Enhancing Human Superorganism Ecosystem Resilience by Holistically ‘Managing Our Microbes’
Abstract
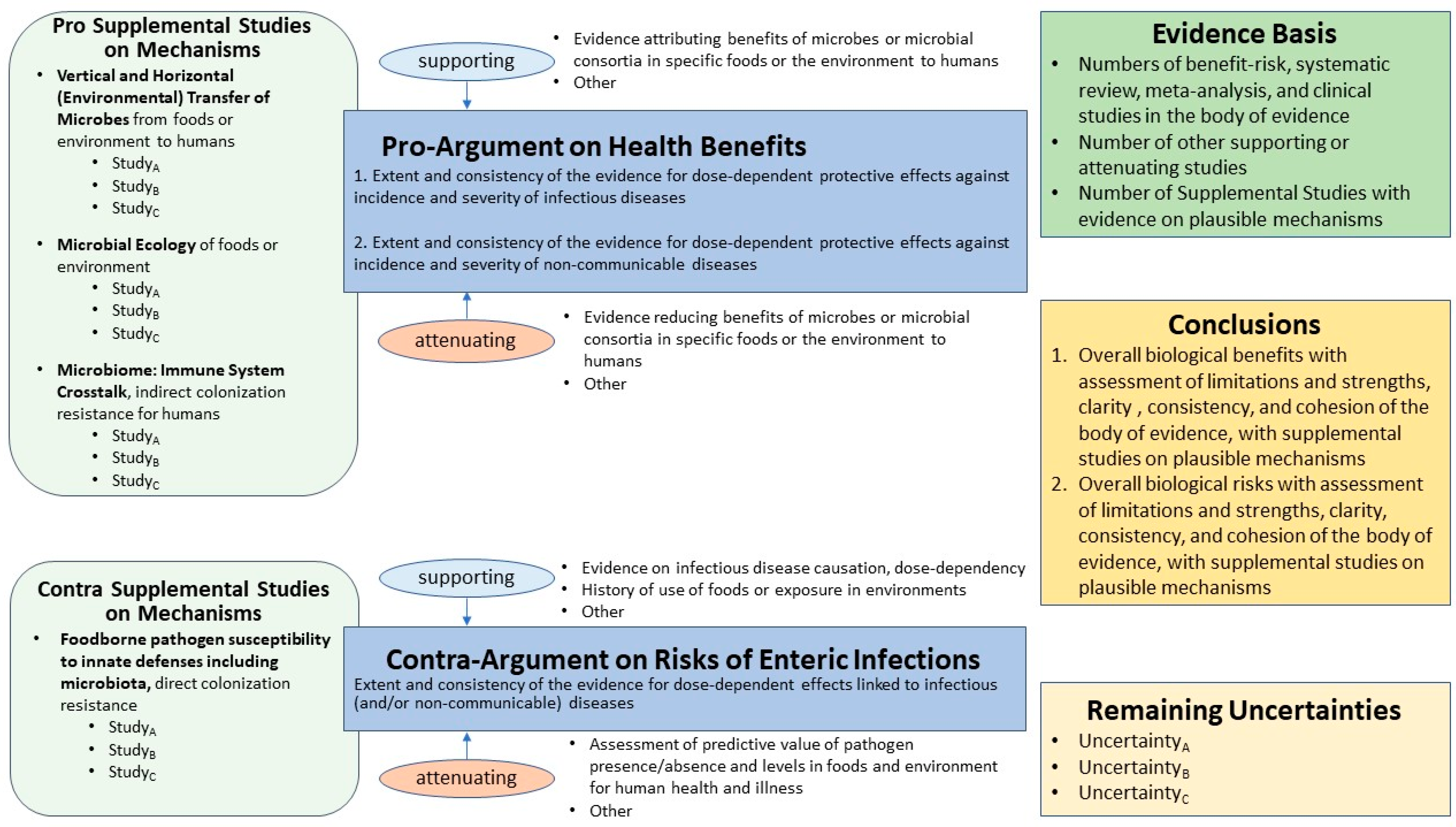
:1. Background
1.1. Risks and Benefits of Managing Gut Microbes and Colonization Resistance
1.2. Expanding Traditional Risk Paradigms for the Microbiota
2. CDI in the Gut
2.1. Using Commensal Bacteria for Enhancing or Restoring Colonization Resistance
2.2. Using the Virome: Phage Therapy
2.3. C. difficile Summary
3. Staph A, Asthma, and Allergic and Infectious Diseases
3.1. Beyond Infection to Asthma and Allergic Diseases
3.2. Staph A and Asthma
3.3. Staph A and IL-36
3.4. Vulnerable Populations
3.5. Using Staph against Staph
3.6. Other Microbes Outcompeting Staph A
3.7. Staph A Summary
4. Managing Stressors along the Gut–Brain Axis for Autism
4.1. Clinical Evidence for Differences in Gut Microbiota for ASD Children
4.2. Clinical Evidence for Interventions That Restore Gut Microbiota Health for ASD Children
4.3. ASD Summary
5. Breastmilk Ecosystem and Benefit–Risk Analysis
5.1. Breastmilk Microbiota
5.2. Microbial Ecology of Breastmilk and Reductionist Dilemma
5.3. Benefit–Risk Methodology Applied to the Breastmilk Ecosystem
5.4. Future Evidence-Based Policies for Donor Breastmilk?
5.5. Summary of Breastmilk Ecosystem Evidence Map
6. Future for ‘Managing Our Microbes’
- Does not require complicated quantitative modeling of exposure assessment and dose–response assessment typical of quantitative microbial risk assessment (QMRA) for data synthesis;
- Presents simple qualitative narrative in a structured format, with a graphical representation of the evidence basis, drawing attention to evidence for both pro- and contra-arguments, with supporting and attenuating data;
- Assists a diverse array of experts and non-experts in paying attention to the entire ‘state of the science’, a visual picture of the evidence basis, quality of evidence, and uncertainty;
- Promotes openness and transparency for evaluating rarely unambiguous scientific evidence for applications in risk analysis;
- Assists risk analysts in avoiding traps such as ‘confirmation bias’ that may distort judgments about weighing and synthesizing evidence from multiple disciplines; and
- Facilitates constructive dialogue between diverse perspectives/opinions of all stakeholders, including decisions makers and the public.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Heiman, M.L.; Greenway, F.L. A Healthy Gastrointestinal Microbiome Is Dependent on Dietary Diversity. Mol. Metab. 2016, 5, 317–320. [Google Scholar] [CrossRef] [PubMed]
- National Institutes of Health (NIH)—Human Microbiome Portfolio Analysis Team. A Review of 10 Years of Human Microbiome Research Activities at the US National Institutes of Health, Fiscal Years 2007–2016. Microbiome 2019, 7, 31. [Google Scholar] [CrossRef] [PubMed]
- Integrative HMP; Proctor, L.M.; Creasy, H.H.; Fettweis, J.M.; Lloyd-Price, J.; Mahurkar, A.; Huttenhower, C. The Integrative Human Microbiome Project. Nature 2019, 569, 641–648. [Google Scholar] [CrossRef] [Green Version]
- Dietert, R.R. The Human Superorganism: How the Microbiome Is Revolutionizing the Pursuit of a Healthy Life; Dutton: New York, NY, USA, 2016. [Google Scholar]
- Dietert, R.R. Safety and Risk Assessment for the Human Superorganism. Hum. Ecol. Risk Assess. 2017, 23, 1819–1829. [Google Scholar] [CrossRef] [Green Version]
- Dietert, R.R.; Dietert, J.M. Twentieth Century Dogmas Prevent Sustainable Healthcare. Am. J. Biomed. Sci. Res. 2021, 13, 409–417. [Google Scholar]
- Coleman, M.; Elkins, C.; Gutting, B.; Mongodin, E.; Solano-Aguilar, G.; Walls, I. Microbiota and Dose Response: Evolving Paradigm of Health Triangle. Risk Anal. 2018, 38, 2013–2028. [Google Scholar] [CrossRef]
- Dietert, R.R.; Silbergeld, E.K. Biomarkers for the 21st Century: Listening to the Microbiome. Toxicol. Sci. 2015, 144, 208–216. [Google Scholar] [CrossRef] [Green Version]
- Dietert, R.R. A Focus on Microbiome Completeness and Optimized Colonization Resistance in Neonatology. NeoReviews 2018, 19, 78–88. [Google Scholar] [CrossRef]
- Sleator, R.D. The Human Superorganism–of Microbes and Men. Med. Hypotheses 2010, 74, 214–215. [Google Scholar] [CrossRef]
- Blaser, M.J. The Microbiome Revolution. J. Clin. Investig. 2014, 124, 4162–4165. [Google Scholar] [CrossRef] [Green Version]
