Abstract
Microbial lipase is one of the major sources of the enzyme that has been broadly exploited in the food, detergent, and pharmaceutical industries due to its high catalytic activity, high yield, and environmental friendliness and cost-effectiveness. Therefore, the aim of this study was to optimize the medium for the submerged fermentation for lipase production by a novel strain, Bacillus salmalaya strain 139SI. The media subjected to lipase production was Luria Bertani (LB) with different carbon sources and surfactants supplemented to determine which would give the highest lipase activity of Bacillus salmalaya. The Lipase activity of the supernatant containing lipase enzyme was ddetermined using the titrimetric method with hydrolysis reaction. Results showed that the olive oil that was used as a carbon source, induced the highest lipase activity (11.0 U/mL) compared to sunflower oil (9.6 U/mL) and cooking oil waste (7.8 U/mL). For surfactants, LB medium supplemented with tween 80 enhanced higher lipase activity (6.8 U/mL) compared to tween 20 (6.0 U/mL) and sodium dodecyl sulphate (SDS) (2.0 U/mL). Thus, it can be concluded that submerged fermentation allows optimization of the culture medium whereby, among carbon sources, olive oil induced the highest lipase production, whereas Tween 80 was the best lipase inducer compared to other surfactants.
1. Introduction
Bacillus is one of the most common bacterial genera reported in soil, and various species have been found in a variety of environmental niches [1]. Bacillus salmalaya 139SI found from an agricultural soil in Malaysia that was identified by Salmah and Dadrasnia (2015) as a member of a novel species belonging to genus bacillus based on 16S rRNA gene sequences. According to Salmah and Dadrasnia (2015) the isolated colonies were large and gray with a rough and irregular edge. In addition, the colonies were 2–3 mm in diameter and exhibited strong hemolytic activity after 16 h of incubation at 37 °C on 5% sheep blood agar. The novel strain showed the closest physical and genetic similarities to Bacillus species after morphological and genetic analysis [2,3]. This isolated strain of Bacillus salmalaya 139SI is excellent for improving the biotreatment of oil in wastewater [3]. Phenotypic traits, phylogenetic analysis, and 16S rRNA G+C characterization were used to classify this strain as a species [4].
Recently, environmental studies have investigated a new approach using lipase enzymes to clean septic tanks, grease traps, and other systems. Research had proven that lipase produced by some bacteria are capable of degrading FOG fat, oil, and grease) [5] Lipases are considered as one of the most important biocatalysts, whereby they can catalyze the reaction of ester hydrolysis, synthesis, esterification and transesterification. Recently, there has been a significant increase of interest in lipase production as it has the advantages of being active in the interfaces of organic compounds, produce high yield, while having low production cost, ease of genetic manipulation and has a broad substrate specificity. Microbial enzymes are also much more preferred than chemicals for hydrolytic reactions because they require less energy yet products are in higher quality [5,6]
Lipases can be obtained from a variety of sources, including animal, vegetable, and microbial sources [1,5]. Lipases, also known as triacylglycerol acyl hydrolases (E.C. 3.1.1.3) are hydrolases that operate on carboxylic ester bonds. They belong to the class of serine hydrolases and do not require any cofactor [6].
Large volumes of fats, oils, and greases (FOG), by-products of food preparation, are received daily by sewage treatment works (STWs) [7]. FOG (has long been a source of environmental concern. When it’s discharged improperly into the sewer systems, many problems arise. It will be deposited on the wall of the sewer, build-up, solidifies, and eventually causing the pipes blockage, hence restricting the wastewater flow. Worse, the blockage may result in sanitary sewer overflow which can lead to other environmental problems [8].
The development and application of novel and environmentally friendly technologies to degrade and/or eliminate these chemical molecules has recently gotten a lot of attention. Many microbial species have been found to be effective at breaking down organic molecules, which these microbes use as energy sources [3]. Biosurfactants, a class of new age surfactants derived from microorganisms, are equally diverse in terms of structure and function, and are gaining popularity due to their biodegradability and eco-friendliness [9].
The purpose of this study is to see whether B. salmalaya strain 139SI can produce lipase enzyme, as well as to see if alternative carbon sources and surfactants may help the strain produce more lipase. This research also aids in the identification of the appropriate carbon source and surfactant to use as a lipase inducer.
2. Material and Methodology
2.1. Preparation of Bacillus salmalaya Strain 139S1
To isolate and obtain a pure culture, the B. salmalaya was inoculated on a blood agar plate using the streak plate method. This step was carried out near flame to avoid contamination during the process. Then, the blood agar was incubated overnight at 37 °C. The next day, single colonies with clear zone was observed, indicating that B. salmalaya possess the haemolytic activity by the lysis of blood in the agar plate. Then, gram staining was performed to the culture of B. salmalaya to further confirm the identity of the bacteria. B. salmalaya is gram positive, which stained purple and rod-shaped. A stock culture of B. salmalaya was then prepared for future use. First, BHI agar [10] was prepared in bijou bottles. Then, the pure culture of B. salmalaya was inoculated on the BHI agar and incubated overnight at 37 °C. After that, the stock culture was stored in a 4 °C refrigerator until further use.
