Model Findings
4.2. Experiment with FPN Architecture and Resent34 Base
4.2.1. Teacher Model Findings
4.2.2. Student Model Findings
4.3. Discussion
4.4. Implications of the Study
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| No. | Year | Model (Architecture) | Description |
|---|---|---|---|
| 1 | 2014 | CRF Deep Learning | Combines a basic Convolutional Neural Network (CNN) with a Conditional Random Field (CRF) for improved image segmentation, enhancing boundary delineation by refining the CNN’s output with the CRF [44]. |
| 2 | 2014 | Fully Convolutional Networks (FCN) | Pioneering architecture in semantic segmentation that uses convolutional layers to process images of any size and outputs segmentation maps [45]. |
| 3 | 2015 | U-Net | A highly effective network for medical image segmentation, featuring a U-shaped architecture that excels in tasks where data are limited [19]. |
| 4 | 2015 | ReSeg | A model based on Recurrent Neural Networks (RNNs) and FCN, designed for semantic image segmentation, leveraging the sequential nature of RNNs for improved segmentation [46]. |
| 5 | 2015 | Deconvolution Network | Uses deconvolutional layers to perform up-sampling of feature maps, enabling precise localization in semantic segmentation tasks [47]. |
| 6 | 2015 | Dilated ConvNet | Incorporates dilated convolutions to expand the receptive field without reducing resolution, enhancing performance in dense prediction tasks like semantic segmentation [48]. |
| 7 | 2015 | ParseNet | Enhances FCNs by adding global context to improve segmentation accuracy, focusing on understanding the whole scene context [49]. |
| 8 | 2015 | SegNet | SegNet was designed for road scene understanding in the context of autonomous driving [50]. |
| 9 | 2016 | DeepLab | DeepLabv1 and its successive versions (v2, v3, v3+, and v4) made significant contributions in semantic segmentation, incorporating dilated convolutions, atrous spatial pyramid pooling, and encoder–decoder structures [51]. |
| 10 | 2016 | PSPNet | Proposed Pyramid Scene Parsing Network for scene parsing tasks [52]. |
| 11 | 2016 | Instance-Aware Segmentaiton | This approach to segmentation is designed to not only classify pixels but also differentiate between separate instances of the same class in the image. It is commonly used in scenarios where identifying individual objects (instances) is crucial, such as in instance segmentation tasks [53]. |
| 12 | 2016 | V-Net | An adaptation of the U-Net model for volumetric (3D) medical image segmentation. It is particularly effective for tasks like organ segmentation in 3D medical scans, using a similar architecture to U-Net but extended to three dimensions [54]. |
| 13 | 2016 | ENet | A lightweight and efficient network designed for real-time semantic segmentation, particularly in mobile or low-power devices. It achieves a good balance between accuracy and speed, making it suitable for applications where computational resources are limited [55]. |
| 14 | 2016 | RefineNet | Utilizes a multi-path refinement network for high-resolution semantic segmentation [56]. |
| 15 | 2017 | Tiramisu | This is also known as The One Hundred Layers Tiramisu; it utilizes DenseNets for semantic segmentation [57]. |
| 16 | 2017 | Mask R-CNN | An extension of Faster R-CNN, Mask R-CNN is effective for instance segmentation tasks [22]. |
| 17 | 2017 | FC-DenseNet | Combines the principles of DenseNets (densely connected convolutional networks) with FCNs, leading to efficient and accurate semantic segmentation, especially in medical imaging [57]. |
| 18 | 2017 | Global Convolutional Net | Designed for semantic segmentation, this network uses large kernels and global convolutional layers to handle both classification and localization tasks effectively [58]. |
| 19 | DeepLab V3 | An advanced version of DeepLab, it employs atrous convolutions and spatial pyramid pooling to effectively segment objects at multiple scales [59]. | |
| 20 | 2017 | FPN—Feature Pyramid Network | A versatile architecture used in both object detection and segmentation, it builds a multi-scale feature pyramid from a single input image for efficient and accurate detection at multiple scales [38]. |
| 21 | 2017 | LinkNet | Utilizes an encoder–decoder architecture for fast and accurate semantic segmentation [26]. |
| 22 | 2018 | ICNet | Designed for real-time semantic segmentation tasks [60]. |
| 23 | 2018 | Attention U-Net | Incorporates attention mechanisms into the U-Net architecture [61]. |
| 24 | 2018 | Nested U-Net | A U-Net architecture with nested and dense skip pathways [60]. |
| 25 | 2018 | Panoptic Segmentation | A unified model that simultaneously performs semantic segmentation [62]. |
| 26 | 2018 | Mask-Lab | A deep learning model that combines semantic segmentation, direction prediction, and instance center prediction for instance segmentation tasks [63]. |
| 27 | 2018 | Path Aggregation Network | Enhances feature hierarchy and representation capability for object detection by enabling efficient multi-scale feature aggregation [64]. |
| 28 | 2018 | Dense-ASSP | A network that integrates dense connections and atrous spatial pyramid pooling for robust semantic image segmentation [65]. |
| 29 | 2018 | Excuse | A model that fuses semantic and boundary information at multiple levels to enhance feature representation and segmentation accuracy [63]. |
| 30 | 2018 | Context Encoding Network | Focuses on capturing global context information for semantic segmentation, often using a context encoding module to improve performance [66]. |
| 31 | 2019 | Panoptic FPN | A framework that combines the Feature Pyramid Network (FPN) with panoptic segmentation, effectively handling both object detection and segmentation tasks [41]. |
| 32 | 2019 | Gated-SCNN | A semantic segmentation network with a gated shape stream that focuses on capturing shape information alongside the usual texture features [67]. |
| 33 | 2019 | UPS-Net | A unified panoptic segmentation network that effectively combines instance and semantic segmentation tasks into a single, coherent framework [68]. |
| 34 | 2019 | TensorMask | A dense prediction model for instance segmentation that uses structured 4D tensors to represent masks, enabling precise spatial understanding [69]. |
| 35 | 2019 | HRNet | Maintains high-resolution representations through the network, enhancing performance in tasks like semantic segmentation and object detection [70]. |
| 36 | 2019 | CC-Net: CrissCross Attention | Employs criss-cross attention to capture long-range contextual information in a computationally efficient manner for semantic segmentation [71]. |