- Marks, H.; Coleman, M. Scientific Data and Theories for Salmonellosis Dose—Response Assessment. Hum. Ecol. Risk Assess. Int. J. 2017, 23, 1857–1876. [Google Scholar] [CrossRef]
- Coleman, M.E.; Dietert, R.R.; North, D.W.; Stephenson, M.M. Examining Evidence of Benefits and Risks for Pasteurizing Donor Breastmilk. Appl. Microbiol. 2021, 1, 408–425. [Google Scholar] [CrossRef]
- Endt, K.; Stecher, B.; Chaffron, S.; Slack, E.; Tchitchek, N.; Benecke, A.; Hardt, W.D. The Microbiota Mediates Pathogen Clearance from the Gut Lumen after Non-Typhoidal Salmonella Diarrhea. PLoS Pathog. 2010, 6, 1001097. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brugiroux, S.; Beutler, M.; Pfann, C.; Garzetti, D.; Ruscheweyh, H.J.; Ring, D.; Stecher, B. Genome-Guided Design of a Defined Mouse Microbiota That Confers Colonization Resistance against Salmonella Enterica Serovar Typhimurium. Nat. Microbiol. 2016, 2, 16215. [Google Scholar] [CrossRef] [PubMed]
- Gazzaniga, F.S.; Camacho, D.M.; Wu, M.; Silva Palazzo, M.F.; Dinis, A.L.; Grafton, F.N.; Ingber, D.E. Harnessing Colon Chip Technology to Identify Commensal Bacteria That Promote Host Tolerance to Infection. Front. Cell. Infect. Microbiol. 2021, 11, 105. [Google Scholar] [CrossRef]
- Rogers, A.W.; Tsolis, R.M.; Bäumler, A.J. Salmonella versus the Microbiome. Microbiol. Mol. Biol. Rev. 2021, 85, e00027-19. [Google Scholar] [CrossRef]
- Sehgal, K.; Khanna, S. Gut Microbiome and Clostridioides Difficile Infection: A Closer Look at the Microscopic Interface. Ther. Adv. Gastroenterol. 2021, 14, 1756284821994736. [Google Scholar] [CrossRef]
- Kim, S.; Covington, A.; Pamer, E.G. The Intestinal Microbiota: Antibiotics, Colonization Resistance, and Enteric Pathogens. Immunol. Rev. 2017, 279, 90–105. [Google Scholar] [CrossRef]
- Palmer, A.D.; Slauch, J.M. Mechanisms of Salmonella Pathogenesis in Animal Models. Hum. Ecol. Risk Assess. Int. J. 2017, 23, 1877–1892. [Google Scholar] [CrossRef]
- Dietert, R.R.; Dietert, J.M. Microbiome First Approaches in Pain Prevention and Management. Am. J. Biomed. Sci. Res. Press 2021, 14, 182–192. [Google Scholar]
- Levy, E.I.; Hoang, D.M.; Vandenplas, Y. The Effects of Proton Pump Inhibitors on the Microbiome in Young Children. Acta Paediatr. 2020, 109, 1531–1538. [Google Scholar] [CrossRef]
- Abenavoli, L.; Scarpellini, E.; Colica, C.; Boccuto, L.; Salehi, B.; Sharifi-Rad, J.; Capasso, R. Gut Microbiota and Obesity: A Role for Probiotics. Nutrients 2019, 11, 2690. [Google Scholar] [CrossRef] [Green Version]
- King, C.H.; Desai, H.; Sylvetsky, A.C.; LoTempio, J.; Ayanyan, S.; Carrie, J.; Crandall, K.A.; Fochtman, B.C.; Gasparyan, L.; Gulzar, N.; et al. Baseline Human Gut Microbiota Profile in Healthy People and Standard Reporting Template. PLoS ONE 2019, 14, e0206484. [Google Scholar] [CrossRef] [Green Version]
- Almonacid, D.E.; Kraal, L.; Ossandon, F.J.; Budovskaya, Y.V.; Cardenas, J.P.; Bik, E.M.; Goddard, A.D.; Richman, J.; Apte, Z.S. 16S RRNA Gene Sequencing and Healthy Reference Ranges for 28 Clinically Relevant Microbial Taxa from the Human Gut Microbiome. PLoS ONE 2017, 12, e0176555. [Google Scholar] [CrossRef] [Green Version]
- Ducarmon, Q.R.; Kuijper, E.J.; Olle, B. Opportunities and Challenges in Development of Live Biotherapeutic Products to Fight Infections. J. Infect. Dis. 2021, 223, S283–S289. [Google Scholar] [CrossRef]
- Hromada, S.; Clark, R.L.; Qian, Y.; Watson, L.; Safdar, N.; Venturelli, O.S. Species Richness Determines, C. Difficile Invasion Outcome in Synthetic Human Gut Communities. bioRxiv 2021, 1–27. [Google Scholar] [CrossRef]
- Maki, J.J.; Klima, C.L.; Sylte, M.J.; Looft, T. The Microbial Pecking Order: Utilization of Intestinal Microbiota for Poultry Health. Microorganisms 2019, 7, 376. [Google Scholar] [CrossRef] [Green Version]
- National Research Council (US) Committee on the Institutional Means for Assessment of Risks to Public Health. Risk Assessment in the Federal Government: Managing the Process; National Academies Press (US): Washington, DC, USA, 1983; ISBN 978-0-309-03349-7. [Google Scholar]
- Marks, H.M.; Coleman, M.E.; Lin, C.T.J.; Roberts, T. Topics in Microbial Risk Assessment: Dynamic Flow Tree Process. Risk Anal. 1998, 18, 309–328. [Google Scholar] [CrossRef]
- Codex Alimentarius Commission (CAC). Principles and Guidelines for the Conduct of Microbiological Risk Assessment. Available online: http://www.fao.org/3/y1579e/y1579e05.htm (accessed on 8 March 2021).