2.2. Preparation of Tween 20 Agar Plate
To screen for lipase production, a Tween 20 agar plate was prepared consisting of 10 g−1 peptone, 5 g−1 Sodium Chloride (NaCl), 0.1 g−1 Calcium Chloride dihydrate (CaCl2∙2H2O), 20 g−1 bacto agar and 10 mL Tween 20 (recipe of 1 Litre media). The agar medium was autoclaved at 121 °C for 20 min then cooled to 60 °C, the agar medium was poured into sterile plates and left to be solidified. The agar plates were refrigerated overnight at 4 °C. The tween-20 agar plates were then ready to be used for the detection of lipolytic activity of B. salmalaya.
2.3. Culture Media and Conditions
Inoculum for lipase production was first prepared by inoculating the isolate in a Luria Bertani (LB) broth [11]. Which consists of peptone 10 g−1, Yeast extract 5 g−1 and Sodium Chloride 5 g−1 at pH 8.0 (recipe of 1 Litre media). The inoculated LB broth was incubated overnight at 37 °C and was kept agitated on the rotary shaker at 150 rpm to ensure aeration in the broth. The overnight culture was then used as inoculum for enzyme production. Seven media were prepared. A 250 mL LB broth without supplementation of oil or surfactants served as the control. Other six media include, i.e.,
- (a)
- Carbon source
- (i)
- 250 mL LB broth + 10 mL cooking oil waste
- (ii)
- 250 mL LB broth + 10 mL olive oil
- (iii)
- 250 mL LB broth + 10 mL sunflower oil
- (b)
- Surfactants
- (i)
- 250 mL LB broth + 10 mL Tween 80
- (ii)
- 250 mL LB broth + 10 mL Tween 20
- (iii)
- 250 mL LB broth + 10 mL Sodium dodecyl sulphate (SDS)
A 250 mL LB broth without supplementation of oil or surfactants served as the control The media were adjusted to pH 7.2 using 0.5 N NaOH and then autoclaved at 121 °C for 20 min. After autoclaving, the media were left to cool down until 60 °C and then were inoculated with 3 mL of the overnight culture. After that, the media were incubated for 48 h at 37 °C on a rotary shaker at 150 rpm.
2.4. Measuring Bacterial Growth
Bacterial growth was measured by the reading of optical density of 600 nm (OD600) using the spectrophotometer. Reading was taken at 0 h, 12 h, 24 h, 36 h and 48 h.
2.5. Extraction of Crude Lipase Enzyme
The seven types of culture broth had undergone centrifugation after 48 h of incubation to extract the extracellular lipase enzyme. Centrifugation was carried out at 8000 rpm (2147 rcf), 4 °C for 20 min, using a refrigerated centrifuge. The supernatant obtained contained the extracellular lipase.
2.6. Titrimetric Lipase Assay
The activity of lipase was determined using the titrimetric method. First, a reaction cocktail was prepared, using olive oil as a substrate at pH 7.0. The reaction cocktail contains 10% olive oil emulsified in 10% (w/v) gum acacia, 2 mL of 0.6% CaCl2 solution and 5 mL of 0.2 mol/L sodium phosphate buffer. Then, 1 mL of each crude lipase enzyme solutions was added to the reaction cocktails separately. They were then incubated for one hour at 35 °C on an orbital shaker at 150 rpm. After one hour of incubation, the reaction was interrupted by adding 20 mL of acetone: ethanol solution (1:1) and the contents were swirled. 2–3 drops of phenolphthalein indicator were added to each of the reaction mixture. The reaction mixtures were then titrated with 0.05 M NaOH solution to an end point of pink color at pH 10.0. Reaction blanks (control) were run in the same way except that the crude enzymes are added after the addition of ethanol and acetone solutions. The lipase activities were carried out in five replicates.
2.7. Lipase Activity Calculation
Micromoles of free fatty acids obtained from olive oil per mL crude lipase enzyme was used to calculate lipase activity:
whereby, VS stands for the volume of NaOH solution used by the enzyme-substrate cocktail (mL). VB stands for the volume of NaOH used in the control cocktail (mL). The molarity of the NaOH solution employed in the titration (0.05 M) is N. The volume of the substrate cocktail solution (10 mL) is denoted by S. One unit (U) is the amount of lipase enzyme necessary to liberate 1 mol of fatty acids from triglycerides [5].
