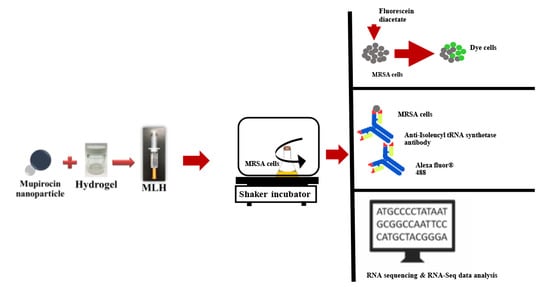
Bioactivity of Mupirocin Nanoparticle-Loaded Hydrogel against Methicillin-Resistant Staphylococcus aureus (MRSA)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Bacterial Strains
2.3. Preparation of MLH
2.4. Minimum Inhibitory Concentration (MIC) and Minimum Biocidal Concentration (MBC)
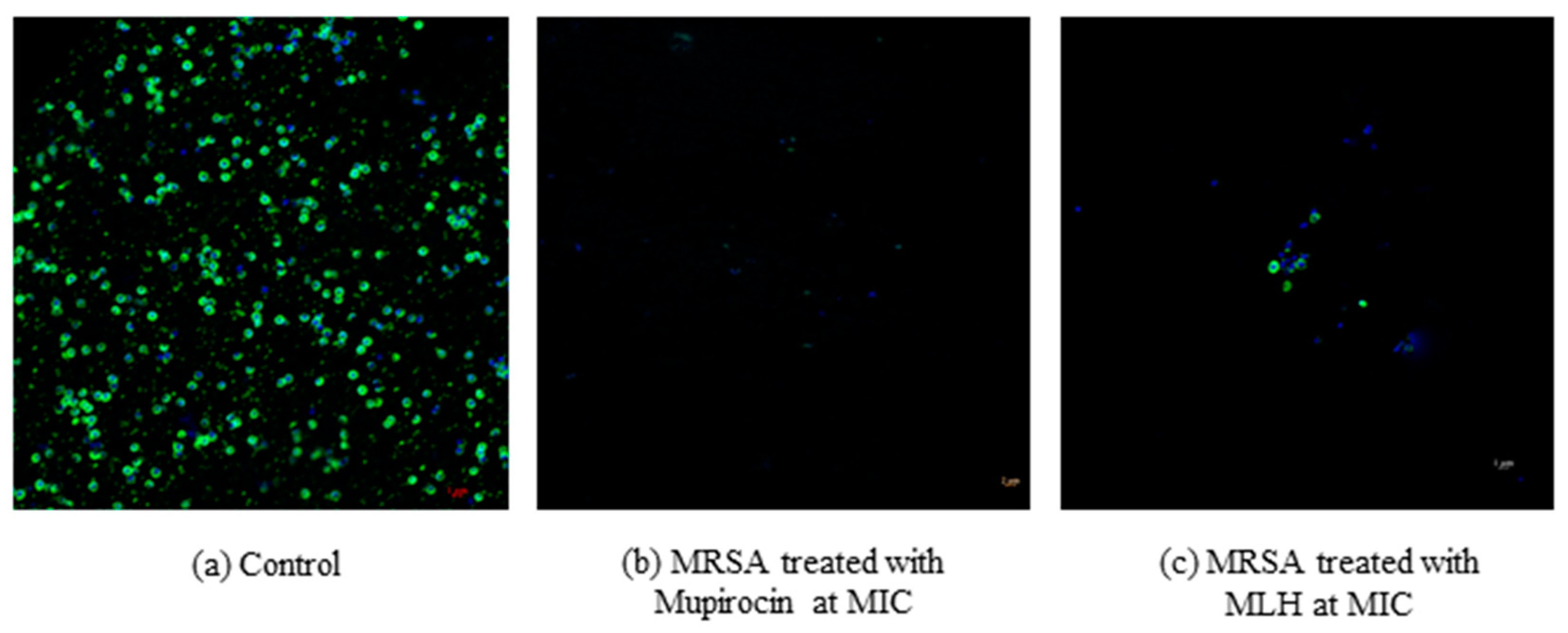
2.5. Bacterial Viability Using Flow Cytometry
2.6. Inhibition of Isoleucyl-tRNA Synthetase in MRSA by MLH
2.7. Effects of MLH on Gene Expression in MRSA
2.8. Data Availability
Statistical Analysis
3. Results
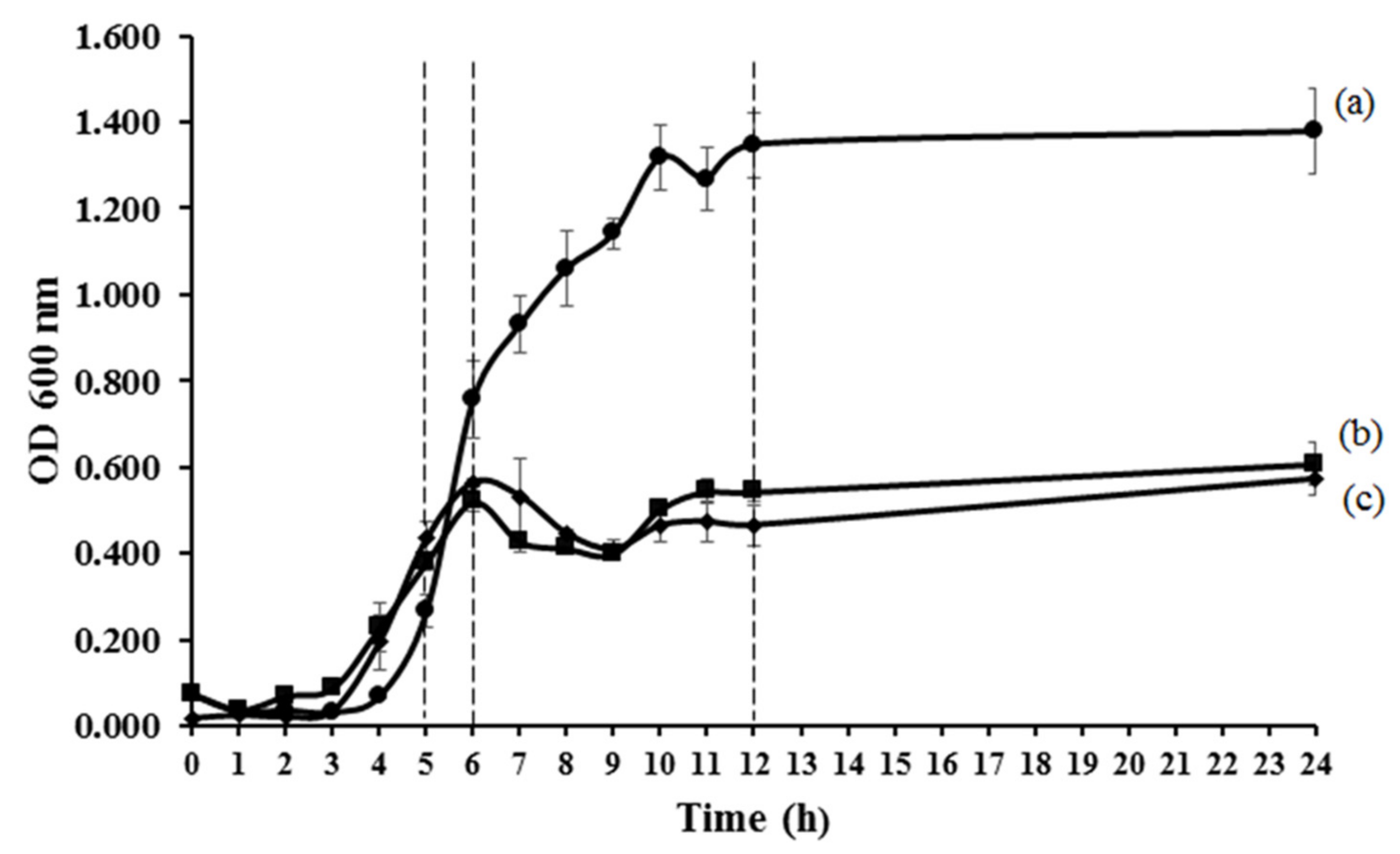
3.1. Antibacterial Activities of MLH
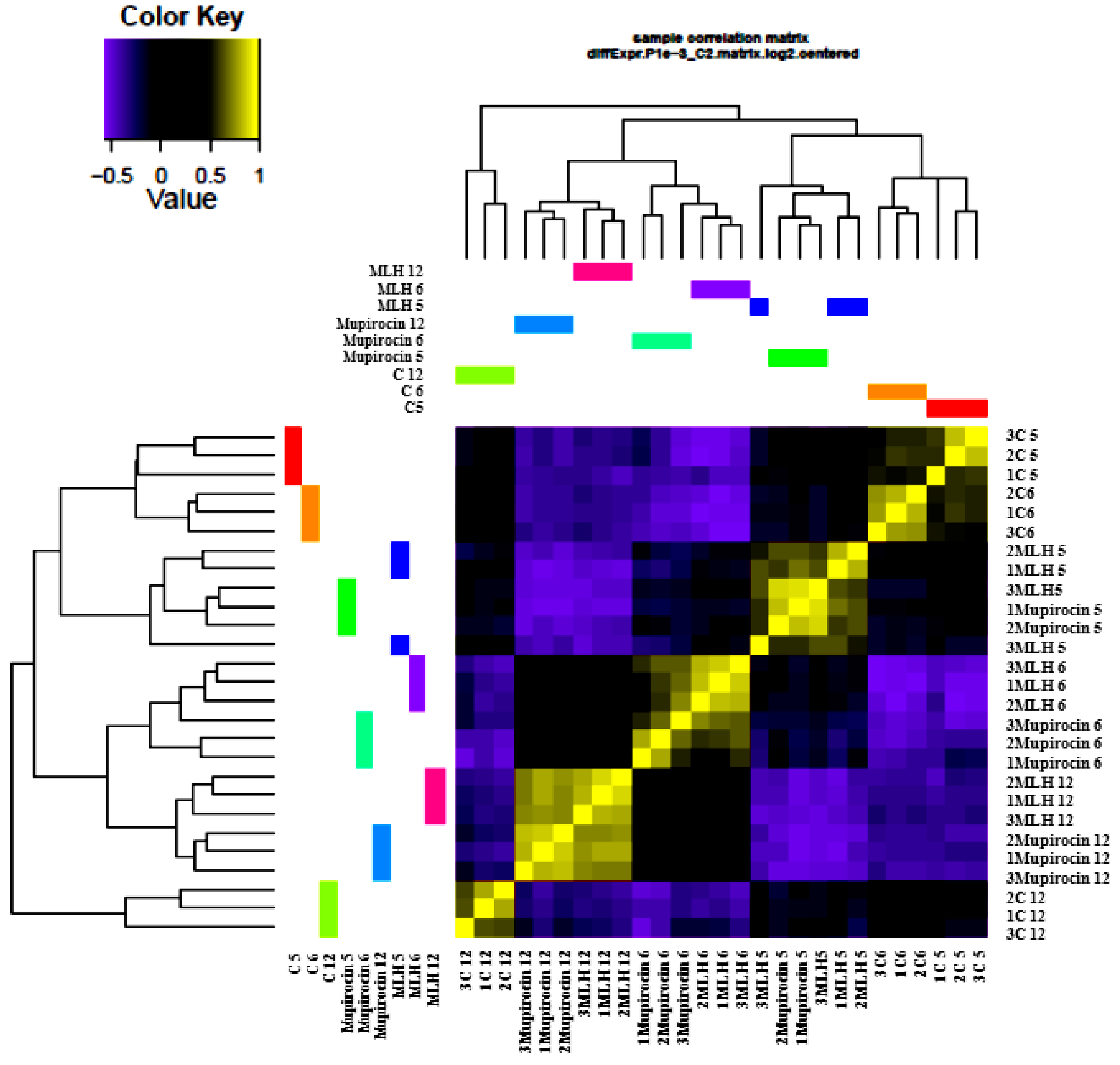
3.2. Gene Expression
3.3. Comparison of Gene Expression in MLH, Mupirocin, and Control Samples
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chong, Y.P.; Park, S.J.; Kim, H.S.; Kim, E.S.; Kim, M.N.; Park, K.H.; Kim, S.H.; Lee, S.O.; Choi, S.H.; Jeong, J.Y.; et al. Persistent Staphylococcus aureus bacteremia: A prospective analysis of risk factors, outcomes, and microbiologic and genotypic characteristics of isolates. Medecine 2013, 92, 98–108. [Google Scholar] [CrossRef] [Green Version]
- Le Moing, V.; Alla, F.; Doco-Lecompte, T.; Delahaye, F.; Piroth, L.; Chirouze, C.; Tattevin, P.; Lavigne, J.P.; Erpelding, M.L.; Hoen, B.; et al. Staphylococcus Aureus bloodstream infection and endocarditis—A prospective cohort study. PLoS ONE 2015, 10, e0127385. [Google Scholar] [CrossRef] [Green Version]