- Smith, A.B.; Soto Ocana, J.; Zackular, J.P. From Nursery to Nursing Home: Emerging Concepts in Clostridioides Difficile Pathogenesis. Infect. Immun. 2020, 88, e00934-19. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention (CDC). Healthcare-Associated Infections—Community Interface (HAIC). Clostridioides Difficile Infection (CDI) Tracking; CDC: Atlanta, GA, USA, 2020.
- Stein, R.R.; Bucci, V.; Toussaint, N.C.; Buffie, C.G.; Rätsch, G.; Pamer, E.G.; Sander, C.; Xavier, J.B. Ecological Modeling from Time-Series Inference: Insight into Dynamics and Stability of Intestinal Microbiota. PLoS Comput. Biol. 2013, 9, e1003388. [Google Scholar] [CrossRef]
- Buffie, C.G.; Bucci, V.; Stein, R.R.; McKenney, P.T.; Ling, L.; Gobourne, A.; Pamer, E.G. Precision Microbiome Reconstitution Restores Bile Acid Mediated Resistance to Clostridium Difficile. Nature 2015, 517, 205–208. [Google Scholar] [CrossRef] [Green Version]
- Yadav, D.; Khanna, S. Safety of Fecal Microbiota Transplantation for Clostridioides Difficile Infection Focusing on Pathobionts and SARS-CoV-2. Ther. Adv. Gastroenterol. 2021, 14, 17562848211009694. [Google Scholar] [CrossRef]
- Khanna, S. Microbiota Restoration for Recurrent Clostridioides Difficile: Getting One Step Closer Every Day! J. Intern. Med. 2021, 290, 294–309. [Google Scholar] [CrossRef]
- Langdon, A.; Schwartz, D.J.; Bulow, C.; Sun, X.; Hink, T.; Reske, K.A.; Jones, C.; Burnham, C.D.; Dubberke, E.R.; Dantas, G. Microbiota Restoration Reduces Antibiotic Resistant Bacteria Gut Colonization in Patients with Recurrent Clostridioides Difficile Infection from the Open-Label PUNCH CD Study. Genome Med. 2021, 13, 28. [Google Scholar] [CrossRef]
- Vakili, B.; Fateh, A.; Aghdaei, H.A.; Sotoodehnejadnematalahi, F.; Siadat, S.D. Intestinal Microbiota in Elderly Inpatients with Clostridioides Difficile Infection. Infect. Drug Resist. 2020, 13, 2723. [Google Scholar] [CrossRef]
- Rodriguez, C.; Taminiau, B.; Korsak, N.; Avesani, V.; Van Broeck, J.; Brach, P.; Daube, G. Longitudinal Survey of Clostridium Difficile Presence and Gut Microbiota Composition in a Belgian Nursing Home. BMC Microbiol. 2016, 16, 229. [Google Scholar] [CrossRef] [Green Version]
- Liang, G.; Bushman, F.D. The Human Virome: Assembly, Composition and Host Interactions. Nat. Rev. Microbiol. 2021, 19, 514–527. [Google Scholar] [CrossRef] [PubMed]
- Martínez, A.; Kuraji, R.; Kapila, Y.L. The Human Oral Virome: Shedding Light on the Dark Matter. Periodontol 2000 2021, 87, 282–298. [Google Scholar] [CrossRef] [PubMed]
- Łusiak-Szelachowska, M.; Weber-Dąbrowska, B.; Żaczek, M.; Borysowski, J.; Górski, A. The Presence of Bacteriophages in the Human Body: Good, Bad or Neutral? Microorganisms 2020, 8, 2012. [Google Scholar] [CrossRef] [PubMed]
- Nayfach, S.; Páez-Espino, D.; Call, L. Metagenomic Compendium of 189,680 DNA Viruses from the Human Gut Microbiome. Nat. Microbiol. 2021, 6, 960–970. [Google Scholar] [CrossRef] [PubMed]
- Śliwka, P.; Ochocka, M.; Skaradzińska, A. Applications of Bacteriophages against Intracellular Bacteria. Crit. Rev. Microbiol. 2021, 47, 1–18. [Google Scholar] [CrossRef]
- Fujimoto, K.; Kimura, Y.; Shimohigoshi, M.; Satoh, T.; Sato, S.; Tremmel, G.; Uematsu, M.; Kawaguchi, Y.; Usui, Y.; Nakano, Y.; et al. Metagenome Data on Intestinal Phage-Bacteria Associations Aids the Development of Phage Therapy against Pathobionts. Cell Host. Microbe 2020, 28, 353–355. [Google Scholar] [CrossRef]
- Adriaenssens, E.M. Phage Diversity in the Human Gut Microbiome: A Taxonomist’s Perspective. mSystems 2021, 6, e00799-21. [Google Scholar] [CrossRef]
- Li, T.; Zhang, Y.; Dong, K.; Kuo, C.J.; Li, C.; Zhu, Y.Q.; Qin, J.; Li, Q.T.; Chang, Y.F.; Guo, X.; et al. Isolation and Characterization of the Novel Phage JD032 and Global Transcriptomic Response during JD032 Infection of Clostridioides Difficile Ribotype 078. MSystems. mSystems 2020, 5, e00017-20. [Google Scholar] [CrossRef]
- Hinc, K.; Kabała, M.; Iwanicki, A.; Martirosian, G.; Negri, A.; Obuchowski, M. Complete Genome Sequence of the Newly Discovered Temperate Clostridioides Difficile Bacteriophage PhiCDKH01 of the Family Siphoviridae. Arch. Virol. 2021, 166, 2305–2310. [Google Scholar] [CrossRef]
- Heuler, J.; Fortier, L.C.; Sun, X. Clostridioides Difficile Phage Biology and Application. FEMS Microbiol. Rev. 2021, 45, 5. [Google Scholar] [CrossRef]
- Selle, K.; Fletcher, J.R.; Tuson, H. In Vivo Targeting of Clostridioides Difficile Using Phage-Delivered CRISPR-Cas3 Antimicrobials. mBio 2020, 11, e00019-20. [Google Scholar] [CrossRef] [Green Version]
- Wu, N.; Dai, J.; Guo, M.; Li, J.; Zhou, X.; Li, F.; Gao, Y.; Qu, H.; Lu, H.; Jin, J.; et al. Pre-Optimized Phage Therapy on Secondary Acinetobacter Baumannii Infection in Four Critical COVID-19 Patients. Emerg. Microbes Infect. 2021, 10, 612–618. [Google Scholar] [CrossRef]
- Cheung, G.Y.C.; Bae, J.S.; Otto, M. Pathogenicity and Virulence of Staphylococcus Aureus. Virulence 2021, 12, 547–569. [Google Scholar] [CrossRef]
- Tong, S.Y.; Davis, J.S.; Eichenberger, E. Staphylococcus Aureus Infections: Epidemiology, Pathophysiology, Clinical Manifestations, and Management. Clin. Microbiol. Rev. 2015, 28, 603–661. [Google Scholar] [CrossRef] [Green Version]
- Food and Drug Administration (FDA). Bad Bug Book; FDA: Washington, DC, USA, 2012.