3. Results
3.1. Screening of Lipase Production
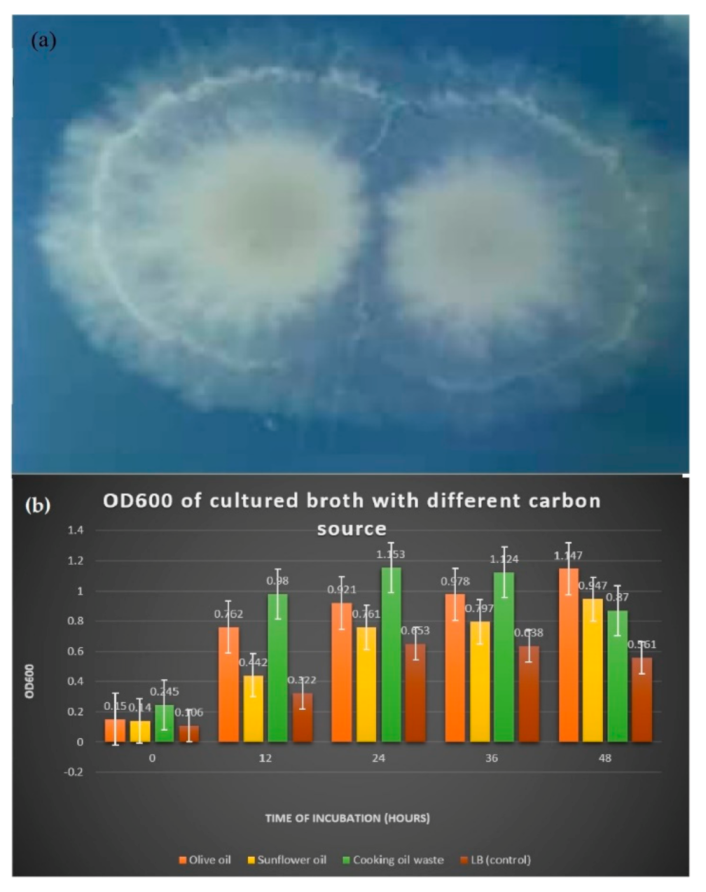
After inoculating B. salmalaya on the tween 20 agar plate and incubated for five days, white milky precipitate appeared around the colonies due to the deposition of calcium salt crystals that were formed by the action of lipase that liberated fatty acid (Figure 1a).
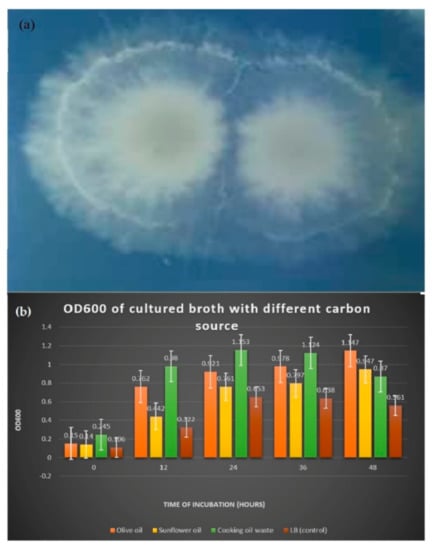
Figure 1.
(a) Formation of white precipitate Supplemented with different carbon sources (b) Comparison of OD600 reading in medium indicated that lipase was released.
3.2. Bacterial Growth
3.2.1. Optical Density (OD600) of Cultured Broth with Different Carbon Source
In Table 1 and Figure 1b, the reading of OD600 of LB cultured broth supplemented with 1% of carbon sources showed differences in bacterial growth during the 48 h incubation period. LB medium that was added with carbon source enhanced the bacterial growth, increasing the OD600. On the 48th h of incubation, bacterial growth was the highest in olive oil medium with OD600 reading of 1147.

Table 1.
The reading of optical density (OD600) of cultured broth incubated for two days.