| 37 | 2017 | Dual Attention Network | Integrates position and channel attention mechanisms to capture rich contextual dependencies for improved scene segmentation [72]. |
| 38 | 2019 | Fast-SCNN | A fast and efficient network design for semantic segmentation on road scenes [73]. |
| 39 | 2020 | DPT | Vision transformer-based architecture for segmentation tasks [74]. |
| 40 | 2020 | SETR | Another Vision Transformer-based method for segmentation shows the effectiveness of transformers in dense prediction tasks [75]. |
| 41 | 2020 | PointRend | Aims at rendering fine-grained detail in segmentation through iterative subdivision [74]. |
| 42 | 2020 | EfficientPS | Combines semantic segmentation and object detection efficiently [76]. |
| 43 | 2019 | FasterSeg | An architecture search-based approach for real-time semantic segmentation [77]. |
| 44 | 2018 | MAnet | Utilizes multi-head attention mechanisms for semantic segmentation [60]. |
| 45 | 2020 | FasterSeg | FasterSeg is an AI-designed segmentation network that outperforms traditional models in speed and accuracy by using advanced neural architecture search and collaborative frameworks [78]. |
| 46 | 2020 | PolarMask, | A novel single-shot instance segmentation method that represents object masks in a polar co-ordinate system; simplifies the instance segmentation process [79]. |
| 47 | 2020 | CenterMask | An efficient anchor-free instance segmentation model that extends the CenterNet object detector by adding a spatial attention-guided mask branch [80]. |
| 48 | 2020 | SC-NAS | Stands for “Semantic-Context Neural Architecture Search”. It is a network architecture search method designed to optimize semantic segmentation networks by considering the semantic context of the task [81]. |
| 49 | 2020 | EffientNet + NAS-FPN | This combines EfficientNet, a scalable and efficient network architecture, with NAS-FPN (Neural Architecture Search Feature Pyramid Network), a method for automatically designing feature pyramid architectures for object detection tasks. This combination aims to optimize both efficiency and accuracy in detection models [82]. |
| 50 | 2020 | Multi-scale Adaptive Feature Fusion Network | Multi-scale Adaptive Feature Fusion Network for Semantic Segmentation in Remote Sensing Images [83]. |
| 51 | 2021 | TUNet | TransUNet [84]. |
| 52 | 2021 | SUnet | Swin-Unet, Swin-Transformer [85]. |
| 53 | 2021 | Segm | Segmenter [86]. |
| 54 | 2021 | MedT | Medical Transformer [87]. |
| 55 | 2021 | BEiT | BERT Image Transformers [88]. |
| 56 | 2023 | CrossFormer | A Hybrid Transformer Architecture for Semantic Segmentation [89] |
| 57 | 2022 | MLP-Mixer | Semantic Segmentation with Transformer and MLP-Mixer [90]. |
| 58 | 2022 | Transformer-Powered Semantic Segmentation | Transformer-Powered Semantic Segmentation with Large-Scale Instance Discrimination [91]. |
| 59 | 2023 | Adaptive Context Fusion | Semantic Segmentation with Adaptive Context Fusion [89]. |
| 60 | 2023 | Multi-Scale Vision Transformers | Semantic Segmentation with Multi-Scale Vision Transformers [92] |
| 61 | 2023 | Hiformer: Hierarchical multi-scale | Semantic Segmentation with Hierarchical Vision Transformers [93] |
References
- Rosenblatt, F. The Perceptron: A probabilistic model for information storage and organization in the brain. Psychol. Rev. 1958, 65, 386–408. [Google Scholar] [CrossRef] [PubMed]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet classification with deep convolutional neural networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef]
- Arnold, M.; Morgan, E.; Rumgay, H.; Mafra, A.; Singh, D.; Laversanne, M.; Vignat, J.; Gralow, J.R.; Cardoso, F.; Sisesling, S.; et al. Current and future burden of breast cancer: Global statistics for 2020 and 2040. Breast 2022, 66, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Pacal, İ. Deep learning approaches for classification of breast cancer in ultrasound (US) images. J. Inst. Sci. Technol. 2022, 12, 1917–1927. [Google Scholar] [CrossRef]
- Bengio, Y.; Courville, A.; Vincent, P. Representation learning: A review and new perspectives. IEEE Trans. Pattern Anal. Mach. Intell. 2013, 35, 1798–1828. [Google Scholar] [CrossRef] [PubMed]
- Schmidhuber, J. Deep learning in neural networks: An overview. Neural Netw. 2015, 61, 85–117. [Google Scholar] [CrossRef]
- Ananthachari, P.; Schutte, S. Big Data Tools, Deep Learning & 3D Objects in the Metaverse. In Digitalization and Management Innovation II: Proceedings of DMI 2023; IOS Press: Amsterdam, The Netherlands, 2023; Volume 376, p. 236. [Google Scholar]
- Alzubaidi, L.; Zhang, J.; Humaidi, A.J.; Al-Dujaili, A.; Duan, Y.; Al-Shamma, O.; Santamaría, J.; Fadhel, M.A.; Al-Amidie, M.; Farhan, L. Review of deep learning: Concepts, CNN architectures, challenges, applications, future directions. J. Big Data 2021, 8, 1–74. [Google Scholar] [CrossRef]
- Rumelhart, D.E.; Hinton, G.E.; Williams, R.J. Learning representations by back-propagating errors. Nature 1986, 323, 533–536. [Google Scholar] [CrossRef]
- Cui, N. Applying gradient descent in convolutional neural networks. In Proceedings of the 2nd International Conference on Machine Vision and Information Technology (CMVIT 2018), Hong Kong, China, 23–25 February 2018; Volume 1004, p. 012027. [Google Scholar]
- Ruder, S. An overview of gradient descent optimization algorithms. arXiv 2016, arXiv:1609.04747. [Google Scholar]
- Kumar, N.; Gupta, R.; Gupta, S. Whole slide imaging (WSI) in pathology: Current perspectives and future directions. J. Digit. Imaging 2020, 33, 1034–1040. [Google Scholar] [CrossRef]
- Pantanowitz, L.; Farahani, N.; Parwani, A. Whole slide imaging in pathology: Advantages, limitations, and emerging perspectives. Pathol. Lab. Med. Int. 2015, 7, 22–23. [Google Scholar] [CrossRef]
- Campanella, G.; Hanna, M.G.; Geneslaw, L.; Miraflor, A.; Werneck Krauss Silva, V.; Busam, K.J.; Brogi, E.; Reuter, V.E.; Klimstra, D.S.; Fuchs, T.J. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat. Med. 2019, 25, 1301–1309. [Google Scholar] [CrossRef] [PubMed]
- Janowczyk, A.; Madabhushi, A. Deep learning for digital pathology image analysis: A comprehensive tutorial with selected use cases. J. Pathol. Inform. 2016, 7, 29. [Google Scholar] [CrossRef] [PubMed]
- Litjens, G.; Sánchez, C.I.; Timofeeva, N.; Hermsen, M.; Nagtegaal, I.; Kovacs, I.; Hulsbergen-van de kaa, C.; Bult, P.; van Ginneken, B.; van der Laak, J. Deep learning as a tool for increased accuracy and efficiency of histopathological diagnosis. Sci. Rep. 2016, 6, 26286. [Google Scholar] [CrossRef] [PubMed]
- Oxford Learner’s Dictionaries. Segmentation Noun—Definition, Pictures, Pronunciation and Usage Notes. Available online: https://www.oxfordlearnersdictionaries.com/us/definition/english/segmentation (accessed on 20 December 2023).