- Strowd, L.C.; Feldman, S. Lead with Your Gut: Research-Implicated Infant Gut Staphylococcal Strains May Predict Development of Atopic Dermatitis. Br. J. Dermatol. 2019, 180, 1296–1297. [Google Scholar] [CrossRef]
- Kang, M.S.; Lim, H.S.; Oh, J.S.; Lim, Y.J.; Wuertz-Kozak, K.; Harro, J.M.; Achermann, Y. Antimicrobial Activity of Lactobacillus Salivarius and Lactobacillus Fermentum against Staphylococcus Aureus. Pathog. Dis. 2017, 75, ftx009. [Google Scholar] [CrossRef]
- Khamash, D.F.; Voskertchian, A.; Milstone, A.M. Manipulating the Microbiome: Evolution of a Strategy to Prevent, S. Aureus Disease in Children. J. Perinatol. 2018, 38, 105–109. [Google Scholar] [CrossRef]
- Rao, C.; Coyte, K.Z.; Bainter, W.; Geha, R.S.; Martin, C.R.; Rakoff-Nahoum, S. Multi-Kingdom Ecological Drivers of Microbiota Assembly in Preterm Infants. Nature 2021, 591, 633–638. [Google Scholar] [CrossRef]
- Nataraj, B.H.; Ramesh, C.; Mallappa, R.H. Extractable Surface Proteins of Indigenous Probiotic Strains Confer Anti-Adhesion Knack and Protect against Methicillin-Resistant Staphylococcus Aureus Induced Epithelial Hyperpermeability in HT-29 Cell Line. Microb. Pathog. 2021, 158, 104974. [Google Scholar] [CrossRef]
- Shi, B.; Leung, D.Y.; Taylor, P.A.; Li, H. MRSA Colonization Is Associated with Decreased Skin Commensal Bacteria in Atopic Dermatitis. J. Investig. Dermatol. 2018, 138, 1668. [Google Scholar] [CrossRef] [Green Version]
- Patrick, G.J.; Liu, H.; Alphonse, M.P.; Dikeman, D.A.; Youn, C.; Otterson, J.C.; Wang, Y.; Ravipati, A.; Mazhar, M.; Denny, G.; et al. Epicutaneous Staphylococcus Aureus Induces IL-36 to Enhance IgE Production and Ensuing Allergic Disease. J. Clin. Investig. 2021, 131, e143334. [Google Scholar] [CrossRef]
- Liu, J.N.; Shin, Y.S.; Yoo, H.S.; Nam, Y.H.; Jin, H.J.; Ye, Y.M.; Nahm, D.H.; Park, H.S. The Prevalence of Serum Specific IgE to Superantigens in Asthma and Allergic Rhinitis Patients. Allergy Asthma Immunol. Res. 2014, 6, 263–266. [Google Scholar] [CrossRef] [Green Version]
- Tomczak, H.; Wróbel, J.; Jenerowicz, D.; Sadowska-Przytocka, A.; Wachal, M.; Adamski, Z.; Czarnecka-Operacz, M.M. The Role of Staphylococcus Aureus in Atopic Dermatitis: Microbiological and Immunological Implications. Postepy Dermatol. Alergol. 2019, 36, 485–491. [Google Scholar] [CrossRef]
- Tsilochristou, O.; Toit, G.; Sayre, P.H.; Roberts, G.; Lawson, K.; Sever, M.L.; Bahnson, H.T.; Radulovic, S.; Basting, M.; Plaut, M.; et al. Immune Tolerance Network Learning Early About Peanut Allergy Study Team. Association of Staphylococcus Aureus Colonization with Food Allergy Occurs Independently of Eczema Severity. J. Allergy Clin. Immunol. 2019, 144, 494–503. [Google Scholar] [CrossRef]
- Kim, Y.C.; Won, H.K.; Lee, J.W.; Sohn, K.H.; Kim, M.H.; Kim, T.B.; Chang, Y.S.; Lee, B.J.; Cho, S.H.; Bachert, C.; et al. Staphylococcus Aureus Nasal Colonization and Asthma in Adults: Systematic Review and Meta-Analysis. J. Allergy Clin. Immunol. Pract. 2019, 615, 606–615. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.H.F.; Lang, A.; Teo, S.M.; Judd, L.M.; Gangnon, R.; Evans, M.D.; Lee, K.E.; Vrtis, R.; Holt, P.G.; Lemanske, R.F., Jr.; et al. Developmental Patterns in the Nasopharyngeal Microbiome during Infancy Are Associated with Asthma Risk. J. Allergy Clin. Immunol. 2021, 147, 1683–1691. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.; Ju, Z.; Zhou, P.K. A Gut Dysbiotic Microbiota-Based Hypothesis of Human-to-Human Transmission of Non-Communicable Diseases. Sci. Total Environ. 2020, 745, 141030. [Google Scholar] [CrossRef] [PubMed]
- Otto, M. Staphylococcus Aureus Toxins. Curr. Opin. Microbiol. 2014, 17, 32–37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oliveira, D.; Borges, A.; Simões, M. Staphylococcus Aureus Toxins and Their Molecular Activity in Infectious Diseases. Toxins 2018, 10, 252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spaulding, A.R.; Salgado-Pabón, W.; Kohler, P.L.; Horswill, A.R.; Leung, D.Y.; Schlievert, P.M. Staphylococcal and Streptococcal Superantigen Exotoxins. Clin. Microbiol. Rev. 2013, 26, 422–447. [Google Scholar] [CrossRef] [Green Version]
- Vickery, T.W.; Ramakrishnan, V.R.; Suh, J.D. The Role of Staphylococcus Aureus in Patients with Chronic Sinusitis and Nasal Polyposis. Curr. Allergy Asthma Rep. 2019, 11, 19–21. [Google Scholar] [CrossRef]
- Muluk, N.B.; Altın, F.; Cingi, C. Role of Superantigens in Allergic Inflammation: Their Relationship to Allergic Rhinitis, Chronic Rhinosinusitis, Asthma, and Atopic Dermatitis. Am. J. Rhinol. Allergy 2018, 32, 502–517. [Google Scholar] [CrossRef]