- Hassoun, A.; Linden, P.K.; Friedman, B. Incidence, Prevalence, and management of mrsa bacteremia across patient populations-a review of recent developments in mrsa management and treatment. Crit. Care 2017, 21, 211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fowler, V.G.; Boucher, H.W.; Corey, G.R.; Abrutyn, E.; Karchmer, A.W.; Rupp, M.E.; Levine, D.P.; Chambers, H.F.; Tally, F.P.; Vigliani, G.A.; et al. Daptomycin versus standard therapy for bacteremia and endocarditis caused by Staphylococcus aureus. N. Engl. J. Med. 2006, 355, 653–665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kullar, R.; Davis, S.L.; Levine, D.P.; Rybak, M.J. Impact of Vancomycin exposure on outcomes in patients with methicillin-resistant Staphylococcus aureus bacteremia: Support for consensus guidelines suggested targets. Clin. Infect. Dis. 2011, 52, 975–981. [Google Scholar] [CrossRef] [Green Version]
- Hurler, J.; Sørensen, K.K.; Fallarero, A.; Vuorela, P.; Škalko-Basnet, N. Liposomes-in-hydrogel delivery system with mupirocin: In vitro antibiofilm studies and in vivo evaluation in mice burn model. BioMed Res. Int. 2013, 2013, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Amrutiya, N.; Bajaj, A.; Madan, M. Development of microsponges for topical delivery of mupirocin. AAAPS Pharmscitech. 2009, 10, 402–409. [Google Scholar] [CrossRef] [Green Version]
- Goldmann, O.; Cern, A.; Müsken, M.; Rohde, M.; Weiss, W.; Barenholz, Y.; Medina, E. Liposomal mupirocin holds promise for systemic treatment of invasive Staphylococcus aureus infections. J. Control. Release 2019, 316, 292–301. [Google Scholar] [CrossRef]