- Chowdhary, C.L.; Acharjya, D.P. Segmentation and feature extraction in medical imaging: A systematic review. Procedia Comput. Sci. 2020, 167, 26–36. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference, Munich, Germany, 5–9 October 2015; pp. 234–241. [Google Scholar]
- Protonotarios, N.E.; Katsamenis, I.; Sykiotis, S.; Dikaios, N.; Kastis, G.A.; Chatziioannou, S.N.; Metaxas, M.; Doulamis, N.; Doulamis, A. A few-shot U-Net deep learning model for lung cancer lesion segmentation via PET/CT imaging. Biomed. Phys. Eng. Express 2022, 8, 025019. [Google Scholar] [CrossRef] [PubMed]
- Falk, T.; Mai, D.; Bensch, R.; Çiçek, Ö.; Abdulkadir, A.; Marrakchi, Y.; Böhm, A.; Deubner, J.; Jäckel, Z.; Seiwald, K.; et al. U-Net: Deep Learning for Cell Counting, Detection, and Morphometry. Nat. Methods 2019, 16, 67–70. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Gkioxari, G.; Dollár, P.; Girshick, R. Mask r-cnn. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 2961–2969. [Google Scholar]
- Soleimani, P.; Farezi, N. Utilizing deep learning via the 3D U-net neural network for the delineation of brain stroke lesions in MRI image. Sci. Rep. 2023, 13, 19808. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, S.; Durairaju, K.; Deeba, K.; Mathivanan, S.K.; Karthikeyan, P.; Shah, M.A. Multimodal Bi-omedical Image Segmentation using Multi-Dimensional U-Convolutional Neural Network. BMC Med. Imaging 2024, 24, 38. [Google Scholar] [CrossRef]
- Yi, X.; Walia, E.; Babyn, P. Generative adversarial network in medical imaging: A review. Med. Image Anal. 2019, 58, 101552. [Google Scholar] [CrossRef]
- Chaurasia, A.; Culurciello, E. Linknet: Exploiting encoder representations for efficient semantic segmentation. In Proceedings of the 2017 IEEE Visual Communications and Image Processing (VCIP), St. Petersburg, FL, USA, 10–13 December 2017; pp. 1–4. [Google Scholar]
- Zhang, H.; Xu, Z.; Yao, D.; Zhang, S.; Chen, J.; Lukasiewicz, T. Multi-Head Feature Pyramid Networks for Breast Mass Detection. In Proceedings of the ICASSP 2023—2023 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Rhodes Island, Greece, 4–10 June 2023; pp. 1–5. [Google Scholar]
- Wang, Y.; Ahsan, U.; Li, H.; Hagen, M. A comprehensive review of modern object segmentation approaches. Found. Trends Comput. Graph. Vis. 2022, 13, 111–283. [Google Scholar] [CrossRef]
- Fathi, A.; Wojna, Z.; Rathod, V.; Wang, P.; Oh Song, H.; Guadarrama, S.; Murphy, K.P. Semantic instance segmentation via deep metric learning. arXiv 2017, arXiv:1703.10277. [Google Scholar]
- Qiu, Z.; Gan, H.; Shi, M.; Huang, Z.; Yang, Z. Self-training with dual uncertainty for semi-supervised medical image segmentation. arXiv 2022, arXiv:2304.04441v2. [Google Scholar]
- Lin, Q.; Ng, H.T. A Semi-Supervised Learning Approach with Two Teachers to Improve Breakdown Identification in Dialogues. In Proceedings of the AAAI Conference on Artificial Intelligence, Washington, DC, USA, 7–14 February 2022; Volume 36, pp. 11011–11019. [Google Scholar] [CrossRef]
- Sun, Z.; Fan, C.; Sun, X.; Meng, Y.; Wu, F.; Li, J. Neural semi-supervised learning for text classification under large-scale pretraining. arXiv 2020, arXiv:2011.08626. [Google Scholar]
- Bertels, J.; Eelbode, T.; Berman, M.; Vandermeulen, D.; Maes, F.; Bisschops, R.; Blaschko, M.B. Optimizing the DICE score and Jaccard index for medical image segmentation: Theory and practice. In Proceedings of the Medical Image Computing and Computer Assisted Intervention—MICCAI 2019, 22nd International Conference, Shenzhen, China, 13–17 October 2019. [Google Scholar] [CrossRef]
- Zou, K.H.; Warfield, S.K.; Bharatha, A.; Tempany, C.M.C.; Kaus, M.R.; Haker, S.J.; Wells, W.M.; Jolesz, F.A.; Kikinis, R. Statistical Validation of Image Segmentation Quality Based on a Spatial Overlap Index. Acad. Radiol. 2004, 11, 178–189. [Google Scholar] [CrossRef] [PubMed]
- Crum, W.R.; Camara, O.; Hill, D.L.G. Generalized overlap measures for evaluation and validation in medical image analysis. IEEE Trans. Med. Imaging 2006, 25, 1451–1461. [Google Scholar] [CrossRef] [PubMed]
- Taha, A.A.; Hanbury, A. Metrics for Evaluating 3D Medical Image Segmentation: Analysis, Selection, and Tool. BMC Med. Imaging 2015, 15, 29. [Google Scholar] [CrossRef]
- Ding, K.; Zhou, M.; Wang, H.; Gevaert, O.; Metaxas, D.; Zhang, S. A Large-Scale Synthetic Pathological Dataset for Deep Learning-Enabled Segmentation of Breast Cancer. Sci. Data 2023, 10, 231. [Google Scholar] [CrossRef]
- Lin, T.-Y.; Dollar, P.; Girshick, R.; He, K.; Hariharan, B.; Belongie, S. Feature Pyramid Networks for Object Detection. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 936–944. [Google Scholar] [CrossRef]
- Badrinarayanan, V.; Kendall, A.; Cipolla, R. SegNet: A Deep Convolutional Encoder-Decoder Architecture for Image Segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 2481–2495. [Google Scholar] [CrossRef]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 1–9. [Google Scholar] [CrossRef]
- Kirillov, A.; Girshick, R.; He, K.; Dollár, P. Panoptic Feature Pyramid Networks. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 16–20 June 2019; pp. 6399–6408. [Google Scholar]
- Guo, C.; Fan, B.; Zhang, Q.; Xiang, S.; Pan, C. AugFPN: Improving Multi-Scale Feature Learning for Object Detection. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Virtual, 14–19 June 2020; pp. 12595–12604. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 26 June–1 July 2016; pp. 1063–6919. [Google Scholar] [CrossRef]
- Chen, L.C.; Papandreou, G.; Kokkinos, I.; Murphy, K.; Yuille, A.L. Semantic image segmentation with deep convolutional nets and fully connected crfs. arXiv 2014, arXiv:1412.7062. [Google Scholar]
- Shelhamer, E.; Long, J.; Darrell, T. Fully convolutional networks for semantic segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 640–651. [Google Scholar] [CrossRef] [PubMed]
- Visin, F.; Romero, A.; Cho, K.; Matteucci, M.; Ciccone, M.; Kastner, K.; Bengio, Y.; Courville, A. ReSeg: A recurrent neural network-based model for semantic segmentation. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 26 June–1 July 2016; pp. 41–48. [Google Scholar] [CrossRef]
- Noh, H.; Hong, S.; Han, B. Learning deconvolution network for semantic segmentation. In Proceedings of the 2015 IEEE International Conference on Computer Vision (ICCV), Santiago, Chile, 13–16 December 2015. [Google Scholar] [CrossRef]