- Bachert, C.; Holtappels, G.; Merabishvili, M.; Meyer, T.; Murr, A.; Zhang, N.; Van Crombruggen, K.; Gevaert, E.; Völker, U.; Bröker, B.M.; et al. Staphylococcus Aureus Controls Interleukin-5 Release in Upper Airway Inflammation. J. Proteom. 2018, 180, 53–60. [Google Scholar] [CrossRef]
- Bachert, C.; Humbert, M.; Hanania, N.A.; Zhang, N.; Holgate, S.; Buhl, R.; Bröker, B.M. Staphylococcus Aureus and Its IgE-Inducing Enterotoxins in Asthma: Current Knowledge. Eur. Respir. J. 2020, 55, 1901592. [Google Scholar] [CrossRef]
- Ueki, S.; Miyabe, Y.; Yamamoto, Y. Charcot-Leyden Crystals in Eosinophilic Inflammation: Active Cytolysis Leads to Crystal Formation. Curr. Allergy Asthma Rep. 2019, 19, 35. [Google Scholar] [CrossRef]
- Tsang, M.S.; Sun, X.; Wong, C.K. The Role of New IL-1 Family Members (IL-36 and IL-38) in Atopic Dermatitis, Allergic Asthma, and Allergic Rhinitis. Curr. Allergy Asthma Rep. 2020, 20, 40. [Google Scholar] [CrossRef]
- Shirai, Y.; Arai, H.; Tamaki, K.; Konishi, H.; Kawase, Y.; Shimizu, N.; Tateda, K.; Yoda, H. Neonatal Methicillin-Resistant Staphylococcus Aureus Colonization and Infection. J. Neonatal. Perinat. Med. 2017, 10, 439–444. [Google Scholar] [CrossRef]
- Khamash, D.F.; Mongodin, E.F.; White, J.R.; Voskertchian, A.; Hittle, L.; Colantuoni, E.; Milstone, A.M. The Association Between the Developing Nasal Microbiota of Hospitalized Neonates and Staphylococcus Aureus Colonization. Open Forum Infect. Dis. 2019, 6, 62. [Google Scholar] [CrossRef] [Green Version]
- Bessesen, M.T.; Kotter, C.V.; Wagner, B.D.; Adams, J.C.; Kingery, S.; Benoit, J.B.; Frank, D.N. MRSA Colonization and the Nasal Microbiome in Adults at High Risk of Colonization and Infection. J. Infect. 2015, 71, 649–657. [Google Scholar] [CrossRef] [Green Version]
- Roghmann, M.C.; Lydecker, A.D.; Langenberg, P.; Mongodin, E.F.; Johnson, J.K. Microbiological Effect of Mupirocin and Chlorhexidine for Staphylococcus Aureus Decolonization in Community and Nursing Home Based Adults. Diagn. Microbiol. Infect. Dis. 2017, 88, 53–57. [Google Scholar] [CrossRef] [Green Version]
- Nordengrün, M.; Abdurrahman, G.; Treffon, J.; Wächter, H.; Kahl, B.C.; Bröker, B.M. Allergic Reactions to Serine Protease-Like Proteins of Staphylococcus Aureus. Front. Immunol. 2021, 12, 913. [Google Scholar] [CrossRef]
- Pollmann, M.; Nordhoff, M.; Pospischil, A.; Tedin, K.; Wieler, L.H. Effects of a Probiotic Strain of Enterococcus Faecium on the Rate of Natural Chlamydia Infection in Swine. Infect. Immun. 2005, 73, 4346–4353. [Google Scholar] [CrossRef] [Green Version]
- Van Hai, N. Research Findings from the Use of Probiotics in Tilapia Aquaculture: A Review. Fish Shellfish Immunol. 2015, 45, 592–597. [Google Scholar]
- Angelakis, E. Weight Gain by Gut Microbiota Manipulation in Productive Animals. Microb. Pathog. 2017, 106, 162–170. [Google Scholar] [CrossRef]
- Wu, Y.; Zhu, C.; Chen, Z.; Chen, Z.; Zhang, W.; Ma, X.; Jiang, Z. Protective Effects of Lactobacillus Plantarum on Epithelial Barrier Disruption Caused by Enterotoxigenic Escherichia Coli in Intestinal Porcine Epithelial Cells. Vet. Immunol. Immunopathol. 2016, 172, 55–63. [Google Scholar] [CrossRef]
- Becker, K.; Heilmann, C.; Peters, G. Coagulase-negative staphylococci. Clin. Microbiol. Rev. 2014, 27, 870–926. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kosecka-Strojek, M.; Sabat, A.J.; Akkerboom, V.; Becker, K.; Zanten, E.; Wisselink, G.; Miedzobrodzki, J.; Koois-tra-Smid, A.; Friedrich, A.W. Development and Validation of a Reference Data Set for Assigning Staphylococcus Species Based on Next-Generation Sequencing of the 16S-23S RRNA Region. Front. Cell. Infect. Microbiol. 2019, 9, 278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chin, D.; Goncheva, M.I.; Flannagan, R.S.; Deecker, G.-O.; Guariglia-Oropeza, V.; Ensminger, A.W.; Heinrichs, D.E. Coagulase-Negative Staphylococci Release a Purine Analog That Inhibits Staphylococcus Aureus Virulence. Nat. Commun. 2021, 12, 1887. [Google Scholar] [CrossRef] [PubMed]
- Iwase, T.; Uehara, Y.; Shinji, H.; Tajima, A.; Seo, H.; Takada, K.; Agata, T.; Mizunoe, Y. Staphylococcus Epidermidis Esp Inhibits Staphylococcus Aureus Biofilm Formation and Nasal Colonization. Nature 2010, 465, 346–349. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, K.; O’Neill, A.M.; Williams, M.R.; Cau, L.; Nakatsuji, T.; Horswill, A.R.; Gallo, R.L. Short Chain Fatty Acids Produced by Cutibacterium Acnes Inhibit Biofilm Formation by Staphylococcus Epidermidis. Sci. Rep. 2020, 10, 21237. [Google Scholar] [CrossRef] [PubMed]