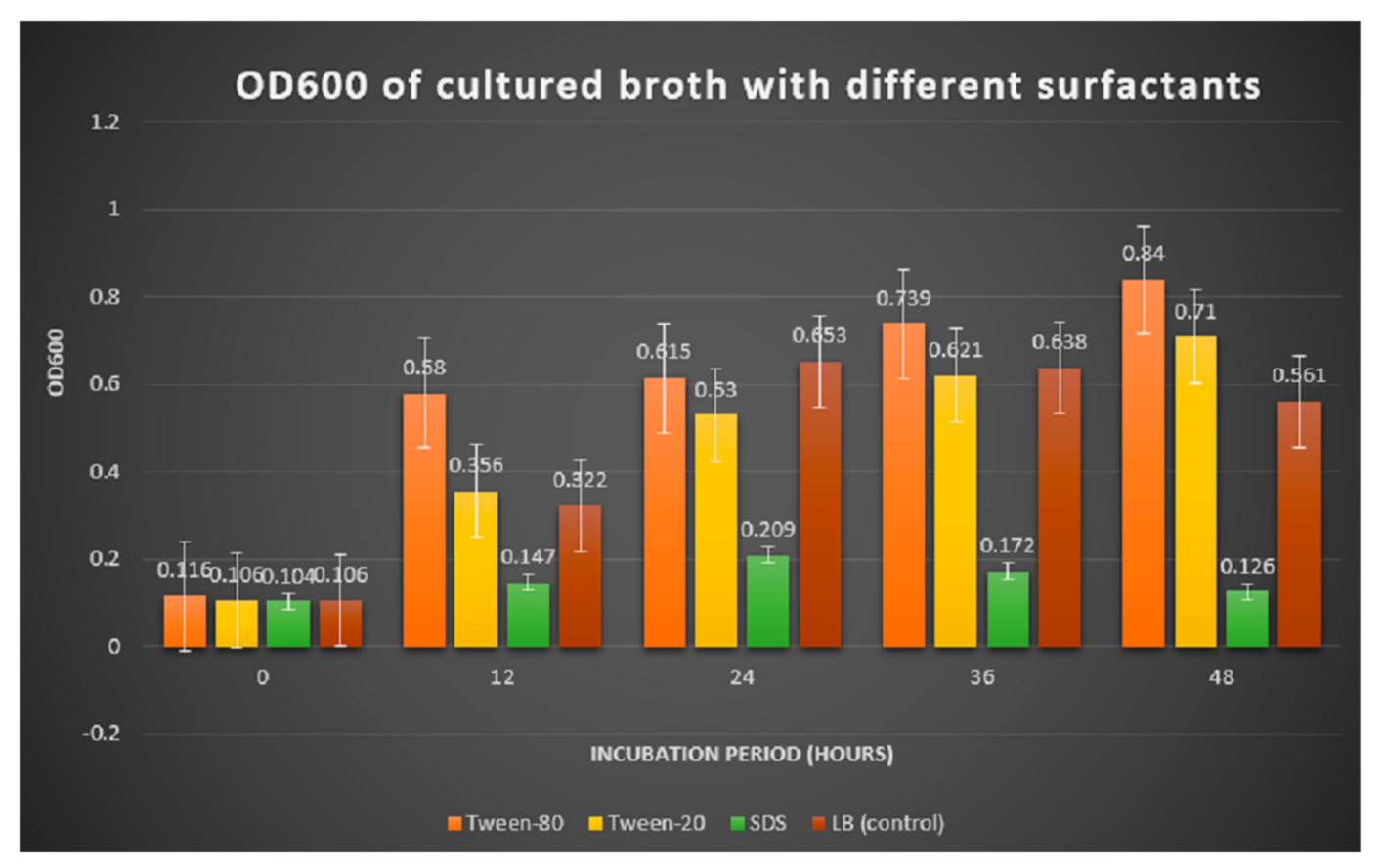
3.2.2. Optical Density of Broth with Different Surfactants
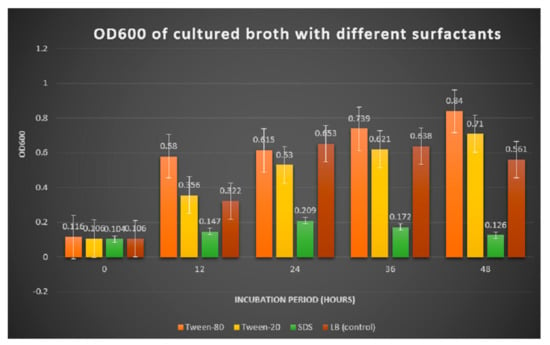
Figure 2.
Comparison of OD600 reading in medium supplemented with different surfactants.

Table 2.
The reading of optical density (OD600) of cultured broth incubated for two days.