- Yu, F.; Koltun, V. Multi-Scale Context Aggregation by Dilated Convolutions. arXiv 2016, arXiv:1511.07122. [Google Scholar]
- Wei, L.; Andrew, R.; Alexander, C.B. ParseNet: Looking Wider to See Better. arXiv 2015, arXiv:1506.04579. [Google Scholar]
- Qiao, J.-J.; Cheng, Z.-Q.; Wu, X.; Li, W.; Zhang, J. Real-time semantic segmentation with parallel multiple views feature augmentation. In Proceedings of the 30th ACM International Conference on Multimedia, Lisbon, Portugal, 10–14 October 2022; pp. 6300–6308. [Google Scholar] [CrossRef]
- Chen, L.-C.; Papandreou, G.; Kokkinos, I.; Murphy, K.; Yuille, A.L. DeepLab: Semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected crfs. IEEE Trans. Pattern Anal. Mach. Intell. 2018, 40, 834–848. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Shi, J.; Qi, X.; Wang, X.; Jia, J. Pyramid Scene Parsing Network. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 2881–2890. [Google Scholar] [CrossRef]
- Li, Y.; Qi, H.; Dai, J.; Ji, X.; Wei, Y. Fully Convolutional Instance-Aware Semantic Segmentation. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 4438–4446. [Google Scholar] [CrossRef]
- Milletari, F.; Navab, N.; Ahmadi, S.-A. V-net: Fully convolutional neural networks for volumetric medical image segmentation. In Proceedings of the 2016 Fourth International Conference on 3D Vision (3DV), Stanford, CA, USA, 25–28 October 2016; pp. 565–571. [Google Scholar] [CrossRef]
- Paszke, A.; Chaurasia, A.; Kim, S.; Culurciello, E. ENet: A Deep neural network architecture for real-time semantic segmentation. arXiv 2016. [Google Scholar] [CrossRef]
- Lin, G.; Milan, A.; Shen, C.; Reid, I. RefineNet: Multi-path refinement networks for high-resolution semantic segmentation. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 5168–5177. [Google Scholar] [CrossRef]
- Jegou, S.; Drozdzal, M.; Vazquez, D.; Romero, A.; Bengio, Y. The one hundred layers tiramisu: Fully convolutional DenseNets for Semantic segmentation. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition Workshops (CVPRW), Honolulu, HI, USA, 21–26 July 2017. [Google Scholar] [CrossRef]
- Peng, C.; Zhang, X.; Yu, G.; Luo, G.; Sun, J. Large kernel matters—Improve semantic segmentation by global convolutional network. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 1743–1751. [Google Scholar] [CrossRef]
- Chen, L.-C.; Papandreou, G.; Schroff, F.; Adam, H. Rethinking atrous convolution for semantic Image segmentation. arXiv 2017. [Google Scholar] [CrossRef]
- Zhao, H.; Qi, X.; Shen, X.; Shi, J.; Jia, J. Icnet for real-time semantic segmentation on high-resolution images. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 405–420. [Google Scholar]
- Han, G.; Zhang, M.; Wu, W.; He, M.; Liu, K.; Qin, L.; Liu, X. Improved U-Net based insulator image segmentation method based on attention mechanism. Energy Rep. 2021, 7, 210–217. [Google Scholar] [CrossRef]
- Xiong, Y.; Liao, R.; Zhao, H.; Hu, R.; Bai, M.; Yumer, E.; Urtasun, R. Upsnet: A unified panoptic segmentation network. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 16–20 June 2019; pp. 8818–8826. [Google Scholar]
- Zhang, G.; Lu, X.; Tan, J.; Li, J.; Zhang, Z.; Li, Q.; Hu, X. Refinemask: Towards high-quality instance segmentation with fine-grained features. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Virtual, 19–25 June 2021; pp. 6861–6869. [Google Scholar]
- Liu, S.; Qi, L.; Qin, H.; Shi, J.; Jia, J. Path aggregation network for instance segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018; pp. 8759–8768. [Google Scholar]
- Eerapu, K.K.; Ashwath, B.; Lal, S.; Dell’Acqua, F.; Dhan, A.N. Dense refinement residual network for road extraction from aerial imagery data. IEEE Access 2019, 7, 151764–151782. [Google Scholar] [CrossRef]
- Gu, Z.; Cheng, J.; Fu, H.; Zhou, K.; Hao, H.; Zhao, Y.; Zhang, T.; Gao, S.; Liu, J. Ce-net: Context encoder network for 2d medical image segmentation. IEEE Trans. Med. Imaging 2019, 38, 2281–2292. [Google Scholar] [CrossRef] [PubMed]
- Takikawa, T.; Acuna, D.; Jampani, V.; Fidler, S. Gated-scnn: Gated shape cnns for semantic segmentation. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Seoul, Republic of Korea, 27 October–2 November 2019; pp. 5229–5238. [Google Scholar]
- Cui, B.; Fei, D.; Shao, G.; Lu, Y.; Chu, J. Extracting raft aquaculture areas from remote sensing images via an improved U-net with a PSE structure. Remote Sens. 2019, 11, 2053. [Google Scholar] [CrossRef]
- Chen, X.; Girshick, R.; He, K.; Dollár, P. Tensormask: A foundation for dense object segmentation. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Seoul, Republic of Korea, 27 October–2 November 2019; pp. 2061–2069. [Google Scholar]
- Seong, S.; Choi, J. Semantic segmentation of urban buildings using a high-resolution network (HRNet) with channel and spatial attention gates. Remote Sens. 2021, 13, 3087. [Google Scholar] [CrossRef]
- Huang, Z.; Wang, X.; Huang, L.; Huang, C.; Wei, Y.; Liu, W. Ccnet: Criss-cross attention for semantic segmentation. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Seoul, Republic of Korea, 27 October–2 November 2019; pp. 603–612. [Google Scholar]
- Nam, H.; Ha, J.W.; Kim, J. Dual attention networks for multimodal reasoning and matching. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 299–307. [Google Scholar]
- Poudel, R.P.; Liwicki, S.; Cipolla, R. Fast-scnn: Fast semantic segmentation network. arXiv 2019, arXiv:1902.04502. [Google Scholar]
- Chen, Z.; Zhu, Y.; Zhao, C.; Hu, G.; Zeng, W.; Wang, J.; Tang, M. Dpt: Deformable patch-based transformer for visual recognition. In Proceedings of the 29th ACM International Conference on Multimedia, Virtual, 20–24 October 2021; pp. 2899–2907. [Google Scholar]
- Zheng, S.; Lu, J.; Zhao, H.; Zhu, X.; Luo, Z.; Wang, Y.; Fu, Y.; Feng, J.; Xiang, T.; Torr, P.H.S.; et al. Rethinking semantic segmentation from a sequence-to-sequence perspective with transformers. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Virtual, 19–25 June 2021; pp. 6881–6890. [Google Scholar]
- Corradetti, M. A Point-Based Rendering Approach for On-Board Instance Segmentation of Non-Cooperative Resident Space Objects. Master’s Thesis, Politecnico di Milano, Milan, Italy, 6 October 2022. Available online: https://hdl.handle.net/10589/195413 (accessed on 6 October 2022).