- Glatthardt, T.; Campos, J.C.M.; Chamon, R.C.; Sá Coimbra, T.F.; Rocha, G.A.; Melo, M.A.F.; Parente, T.E.; Lobo, L.A.; Antunes, L.C.M.; Dos Santos, K.R.N.; et al. Small Molecules Produced by Commensal Staphylococcus Epidermidis Disrupt Formation of Biofilms by Staphylococcus Aureus. Appl. Environ. Microbiol. 2020, 86, e02539-19. [Google Scholar] [CrossRef]
- Cole, A.L.; Sundar, M.; Lopez, A.; Forsman, A.; Yooseph, S.; Cole, A.M. Identification of Nasal Gammaproteobacteria with Potent Activity against Staphylococcus Aureus: Novel Insights into the “Noncarrier” State. Msphere 2021, 6, e01015–e01020. [Google Scholar] [CrossRef]
- Vuong, H.E.; Hsiao, E.Y. Emerging Roles for the Gut Microbiome in Autism Spectrum Disorder. Biol. Psychiatry 2017, 81, 411–423. [Google Scholar] [CrossRef] [Green Version]
- Adams, J.B.; Johansen, L.J.; Powell, L.D.; Quig, D.; Rubin, R.A. Gastrointestinal Flora and Gastrointestinal Status in Children with Autism–Comparisons to Typical Children and Correlation with Autism Severity. BMC Gastroenterol. 2011, 11, 22. [Google Scholar] [CrossRef] [Green Version]
- Kang, D.W.; Adams, J.B.; Coleman, D.M.; Pollard, E.L.; Maldonado, J.; McDonough-Means, S.; Krajmalnik-Brown, R. Long-Term Benefit of Microbiota Transfer Therapy on Autism Symptoms and Gut Microbiota. Sci. Rep. 2019, 9, 5821. [Google Scholar] [CrossRef]
- Abdellatif, B.; McVeigh, C.; Bendriss, G.; Chaari, A. The Promising Role of Probiotics in Managing the Altered Gut in Autism Spectrum Disorders. Int. J. Mol. Sci. 2020, 21, 4159. [Google Scholar] [CrossRef]
- Johnson, D.; Letchumanan, V.; Thurairajasingam, S.; Lee, L.H. A Revolutionizing Approach to Autism Spectrum Disorder Using the Microbiome. Nutrients 2020, 12, 1983. [Google Scholar] [CrossRef]
- Salamone, D.; Rivellese, A.A.; Vetrani, C. The Relationship between Gut Microbiota, Short-Chain Fatty Acids and Type 2 Diabetes Mellitus: The Possible Role of Dietary Fibre. Acta Diabetol. 2021, 58, 1131–1138. [Google Scholar] [CrossRef]
- Ye, F.; Gao, X.; Wang, Z.; Cao, S.; Liang, G.; He, D.; Zhang, Q. Comparison of Gut Microbiota in Autism Spectrum Disorders and Neurotypical Boys in China: A Case-Control Study. Synth. Syst. Biotechnol. 2021, 6, 120–126. [Google Scholar] [CrossRef]
- Shaaban, S.Y.; El Gendy, Y.G.; Mehanna, N.S.; El-Senousy, W.M.; El-Feki, H.S.; Saad, K.; El-Asheer, O.M. The Role of Probiotics in Children with Autism Spectrum Disorder: A Prospective, Open-Label Study. Nutr. Neurosci. 2018, 21, 676–681. [Google Scholar] [CrossRef]
- Qureshi, F.; Adams, J.; Hanagan, K.; Kang, D.W.; Krajmalnik-Brown, R.; Hahn, J. Multivariate Analysis of Fecal Metabolites from Children with Autism Spectrum Disorder and Gastrointestinal Symptoms before and after Microbiota Transfer Therapy. J. Pers. Med. 2020, 10, 152. [Google Scholar] [CrossRef]
- Ho, L.; Tong, V.J.W.; Syn, N.; Nagarajan, N.; Tham, E.H.; Tay, S.K.; Shorey, S.; Tambyah, P.A.; Law, E.C.N. Gut Microbiota Changes in Children with Autism Spectrum Disorder: A Systematic Review. Gut Pathog. 2020, 12, 6. [Google Scholar] [CrossRef] [Green Version]
- Oikonomou, G.; Addis, M.F.; Chassard, C.; Nader-Macias, M.E.F.; Grant, I.; Delbès, C.; Even, S. Milk Microbiota: What Are We Exactly Talking About? Front. Microbiol. 2020, 11, 60. [Google Scholar] [CrossRef] [Green Version]
- American Academy of Pediatrics (AAP). Committee Donor Human Milk for the High-Risk Infant: Preparation, Safety, and Usage Options in the United States. Pediatrics 2017, 139, e20163440. [Google Scholar] [CrossRef] [Green Version]
- Klotz, D.; Jansen, S.; Gebauer, C.; Fuchs, H. Handling of Breast Milk by Neonatal Units: Large Differences in Current Practices and Beliefs. Front. Pediatr. 2018, 6, 235. [Google Scholar] [CrossRef]
- Picaud, J.C.; Buffin, R.; Gremmo-Feger, G.; Rigo, J.; Putet, G.; Casper, C.; Working group of the French Neonatal Society on fresh human milk use in preterm infants. Review Concludes That Specific Recommendations Are Needed to Harmonise the Provision of Fresh Mother’s Milk to Their Preterm Infants. Acta Paediatr. 2018, 107, 1145–1155. [Google Scholar] [CrossRef]
- Moro, G.E.; Billeaud, C.; Rachel, B.; Calvo, J.; Cavallarin, L.; Christen, L.; Picaud, J.C. Processing of Donor Human Milk: Update and Recommendations from the European Milk Bank Association (EMBA). Front. Pediatr. 2019, 7, 49. [Google Scholar] [CrossRef] [Green Version]
- World Health Organization (WHO). Donor Human Milk for Low-Birth-Weight Infants; Last Update; World Health Organization: Geneva, Switzerland, 2019. [Google Scholar]
- Zimmermann, P.; Curtis, N. Breast Milk Microbiota: A Review of the Factors That Influence Composition. J. Infect. 2020, 81, 17–47. [Google Scholar] [CrossRef] [PubMed]