3.3. Quantification of Fatty Acids Liberated
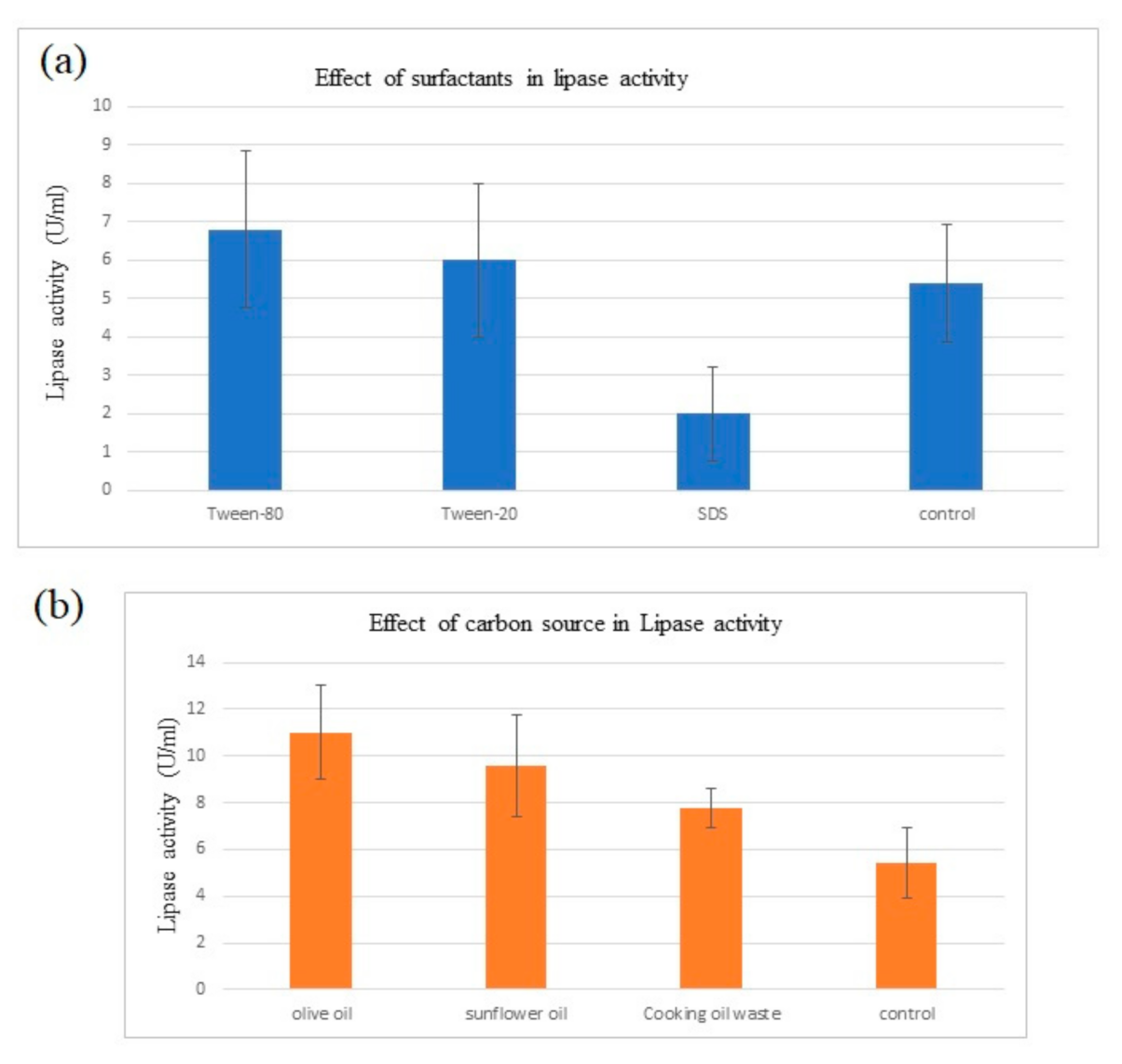
Effect of carbon source on lipase production: Lipase activity that was determined titrimetrically by olive oil hydrolysis showed the effect of using different oils as carbon source in fermentation medium. From Table 3 and Figure 3b, results showed that the presence of olive oil in the fermentation medium gave the maximum lipase activity (11.00 ± 2.00 U/mL), followed by sunflower oil and cooking oil waste.

Table 3.
Lipase activity of Bacillus salmalaya grew in different production media.
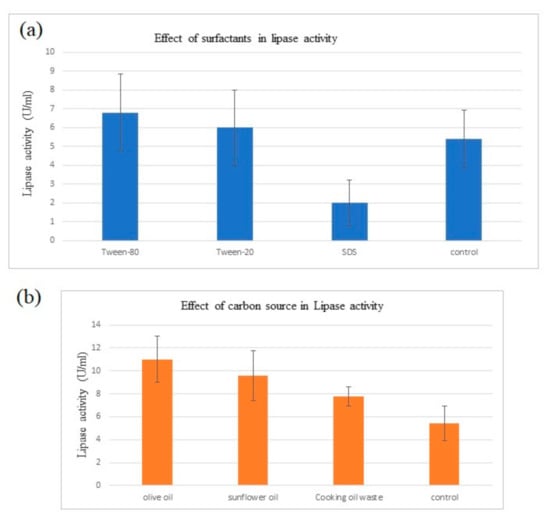
Figure 3.
(a) Comparison of lipase activity from medium of different surfactants. (b) Comparison of lipase activity from medium of d/f carbon sources.
3.4. The Effect of Surfactants in Lipase Production
Extracellular lipase that was produced from submerged fermentation with different surfactants were quantified for its lipase activity as shown in Table 4 and Figure 3. Amongst the media that were supplemented with different surfactants, lipase activity was the highest with tween 80 as surfactant compared to tween 20, SDS and control without surfactant.

Table 4.