- Mohan, R.; Valada, A. Efficientps: Efficient panoptic segmentation. Int. J. Comput. Vis. 2021, 129, 1551–1579. [Google Scholar] [CrossRef]
- Chen, W.; Gong, X.; Liu, X.; Zhang, Q.; Li, Y.; Wang, Z. Fasterseg: Searching for faster real-time semantic segmentation. arXiv 2019, arXiv:1912.10917. [Google Scholar]
- Xie, E.; Sun, P.; Song, X.; Wang, W.; Liu, X.; Liang, D.; Shen, C.; Luo, P. Polarmask: Single shot instance segmentation with polar representation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Virtual, 14–19 June 2020; pp. 12193–12202. [Google Scholar]
- Lee, Y.; Park, J. Centermask: Real-time anchor-free instance segmentation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Virtual, 14–19 June 2020; pp. 13906–13915. [Google Scholar]
- Song, Y.; Sha, E.H.-M.; Zhuge, Q.; Xu, R.; Xu, X.; Li, B.; Yang, L. Hardware-aware neural architecture search for stochastic computing-based neural networks on tiny devices. J. Syst. Archit. 2023, 135, 102810. [Google Scholar] [CrossRef]
- Ghiasi, G.; Cui, Y.; Srinivas, A.; Qian, R.; Lin, T.Y.; Cubuk, E.D.; Le, Q.V.; Zoph, B. Simple copy-paste is a strong data augmentation method for instance segmentation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Virtual, 19–25 June 2021; pp. 2918–2928. [Google Scholar]
- Shang, R.; Zhang, J.; Jiao, L.; Li, Y.; Marturi, N.; Stolkin, R. Multi-scale adaptive feature fusion network for semantic segmentation in remote sensing images. Remote Sens. 2020, 12, 872. [Google Scholar] [CrossRef]
- Nguyen, V.A.; Nguyen, A.H.; Khong, A.W. Tunet: A block-online bandwidth extension model based on transformers and self-supervised pretraining. In Proceedings of the ICASSP 2022-2022 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Singapore, 23–27 May 2022; pp. 161–165. [Google Scholar]
- Fan, C.M.; Liu, T.J.; Liu, K.H. SUNet: Swin transformer UNet for image denoising. In Proceedings of the 2022 IEEE International Symposium on Circuits and Systems (ISCAS), Austin, TX, USA, 27 May–1 June 2022; pp. 2333–2337. [Google Scholar]
- Strudel, R.; Garcia, R.; Laptev, I.; Schmid, C. Segmenter: Transformer for semantic segmentation. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Virtual, 19–25 June 2021; pp. 7262–7272. [Google Scholar]