- Carr, L.E.; Elolimy, A.; Rosa, F.; Virmani, M.D.; Munblit, D.; Yeruva, L. Role of Human Milk Bioactives on Infants’ Gut and Immune Health. Front. Immunol. 2021, 12, 290. [Google Scholar] [CrossRef] [PubMed]
- Witkowska-Zimny, M.; Kaminska-El-Hassan, E. Cells of Human Breast Milk. Cell. Mol. Biol. Lett. 2017, 22, 11. [Google Scholar] [CrossRef] [PubMed]
- Lyons, K.E.; Ryan, C.A.; Dempsey, E.M.; Ross, R.P.; Stanton, C. Breast Milk, a Source of Beneficial Microbes and Associated Benefits for Infant Health. Nutrients 2020, 12, 1039. [Google Scholar] [CrossRef] [PubMed]
- García-González, I.; Corona-Cervantes, K.; Hernández-Quiroz, F.; Villalobos-Flores, L.E.; Galván-Rodríguez, F.; Romano, M.C.; Miranda-Brito, C.; Piña-Escobedo, A.; Borquez-Arreortúa, F.G.; Rangel-Calvillo, M.N.; et al. The Effect of Holder Pasteurization on the Diversity of the Human Milk Bacterial Microbiota Using High-Throughput DNA Sequencing. J. Hum. Lact. 2021, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Meltzer, H.M.; Brandtzæg, P.; Knutsen, H.K.; Løland, B.F.; Odland, J.Ø.; Skåre, J.U.; Torheim, L.E. Benefit and Risk Assessment of Breastmilk for Infant Health in Norway. Opinion of the Steering of the Norwegian Scientific Committee for Food Safety; Norwegian Scientific Committee for Food Safety (VKM): Nydalen, Norway, 2013. [Google Scholar]
- Meltzer, H.M.; Brandtzæg, P.; Knutsen, H.K.; Løland, B.F.; Odland, J.Ø.; Skåre, J.U.; Torheim, L.E. Benefit and Risk Assessment of Breastmilk for Infant Health in Norway. Eur. J. Nutr. Food Saf. 2016, 6, 101–110. [Google Scholar] [CrossRef] [Green Version]
- Miller, J.; Tonkin, E.; Damarell, R.; McPhee, A.; Suganuma, M.; Suganuma, H.; Collins, C. A Systematic Review and Meta-Analysis of Human Milk Feeding and Morbidity in Very Low Birth Weight Infants. Nutrients 2018, 10, 707. [Google Scholar] [CrossRef] [Green Version]
- Villamor-Martínez, E.; Pierro, M.; Cavallaro, G.; Mosca, F.; Kramer, B.W.; Villamor, E. Donor Human Milk Protects against Bronchopulmonary Dysplasia: A Systematic Review and Meta-Analysis. Nutrients 2018, 10, 238. [Google Scholar] [CrossRef] [Green Version]
- Sun, H.; Han, S.; Cheng, R.; Hei, M.; Kakulas, F.; Lee, S.K. Testing the Feasibility and Safety of Feeding Preterm Infants Fresh Mother’s Own Milk in the NICU: A Pilot Study. Sci. Rep. 2019, 9, 941. [Google Scholar] [CrossRef]
- Ford, S.L.; Lohmann, P.; Preidis, G.A.; Gordon, P.S.; O’Donnell, A.; Hagan, J.; Hair, A.B. Improved Feeding Tolerance and Growth Are Linked to Increased Gut Microbial Community Diversity in Very-Low-Birth-Weight Infants Fed Mother’s Own Milk Compared with Donor Breast Milk. Am. J. Clin. Nutr. 2019, 109, 1088–1097. [Google Scholar] [CrossRef]
- Henrick, B.M.; Rodriguez, L.; Lakshmikanth, T.; Pou, C.; Henckel, E.; Arzoomand, A.; Olin, A.; Wang, J.; Mikes, J.; Tan, Z.; et al. Bifidobacteria-Mediated Immune System Imprinting Early in Life. Cell 2021, 184, 3884–3898. [Google Scholar] [CrossRef]
- Salminen, S.; Stahl, B.; Vinderola, G.; Szajewska, H. Infant Formula Supplemented with Biotics: Current Knowledge and Future Perspectives. Nutrients 2020, 12, 1952. [Google Scholar] [CrossRef]
- Almeida, C.C.; Mendonça Pereira, B.F.; Leandro, K.C.; Costa, M.P.; Spisso, B.F.; Conte-Junior, C.A. Bioactive Compounds in Infant Formula and Their Effects on Infant Nutrition and Health: A Systematic Literature Review. Int. J. Food Sci. 2021, 2021, 8850080. [Google Scholar] [CrossRef]
- Hu, Y.; Chen, Y.; Liu, S.; Jiang, F.; Wu, M.; Yan, C.; Tong, S. Breastfeeding Duration Modified the Effects of Neonatal and Familial Risk Factors on Childhood Asthma and Allergy: A Population-Based Study. Respir. Res. 2021, 22, 41. [Google Scholar] [CrossRef]
- Wiedemann, P.; Schütz, H.; Spangenberg, A.; Krug, H.F. Evidence Maps: Communicating Risk Assessments in Societal Controversies: The Case of Engineered Nanoparticles. Risk Anal. Int. J. 2011, 31, 1770–1783. [Google Scholar] [CrossRef]
- Grøvslien, A.H.; Grønn, M. Donor Milk Banking and Breastfeeding in Norway. J. Hum. Lact. 2009, 25, 206–210. [Google Scholar] [CrossRef]
- Mizuno, K.; Sakurai, M.; Itabashi, K. Necessity of Human Milk Banking in Japan: Questionnaire Survey of Neonatologists. Pediatr. Int. 2015, 57, 639–644. [Google Scholar] [CrossRef]
- Grøvslien, A.H. Personal Communication. Human Milk Bank Monitoring for Norway; University of Oslo: Oslo, Norway, 2020. [Google Scholar]
- World Health Organization (WHO). WHO/UNICEF Statement on the 40th Anniversary of the International Code of Marketing Breastmilk Substitutes; WHO: Geneva, Switzerland, 2021. [Google Scholar]
- Beam, A.; Clinger, E.; Hao, L. Effect of Diet and Dietary Components on the Composition of the Gut Microbiota. Nutrients 2021, 13, 2795. [Google Scholar] [CrossRef]