- Ishida, M.; Nambu, N.; Nagai, T. Ointment-type oral mucosal dosage form of carbopol containing prednisolone for treatment of aphtha. Chem. Pharm. Bull. 1983, 31, 1010–1014. [Google Scholar] [CrossRef] [Green Version]
- Cern, A.; Nativ-Roth, E.; Goldblum, A.; Barenholz, Y. Effect of solubilizing agents on mupirocin loading into and release from pegylated nanoliposomes. J. Pharm. Sci. 2014, 103, 2131–2138. [Google Scholar] [CrossRef]
- Torchilin, V.; Weissig, V. Liposomes: A Practical Approach; R.R.C. New, Ed.; Oxford University Press: Oxford, UK, 2003. [Google Scholar]
- Kamlungmak, S.; Rugmai, S.; Tinpun, K.; Nakpheng, T.; Srichana, T. Phase Behavior, in vitro drug release, and antibacterial activity of thermoresponsive poloxamer– polyvinyl alcohol hydrogel-loaded mupirocin nanoparticles. J. Appl. Polym. Sci. 2020, 137, 49325. [Google Scholar] [CrossRef]
- Lundgren, B. Fluorescein diacetate as a stain of metabolically active bacteria in soil. Oikos 1981, 36, 17–22. [Google Scholar] [CrossRef]
- Boyd, V.; Cholewa, O.M.; Papas, K.K. Limitations in the use of fluorescein diacetate/propidium iodide (fda/pi) and cell permeable nucleic acid stains for viability measurements of isolated islets of langerhans. Curr. Trends Biotechnol. Pharm. 2008, 2, 66–84. [Google Scholar]
- Simms, D.; Cizdziel, P.; Chomczynski, P. TRIzol: A new reagent for optimal single-step isolation of RNA. Focus 1993, 15, 99–102. [Google Scholar] [CrossRef]