- Valanarasu, J.M.J.; Oza, P.; Hacihaliloglu, I.; Patel, V.M. Medical transformer: Gated axial-attention for medical image segmentation. In Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2021: 24th International Conference, Strasbourg, France, 27 September–1 October 2021; pp. 36–46. [Google Scholar]
- Bao, H.; Dong, L.; Piao, S.; Wei, F. Beit: Bert pre-training of image transformers. arXiv 2021, arXiv:2106.08254. [Google Scholar]
- He, A.; Wang, K.; Li, T.; Du, C.; Xia, S.; Fu, H. H2Former: An efficient hierarchical hybrid transformer for medical image segmentation. IEEE Trans. Med. Imaging 2023, 42, 2763–2775. [Google Scholar] [CrossRef]
- Lai, H.P.; Tran, T.T.; Pham, V.T. Axial attention mlp-mixer: A new architecture for image segmentation. In Proceedings of the 2022 IEEE Ninth International Conference on Communications and Electronics (ICCE), Nha Trang, Vietnam, 27–29 July 2022; pp. 381–386. [Google Scholar]
- Razumovskaia, E.; Glavas, G.; Majewska, O.; Ponti, E.M.; Korhonen, A.; Vulic, I. Crossing the conversational chasm: A primer on natural language processing for multilingual task-oriented dialogue systems. J. Artif. Intell. Res. 2022, 74, 1351–1402. [Google Scholar] [CrossRef]
- Mkindu, H.; Wu, L.; Zhao, Y. 3D multi-scale vision transformer for lung nodule detection in chest CT images. Signal Image Video Process. 2023, 17, 2473–2480. [Google Scholar] [CrossRef]
- Heidari, M.; Kazerouni, A.; Soltany, M.; Azad, R.; Aghdam, E.K.; Cohen-Adad, J.; Merhof, D. Hiformer: Hierarchical multi-scale representations using transformers for medical image segmentation. In Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision, Waikoloa, HI, USA, 3–7 January 2023; pp. 6202–6212. [Google Scholar]




| New Cases | Deaths | |||||
|---|---|---|---|---|---|---|
| Country | N | ASR | Cum. Risk | N | ASR | Cum. Risk |
| Eastern Africa | 45,709 | 33 | 3.6 | 24,047 | 17.9 | 2 |
| Middle Africa | 17,896 | 32.7 | 3.4 | 9500 | 18 | 1.9 |
| Northern Africa | 57,128 | 49.6 | 5.1 | 21,524 | 18.8 | 1.9 |