Lipase activity Bacillus salmalaya grew in production media with different surfactants.
3.5. Statistical Analysis
3.5.1. Carbon Source
The statistical analysis of variance depicted that the increase in lipase activity induced by carbon sources were significant (p < 0.001) (Table 5). From the analysis of variance in Table 6, significance in lipase activity between carbon sources were represented. Every medium supplemented with oil as a carbon source showed significantly higher lipase activity compared to the control p < 0.05. Meanwhile, among carbon sources, lipase activity was the highest in olive oil (11.00 ± 0.72) where it depicts significantly higher reading than cooking oil waste (7.80 ± 0.72) and slightly higher than sunflower oil (9.60 ± 0.72). Meanwhile, sunflower oil showed a small significant difference with cooking oil waste in lipase activity.

Table 5.
Analysis of Variance (ANOVA) for lipase activity induced by carbon source (waste cooking oil, olive oil and sunflower oil) and lipase activity induced by surfactants (tween 80, tween 20 and SDS).

Table 6.
Analysis of Variance for comparing lipase activity between carbon sources of waste cooking oil, olive oil and sunflower oil.
3.5.2. Surfactants
Referring to Table 5, the effect of different surfactants to lipase activity were non-significant (p > 0.001). The analysis of variance to determine the significant differences of lipase activity between surfactants were depicted from Table 7. The lipase activity between tween 80, between 20 and control showed no significant differences (6.80 ± 1.01, 6.00 ± 1.01 and 5.40 ± 1.01), respectively. Tween 80 represents a non-significant slightly higher lipase activity from tween 20, SDS and control. However, SDS showed a non-significant slightly lower lipase activity compared to the other media (2.00 ± 1.01).

Table 7.
Analysis of Variance for comparing lipase activity between surfactants of tween 80, tween 20 and SD.
4. Discussion
Based on the screening for lipase production by B. salmalaya, a white milky precipitate was formed on the tween 20 agar plate. Tween 20 is a polyoxyethylene sorbitol ester containing lauric acid as the primary fatty acid. The lipase produced the B. salmalaya liberated fatty acid from the tween 20 which reacted with the calcium contained in the agar. This resulted on the formation of calcium salt crystals, thus forming the precipitate. Furthermore, a study by Kumar et al. isolated a Bacillus sp. strain DVL2 from soil samples to evaluate its enzymatic potential using several assays including the tween 20 hydrolysis method. The observed result showed similarity with the present study, signifying its ability to produce lipase. This method was also practiced in screening for lipase-producing fungal species [12]. In addition, Kanimozhi and Perinbam (2005) also isolated bacteria from oil contaminated sites. Only 8 of the 52 isolates revealed indications of lipase production, such as as white precipitate around the colony. The isolate with the highest precipitation zone was recognized as the main candidate for lipase production. The isolate was identified as Pseudomonas sp. strain Lp1 based on its biochemical, cultural and morphological characteristics [13]. To support this finding, a study by Ramnath et al. (2017) isolated three different Bacillus spp. from Eucalyptus wood as shown in Table 8 and screened for their lipase activity by using the agar well diffusion method. An olive oil/ rhodamine agar plates were used at first, but a difficulty encountered with the visualization of the result, leading to additional screening method; assay with phenol red and tween 20 [14].

Table 8.
Lipase and esterase activity of bacteria isolated from a mixed Eucalyptus wood chip pile [14].
The phenol red agar plate assay works on the idea that a little drop in pH from 7.3 (the end point of the phenol red) to a more acidic pH causes the red color to turn orange, indicating that fatty acid was released via hydrolysis. Then, the precipitation test using tween 20 was carried out to confirm lipase activity [14].
For bacteria fermented in the medium supplemented with tween 80 and tween 20, the OD600 reading showed an increasing growth pattern up until the 48th h. The enhancement of bacterial growth with the presence of carbon sources and surfactants were mainly because bacteria from the genus Bacillus are generally heterotrophs, which oxidize organic matter such as carbohydrates, lipids and protein to feed on and obtain energy. A study by Pallavi et al. (2014) recorded the growth of B. subtilis Y-IVI to reach 1100 with a maximum production of lipase (9.5) when olive oil was used as a substrate [15]. The toxic action of SDS may also cause depolarization of the cell membrane that resulted in the decrease in absorption of essential nutrients and oxygen consumption [16].