- O’Neill, M.; McPartlin, J.; Arthure, K.; Riedel, S.; McMillan, N.D. Comparison of the TLDA with the nanodrop and the reference qubit system. J. Phys. Conf. Ser. 2011, 307, 102047. [Google Scholar] [CrossRef]
- Pertea, M.; Kim, D.; Pertea, G.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with hisat, stringtie, and ballgown. Nat. Protoc. 2019, 176, 139–148. [Google Scholar] [CrossRef]
- Zwietering, M.H.; Jongenburger, I.; Rombouts, F.M.; Riet, K.V. Modeling of the bacterial growth curve. Appl. Environ. Microbiol. 1990, 56, 1875–1881. [Google Scholar] [CrossRef] [Green Version]
- Geiger, T.; Goerke, C.; Fritz, M.; Schäfer, T.; Ohlsen, K.; Liebeke, M.; Lalk, M.; Wolz, C. Role of the (p)PpGpp Synthase RSH, a RelA/SpoT Homolog, in stringent response and virulence of Staphylococcus aureus. Infect. Immun. 2010, 78, 1873–1883. [Google Scholar] [CrossRef] [Green Version]
- Spann, C.T.; Taylor, S.C.; Weinberg, J.M. Topical antimicrobial agents in dermatology. Clin. Dermatol. 2003, 21, 70–77. [Google Scholar] [CrossRef]
- Tenover, F.C. Mechanisms of antimicrobial resistance in bacteria. Am. J. Infect. Control 2006, 34, S3–S10. [Google Scholar] [CrossRef]
- Crosse, A.M.; Greenway, D.L.A.; England, R.R. Accumulation of PpGpp and PpGp in Staphylococcus aureus 8325-4 Following Nutrient Starvation. Lett. Appl. Microbiol. 2000, 31, 332–337. [Google Scholar] [CrossRef] [Green Version]
- Sonenshein, A.L. CodY, a Global regulator of stationary phase and virulence in gram-positive bacteria. Curr. Opin. Microbiol. 2005, 8, 203–207. [Google Scholar] [CrossRef]