| Southern Africa | 16,526 | 50.4 | 5.4 | 5090 | 15.7 | 1.7 |
| Western Africa | 49,339 | 41.5 | 4.5 | 25,626 | 22.3 | 2.5 |
| Caribbean | 14,712 | 51 | 5.5 | 5874 | 18.9 | 2 |
| Central America | 38,916 | 39.5 | 4.2 | 10,429 | 10.4 | 1.2 |
| South America | 156,472 | 56.4 | 6.1 | 41,681 | 14 | 1.5 |
| Northern America | 281,591 | 89.4 | 9.7 | 48,407 | 12.5 | 1.4 |
| Eastern Asia | 551,636 | 43.3 | 4.6 | 141,421 | 9.8 | 1.1 |
| All but China | 135,265 | 66.9 | 7 | 24,247 | 9.4 | 1 |
| China | 416,371 | 39.1 | 4.2 | 117,174 | 10 | 1.2 |
| South-Eastern Asia | 158,939 | 41.2 | 4.5 | 58,670 | 15 | 1.7 |
| South-Central Asia | 254,881 | 26.2 | 2.9 | 124,975 | 13.1 | 1.5 |
| All but India | 76,520 | 27.5 | 3.1 | 34,567 | 12.9 | 1.5 |
| India | 178,361 | 25.8 | 2.8 | 90,408 | 13.2 | 1.5 |
| Western Asia | 60,715 | 46.6 | 5 | 20,943 | 16 | 1.7 |
| Central-Eastern Europe | 158,708 | 57.1 | 6.3 | 51,488 | 15.3 | 1.8 |
| Northern Europe | 83,177 | 86.4 | 9.4 | 17,964 | 13.7 | 1.5 |
| Southern Europe | 120,185 | 79.6 | 8.5 | 28,607 | 13.3 | 1.4 |
| Western Europe | 169,016 | 90.7 | 9.7 | 43,706 | 15.6 | 1.7 |
| Australia/New Zealand | 23,277 | 95.5 | 10.4 | 3792 | 12.1 | 1.3 |
| Melanesia | 2215 | 50.5 | 5.4 | 1121 | 27.5 | 2.9 |
| Micronesia/Polynesia | 381 | 58.2 | 6 | 131 | 19.6 | 2.1 |
| Low HDI | 109,572 | 36.1 | 3.9 | 58,586 | 20.1 | 2.2 |
| Medium HDI | 307,658 | 27.8 | 3 | 147,427 | 13.6 | 1.5 |
| High HDI | 825,438 | 42.7 | 4.6 | 247,486 | 12.1 | 1.4 |
| Very high HDI | 75.7 | 8.2 | 231,093 | 13.4 | 1.5 | |
| World | 47.8 | 5.2 | 684,996 | 13.6 | 1.5 | |
| Seed | Training Loss | Validation Dice | Validation Jaccard | Optimum Epoch | Figure Illustration |
|---|---|---|---|---|---|
| 21 | 0.0102 | 0.9432 | 0.8930 | 92 | N/A |
| 42 | 0.0098 | 0.9425 | 0.8926 | 98 | N/A |
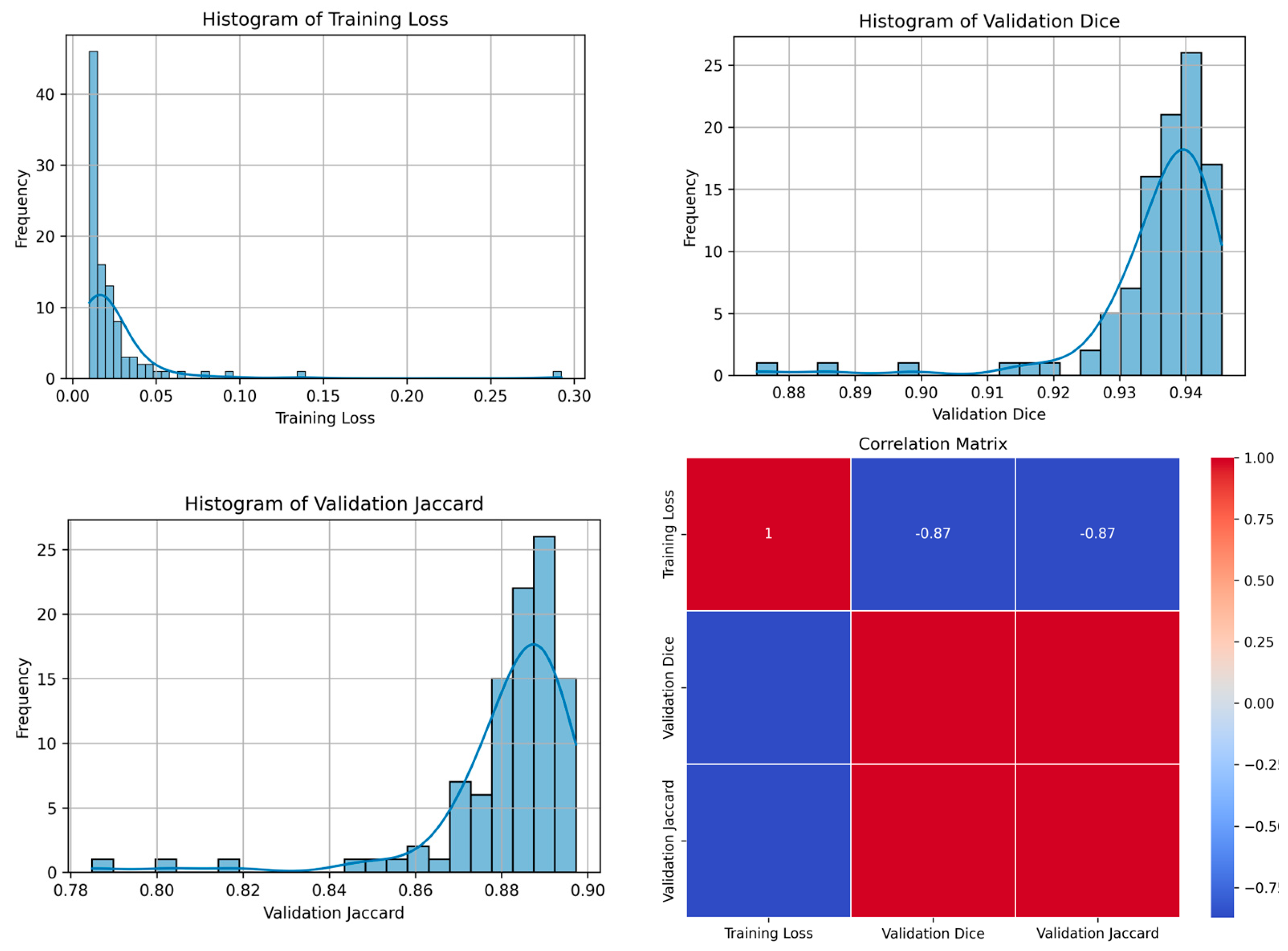
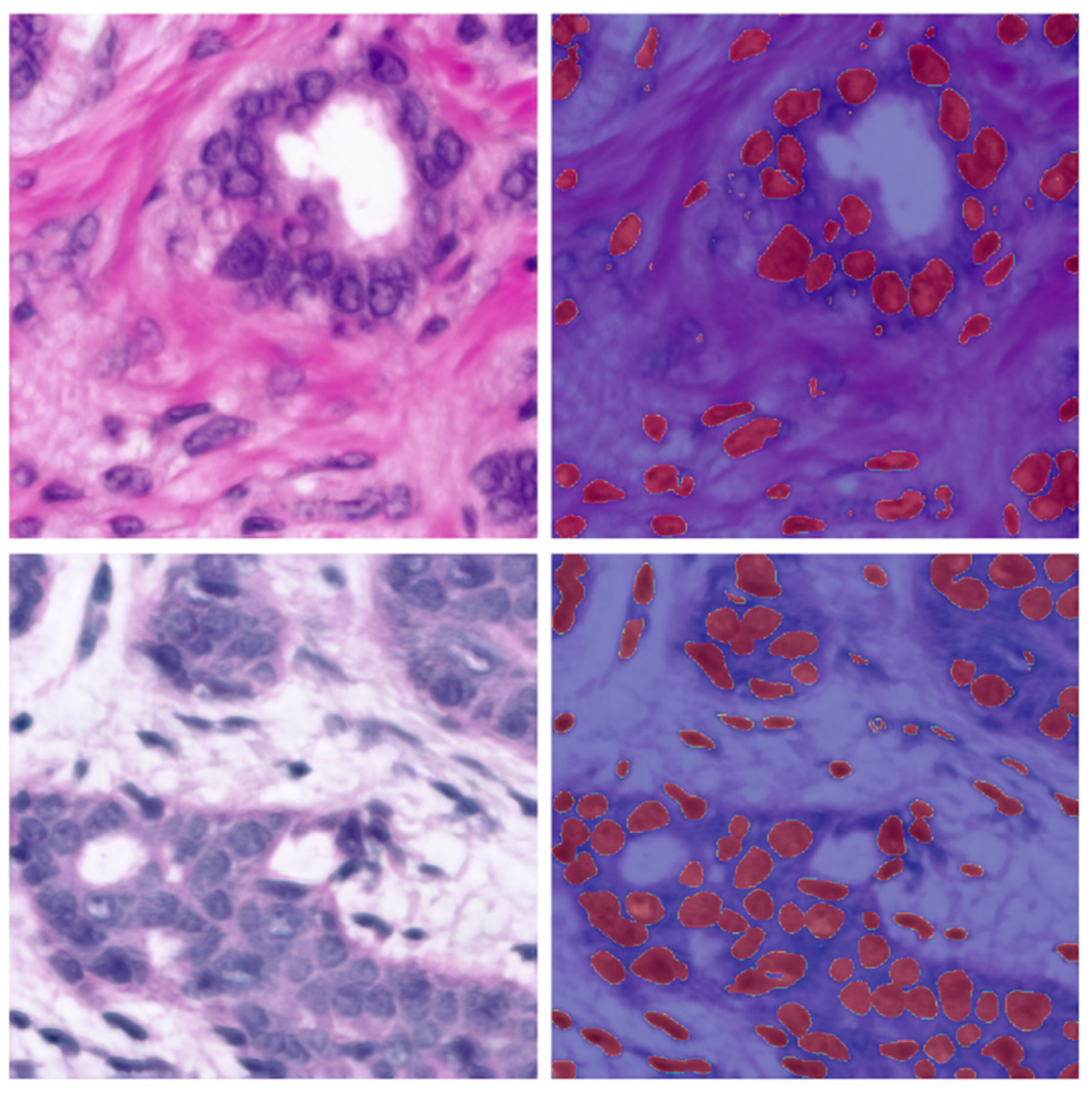
| 84 | 0.0114 | 0.9454 | 0.8972 | 85 | Figure 7 |
| Seed | Training Loss | Validation Dice | Validation Jaccard | Optimum Epoch | Figure Illustration |
|---|---|---|---|---|---|
| 21 | 0.0061 | 0.8938 | 0.8081 | 8 | N/A |
| 42 | 0.0066 | 0.8084 | 0.6784 | 9 | N/A |
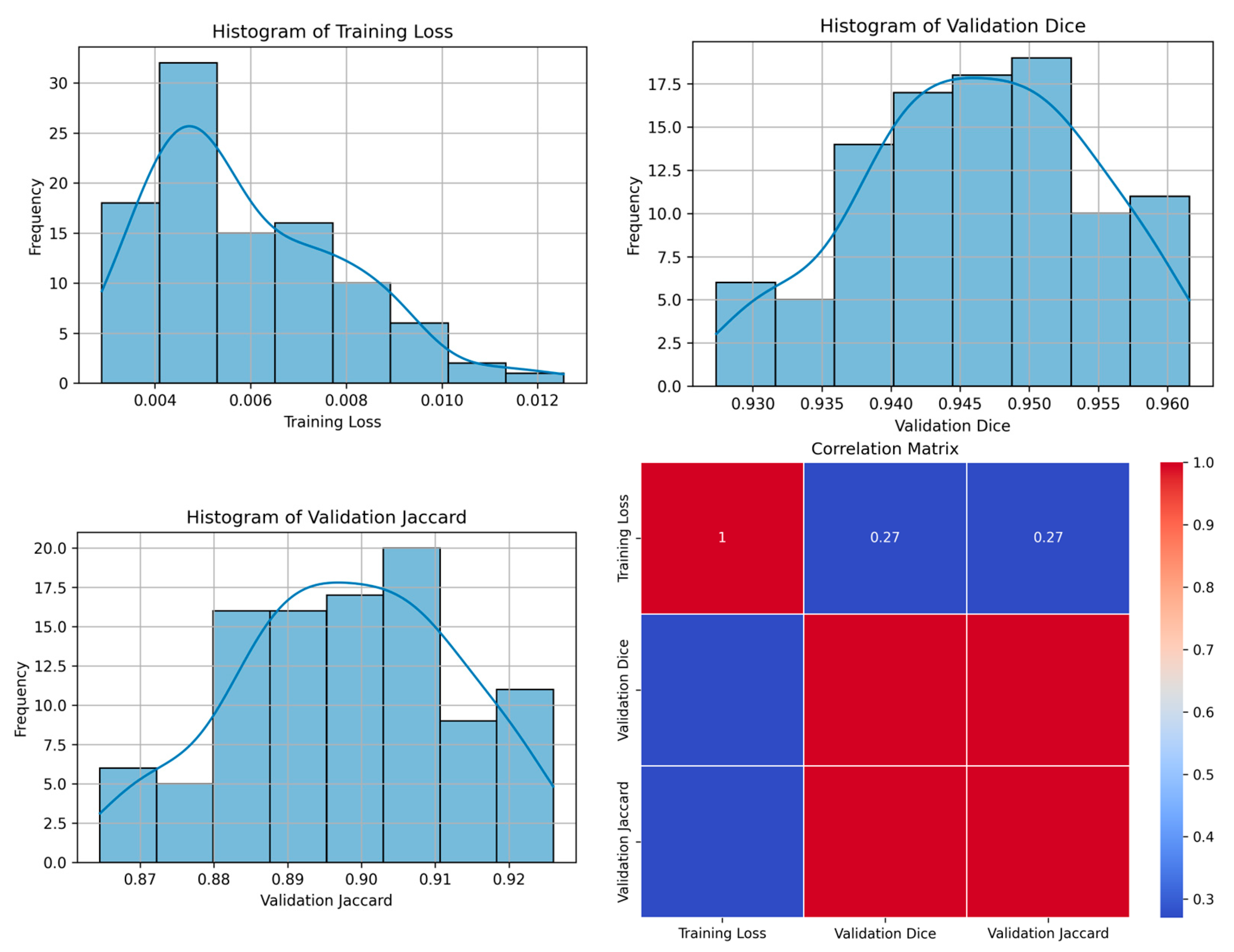