Result showed that the supplementation of different types of oil as a carbon source induced higher lipase production. B. salmalaya that was fermented in LB broth only recorded a lipase activity of 5.4 ± 1.52 U/mL. Meanwhile, the highest lipase activity (11.0 ± 2.00 U/mL) was attained when B. salmalaya was fermented in a medium supplemented with olive oil. Sunflower oil and cooking oil waste induced lipase production at 9.6 ± 2.19 U/mL and 7.8 ± 0.84 U/mL, respectively. Olive oil is consisted of oleic acid (C18:1) which makes up 55 to 83% of olive oil, 3.5 to 21% of linoleic acid (C18:2), 0 to 1.5% linolenic acid (C18:3) and 0.5 to 5% stearic acid (C18:0) [17]. On the other hand, sunflower oil possesses incorrect ref order, 16 detected after 14. You jumped the numbers in between2.8% of stearic acid, 28.0% of oleic acid, 62.0% of linoleic acid and 0.16% linolenic acid [17]. Hence the high composition of C18:n in lipid substrate served as the best carbon source for producing lipase. Meanwhile, it was observed that cooking oil waste induced the lowest amount of lipase among the other carbon sources. Cooking oil waste generally has higher free fatty acid (FFA) content than fresh cooking oil. When oils are fried, the combination of high temperature and water from food lead to the formation of FFA. Lipase production may have been inhibited due to the high concentration of FFA in the fatty acid residue.
The result showed that lipase production was induced with the supplementation of surfactants where highest amount of lipase recorded at 6.8 ± 2.05 U/mL in tween 80 while tween 20 enhanced lipase production only up until 6 ± 2.00 U/mL. it is possible for every microorganism possess specific characteristics to different substrates, even surfactants. Apart from that, tween 80 was believed to be composed of oleic acid (C18:1) while tween 20 are primarily composed of lauric acid (C12). As lipase were induced more in tween 80, this supports the previous suggestion for C18 enhancing lipase production. Meanwhile, SDS recorded a low reading of lipase activity by 2 ± 1.22 U/mL which indicates the inhibition of lipase production. As the growth of bacteria was inhibited during the fermentation process, viable cells were less to be able to produce high amount of lipase. The order of lipase activity is similar to the OD600 reading, suggesting that lipase activity is probably related to the total number of viable cells present. The significant inhibition by SDS could potentially be due to local conformation changes in the enzyme’s active region, which cause inhibition, partly reversible unfolding, and subsequent deactivation [18]. Furthermore, the presence of surfactants increases cell permeability, allowing for the export of a variety of molecules across the cell membrane, including protein secretion. Surfactants also make it easier for the enzyme to make contact with the substrate [19]. Hence, this may be the principle behind the enhancement of lipase production by surfactants.
5. Conclusions
In this study, Bacillus salmalaya strain 139SI was proven to produce lipase enzyme using the precipitation test on tween 20 agar plate. Bacterial growth of B. salmalaya cultivated and fermented in broth of different substrates showed that olive oil induced the highest growth based on the OD600 reading after 48 h. Similarly, for lipase activity, the supplementation of olive oil enhanced the highest lipase activity. Meanwhile, results depicted that amongst surfactants, tween 80 induced the highest bacterial growth and lipase activity. This study concluded that the supplementation of carbon sources and surfactants in the growth medium enhanced the lipase activity of B. salmalaya strain 139SI. While, as B. salmalaya strain 139SI is a novel species, there has been no official published research on the characterization for lipase enzyme produced by this bacterium.
Author Contributions
Conceptualization, A.F. and M.M.; data curation, M.A.M. and M.A.A.H. investigation, M.A.M., A.F. and A.D.; methodology, A.J.A. and S.I.; project administration, S.I.; software, M.M., A.D. and S.G.; supervision, S.I.; validation, A.J.A. and Y.N.A.; writing—original draft, M.A.M. and A.F.; writing—review and editing, M.M., Y.N.A., S.G. and S.I. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Requests to access the datasets should be directed to the corresponding author.