- Barber, T.M.; Valsamakis, G.; Mastorakos, G.; Hanson, P.; Kyrou, I.; Randeva, H.S.; Weickert, M.O. Dietary Influences on the Microbiota-Gut-Brain Axis. Int. J. Mol. Sci. 2021, 22, 3502. [Google Scholar] [CrossRef]
- Alcock, J.; Maley, C.C.; Aktipis, C.A. Is Eating Behavior Manipulated by the Gastrointestinal Microbiota? Evolutionary Pressures and Potential Mechanisms. Bioessays 2014, 36, 940–949. [Google Scholar] [CrossRef]
- Esberg, A.; Haworth, S.; Hasslöf, P.; Lif Holgerson, P.; Johansson, I. Oral Microbiota Profile Associates with Sugar Intake and Taste Preference Genes. Nutrients 2020, 12, 681. [Google Scholar] [CrossRef] [Green Version]
- Khan, A.S.; Keast, R.; Khan, N.A. Preference for Dietary Fat: From Detection to Disease. Prog. Lipid. Res. 2020, 78, 101032. [Google Scholar] [CrossRef]
- Han, H.; Yi, B.; Zhong, R.; Wang, M.; Zhang, S.; Ma, J.; Yin, Y.; Yin, J.; Chen, L.; Zhang, H. From Gut Microbiota to Host Appetite: Gut Microbiota-Derived Metabolites as Key Regulators. Microbiome 2021, 9, 162. [Google Scholar] [CrossRef]
- Leung, R.; Covasa, M. Do Gut Microbes Taste? Nutrients 2021, 13, 2581. [Google Scholar] [CrossRef]
- Bienenstock, J.; Kunze, W.A.; Forsythe, P. Disruptive Physiology: Olfaction and the Microbiome-Gut-Brain Axis. Biol. Rev. Camb. Philos. Soc. 2018, 93, 390–403. [Google Scholar] [CrossRef]
- Schwartz, M.; Canon, F.; Feron, G.; Neiers, F.; Gamero, A. Impact of Oral Microbiota on Flavor Perception: From Food Processing to In-Mouth Metabolization. Foods 2021, 10, 2006. [Google Scholar] [CrossRef]
- Jurczak, A.; Jamka-Kasprzyk, M.; Bębenek, Z.; Staszczyk, M.; Jagielski, P.; Kościelniak, D.; Gregorczyk-Maga, I.; Kołodziej, I.; Kępisty, M.; Kukurba-Setkowicz, M.; et al. Differences in Sweet Taste Perception and Its Association with the Streptococcus Mutans Cariogenic Profile in Preschool Children with Caries. Nutrients 2020, 12, 2592. [Google Scholar] [CrossRef]
- Novelle, M.G. Decoding the Role of Gut-Microbiome in the Food Addiction Paradigm. Int. J. Environ. Res. Public Health 2021, 18, 6825. [Google Scholar] [CrossRef] [PubMed]
- Herman, A.; Bajaka, A. The Role of the Intestinal Microbiota in Eating Disorders—Bulimia Nervosa and Binge Eating Disorder. Psychiatry Res. 2021, 300, 113923. [Google Scholar] [CrossRef] [PubMed]
- Pocheron, A.L.; Le Dréan, G.; Billard, H.; Moyon, T.; Pagniez, A.; Heberden, C.; Le Chatelier, E.; Darmaun, D.; Michel, C.; Parnet, P. Maternal Microbiota Transfer Programs Offspring Eating Behavior. Front. Microbiol. 2021, 12, 672224. [Google Scholar] [CrossRef] [PubMed]
- Shiro, Y.; Arai, Y.C.; Ikemoto, T.; Ueda, W.; Ushida, T. Correlation Between Gut Microbiome Composition and Acute Pain Perception in Young Healthy Male Subjects. Pain Med. 2021, 22, 1522–1531. [Google Scholar] [CrossRef] [PubMed]
- Hill, C. RDA for Microbes—Are You Getting Your Daily Dose? Biochemist 2018, 40, 22–25. [Google Scholar] [CrossRef]
- Marco, M.L.; Hill, C.; Hutkins, R.; Slavin, J.; Tancredi, D.J.; Merenstein, D.; Sanders, M.E. Should There Be a Recommended Daily Intake of Microbes? J. Nutr. 2020, 150, 3061–3067. [Google Scholar] [CrossRef] [PubMed]



| Predominant Phyla | King et al., 2019 | Abenavoli et al., 2019 |
|---|---|---|
| Actinobacteria | 2% | 3% |
| Bacteriodetes | 73% | 23% |
| Firmicutes | 22% | 64% |
| Proteobacter | 2% | 8% |
| Age Group | Incidence Per 100,000 for Community-Associated CDI | Incidence Per 100,000 for Healthcare-Associated CDI |
|---|---|---|
| 1–17 | 27 | 9 |
| 18–44 | 42 | 18 |
| 45–64 | 79 | 72 |
| >65 | 169 | 262 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Coleman, M.E.; Dietert, R.R.; North, D.W.; Stephenson, M.M. Enhancing Human Superorganism Ecosystem Resilience by Holistically ‘Managing Our Microbes’. Appl. Microbiol. 2021, 1, 471-497. https://doi.org/10.3390/applmicrobiol1030031
Coleman ME, Dietert RR, North DW, Stephenson MM. Enhancing Human Superorganism Ecosystem Resilience by Holistically ‘Managing Our Microbes’. Applied Microbiology. 2021; 1(3):471-497. https://doi.org/10.3390/applmicrobiol1030031
Chicago/Turabian StyleColeman, Margaret E., Rodney R. Dietert, D. Warner North, and Michele M. Stephenson. 2021. "Enhancing Human Superorganism Ecosystem Resilience by Holistically ‘Managing Our Microbes’" Applied Microbiology 1, no. 3: 471-497. https://doi.org/10.3390/applmicrobiol1030031
APA StyleColeman, M. E., Dietert, R. R., North, D. W., & Stephenson, M. M. (2021). Enhancing Human Superorganism Ecosystem Resilience by Holistically ‘Managing Our Microbes’. Applied Microbiology, 1(3), 471-497. https://doi.org/10.3390/applmicrobiol1030031