| 84 | 0.0043 | 0.9529 | 0.9100 | 9 | Figure 8 |
| Teacher | Student | |||||
|---|---|---|---|---|---|---|
| Seeds | TL | VD | VJ | TL | VD | VJ |
| 21 | 0.0102 | 0.9430 | 0.893 | 0.0061 | 0.893 | 0.8081 |
| 42 | 0.0098 | 0.9425 | 0.8926 | 0.0066 | 0.8084 | 0.6784 |
| 84 | 0.0114 | 0.9450 | 0.8972 | 0.004 | 0.9529 | 0.9100 |
| Average | 0.0104 | 0.9435 | 0.8942 | 0.0055 | 0.8847 | 0.7988 |
| Seed | Training Loss | Validation Dice | Validation Jaccard | Optimum Epoch | Figure Illustration |
|---|---|---|---|---|---|
| 21 | 0.0173 | 0.9331 | 0.8760 | 78 | N/A |
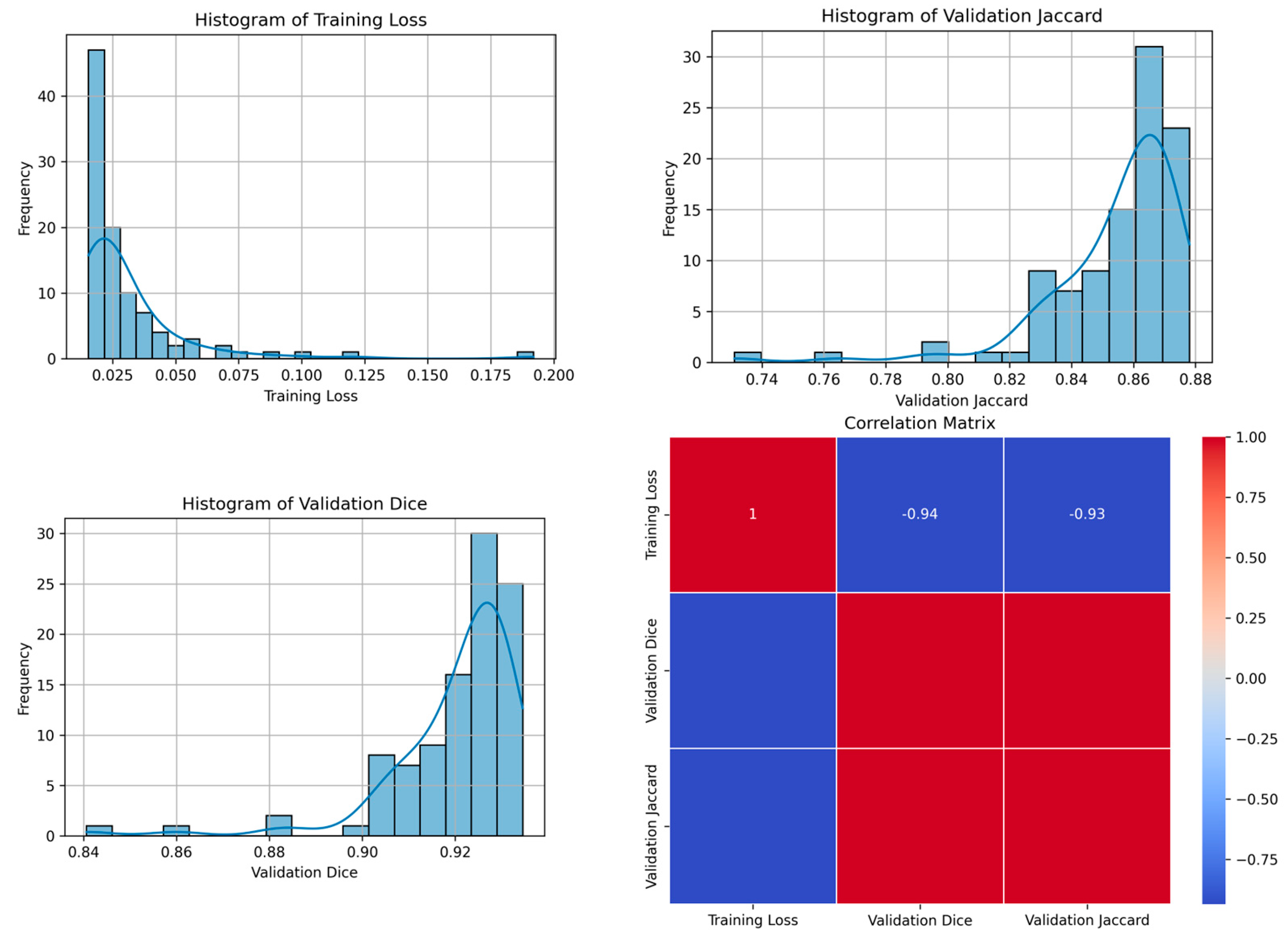
| 42 | 0.0154 | 0.9345 | 0.8780 | 100 | Figure 9 |
| 84 | 0.0160 | 0.9326 | 0.8795 | 94 | N/A |
| Seed | Training Loss | Validation Dice | Validation Jaccard | Optimum Epoch | Figure Illustration |
|---|---|---|---|---|---|
| 21 | 0.0183 | 0.8956 | 0.8109 | 2 | N/A |
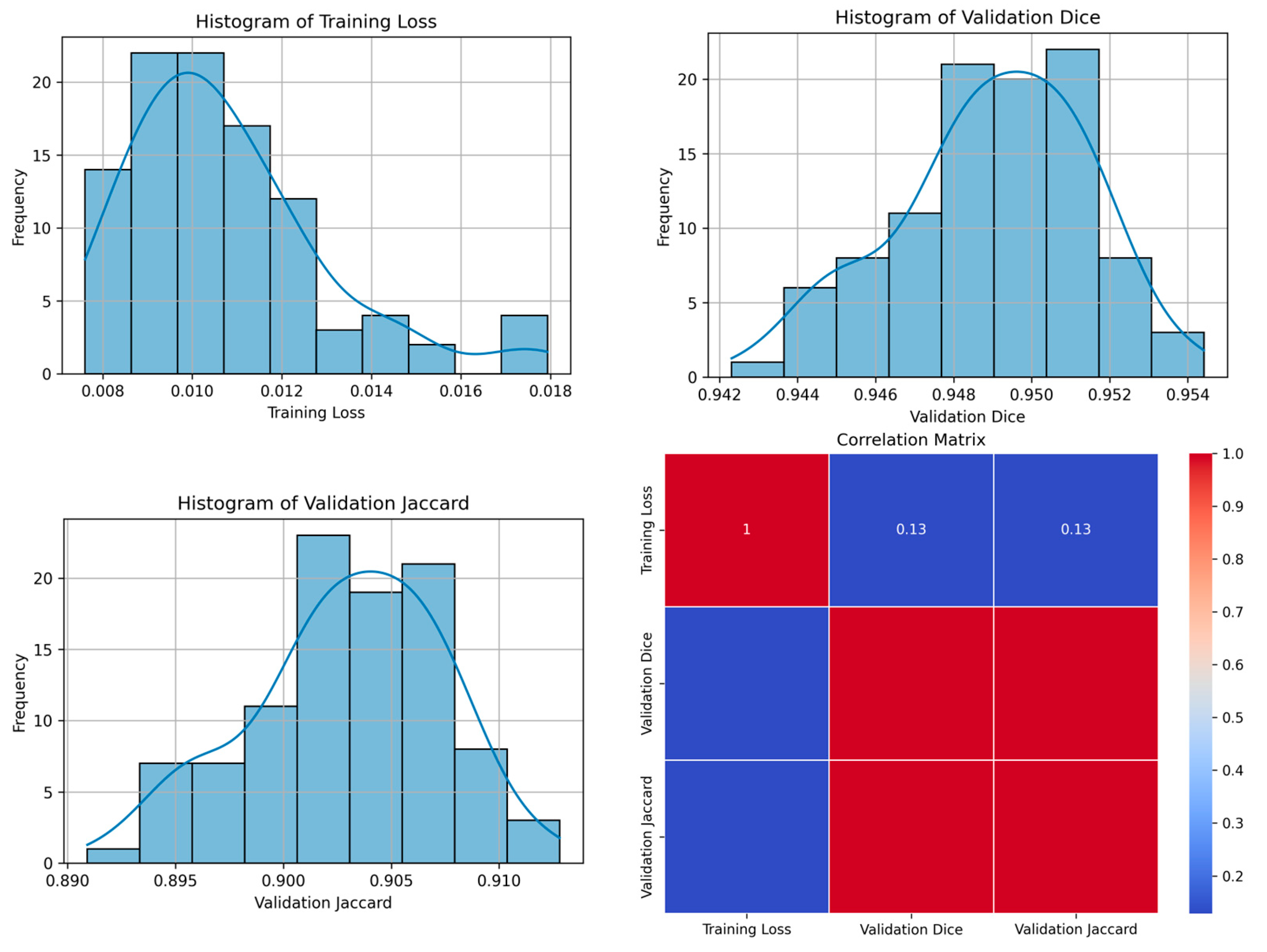
| 42 | 0.0113 | 0.9544 | 0.9128 | 83 | Figure 10 |
| 84 | 0.0227 | 0.9378 | 0.8830 | 2 | N/A |
| Teacher | Student | |||||
|---|---|---|---|---|---|---|
| Seeds | TL | VD | VJ | TL | VD | VJ |
| 21 | 0.0173 | 0.9331 | 0.0173 | 0.0183 | 0.8956 | 0.8109 |
| 42 | 0.0154 | 0.9345 | 0.0154 | 0.0113 | 0.9544 | 0.9128 |
| 84 | 0.0160 | 0.9326 | 0.0160 | 0.0227 | 0.9378 | 0.8830 |
| Average | 0.0162 | 0.9334 | 0.0162 | 0.0174 | 0.9292 | 0.8689 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schutte, S.; Uddin, J. Deep Segmentation Techniques for Breast Cancer Diagnosis. BioMedInformatics 2024, 4, 921-945. https://doi.org/10.3390/biomedinformatics4020052
Schutte S, Uddin J. Deep Segmentation Techniques for Breast Cancer Diagnosis. BioMedInformatics. 2024; 4(2):921-945. https://doi.org/10.3390/biomedinformatics4020052
Chicago/Turabian StyleSchutte, Storm, and Jia Uddin. 2024. "Deep Segmentation Techniques for Breast Cancer Diagnosis" BioMedInformatics 4, no. 2: 921-945. https://doi.org/10.3390/biomedinformatics4020052
APA StyleSchutte, S., & Uddin, J. (2024). Deep Segmentation Techniques for Breast Cancer Diagnosis. BioMedInformatics, 4(2), 921-945. https://doi.org/10.3390/biomedinformatics4020052