Acknowledgments
The authors gratefully acknowledge Nur Huza Aziera Binti Mohamad Huzairo (Institute of Biological Sciences, Faculty of Science, University of Malaya, Malaysia) for experimentation assistance and Institute of Biological Sciences, Faculty of Science, University of Malaya, Malaysia, for providing financial support and lab facility for this research work.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Saxena, A.K.; Kumar, M.; Chakdar, H.; Anuroopa, N.; Bagyaraj, D. Bacillus species in soil as a natural resource for plant health and nutrition. J. Appl. Microbiol. 2020, 128, 1583–1594. [Google Scholar] [CrossRef] [PubMed]
- Dadrasnia, A.; Usman, M.M.; Wei, K.S.C.; Velappan, R.D.; Jamali, H.; Mohebali, N.; Ismail, S. Native soil bacterial isolate in Malaysia exhibit promising supplements on degrading organic pollutants. Process Saf. Environ. Prot. 2016, 100, 264–271. [Google Scholar] [CrossRef]
- Dadrasnia, A.; Ismail, S. Biosurfactant Production by Bacillus salmalaya for Lubricating Oil Solubilization and Biodegradation. Int. J. Environ. Res. Public Health 2015, 12, 9848–9863. [Google Scholar] [CrossRef] [PubMed]
- Azri, M.H.; Ismail, S.; Abdullah, R. Effects of Bacillus salmalaya strain 139SI inoculation on yield and nutrients uptake of oil palm. Int. J. Agric. Biol. 2018, 20, 499–506. [Google Scholar] [CrossRef]
- Baltaci, M.O.; Orak, T.; Taskin, M.; Adiguzel, A.; Ozkan, H. Enhancement of Amylase and Lipase Production from Bacillus licheniformis 016 Using Waste Chicken Feathers as Peptone Source. Waste Biomass-Valorizat. 2018, 11, 1809–1819. [Google Scholar] [CrossRef]
- Chandra, P.; Enespa; Singh, R.; Arora, P.K. Microbial lipases and their industrial applications: A comprehensive review. Microb. Cell Fact. 2020, 19, 169. [Google Scholar] [CrossRef] [PubMed]
- Collin, T.; Cunningham, R.; Asghar, M.Q.; Villa, R.; MacAdam, J.; Jefferson, B. Assessing the potential of enhanced primary clarification to manage fats, oils and grease (FOG) at wastewater treatment works. Sci. Total Environ. 2020, 728, 138415. [Google Scholar] [CrossRef] [PubMed]
- Wallace, T.; Gibbons, D.; O’Dwyer, M.; Curran, T.P. International evolution of fat, oil and grease (FOG) waste management—A review. J. Environ. Manag. 2017, 187, 424–435. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Patil, Y.; Rale, V. Biosurfactant production: Emerging trends and promising strategies. J. Appl. Microbiol. 2019, 126, 2–13. [Google Scholar] [CrossRef] [PubMed]
- Asna, S.M.Z.H.; Karmaker, M.; Sarker, U.J. Brain Heart Infusion Agar: A Surrogate of Agar Blood. Bangladesh J. Med. Microbiol. 2018, 12, 24–26. [Google Scholar] [CrossRef]
- Jira, J.; Rezek, B.; Kriha, V.; Artemenko, A.; Matolínová, I.; Skakalova, V.; Stenclova, P.; Kromka, A. Inhibition of E. coli Growth by Nanodiamond and Graphene Oxide Enhanced by Luria-Bertani Medium. Nanomaterials 2018, 8, 140. [Google Scholar] [CrossRef] [PubMed]
- Kumar, D.; Kumar, L.; Nagar, S.; Raina, C.; Parshad, R.; Gupta, V.K. Screening, isolation and production of lipase/esterase producing Bacillus sp. strain DVL2 and its potential evaluation in esterification and resolution reactions. Arch. Appl. Sci. Res. 2018, 4, 1763–1770. [Google Scholar]
- Kanimozhi, S.; Perinbam, K. Optimization of media components and growth conditions to enhance lipase production by Pseudomonas sp. Lp1. Biomed. Pharmacol. J. 2015, 3, 329–338. [Google Scholar]
- Ramnath, L.; Sithole, B.; Govinden, R. Identification of lipolytic enzymes isolated from bacteria indigenous to Eucalyptus wood species for application in the pulping industry. Biotechnol. Rep. 2017, 15, 114–124. [Google Scholar] [CrossRef] [PubMed]
- Pallavi, P.; Chandra, S.J.; Reddy, V.K.; Reddy, S.R. Optimization of cultural parameters for lipase production by Bacillus subtilis Y-IVI. Int. J. Curr. Microbiol. Appl. Sci. 2014, 3, 194–200. [Google Scholar]
- Rebello, S.; Asok, A.K.; Mundayoor, S.; Jisha, M. Surfactants: Chemistry, toxicity and remediation. In Pollutant Diseases, Remediation and Recycling; Springer International Publishing: Cham, Switzerland, 2013; pp. 277–320. [Google Scholar]
- Orsavova, J.; Misurcova, L.; Ambrozova, J.V.; Vicha, R.; Mlcek, J. Fatty Acids Composition of Vegetable Oils and Its Contribution to Dietary Energy Intake and Dependence of Cardiovascular Mortality on Dietary Intake of Fatty Acids. Int. J. Mol. Sci. 2015, 16, 12871–12890. [Google Scholar] [CrossRef] [PubMed]
- Bora, L.; Bora, M. Optimization of extracellular thermophilic highly alkaline lipase from thermophilic Bacillus sp. isolated from Hotspring of Arunachal Pradesh, India. Braz. J. Microbiol. 2012, 43, 30–42. [Google Scholar] [CrossRef] [PubMed]
- Latha, J.N.L.; Lanka, S.; Pydipalli, M. Optimization of Process Variables for Extracellular Lipase Production from Emericella nidulans NFCCI 3643 Isolated from Palm Oil Mill Effluent (POME) Dump Sites Using OFAT Method. Res. J. Microbiol. 2015, 10, 38–53. [Google Scholar] [CrossRef][Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).