- Hughes, J.; Mellows, G. On the mode of action of pseudomonic acid: Inhibition of protein synthesis in Staphylococcus aureus. J. Antibiot. 1978, 31, 330–335. [Google Scholar] [CrossRef] [Green Version]
- Alhoufie, S.T.S.; Foster, H.A. Effects of sub-lethal concentrations of mupirocin on global transcription in Staphylococcus aureus 8325-4 and a model for the escape from inhibition. J. Med. Microbiol. 2016, 65, 858–866. [Google Scholar] [CrossRef]


| Antibacterial Activities | Sample | ||||
|---|---|---|---|---|---|
| Hydrogel | MLH | Mupirocin Ointment | Vancomycin | Mupirocin (Pure Drug) | |
| MIC | - | 0.31 | 0.61 | 1.56 | 0.31 |
| MBC | - | 2.44 | 9.76 | 1.56 | 2.44 |
| Treatment | % Down-Regulated | % Up-Regulated | ||||
|---|---|---|---|---|---|---|
| 5 h | 6 h | 12 h | 5 h | 6 h | 12 h | |
| MLH | 50.5 | 52.6 | 42.2 | 49.5 | 47.4 | 57.8 |
| Mupirocin | 52.0 | 56.0 | 40.7 | 48.0 | 44.0 | 59.3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kamlungmak, S.; Wonglapsuwan, M.; Surachat, K.; Deachamag, P.; Srichana, T. Bioactivity of Mupirocin Nanoparticle-Loaded Hydrogel against Methicillin-Resistant Staphylococcus aureus (MRSA). Appl. Microbiol. 2022, 2, 260-271. https://doi.org/10.3390/applmicrobiol2010019
Kamlungmak S, Wonglapsuwan M, Surachat K, Deachamag P, Srichana T. Bioactivity of Mupirocin Nanoparticle-Loaded Hydrogel against Methicillin-Resistant Staphylococcus aureus (MRSA). Applied Microbiology. 2022; 2(1):260-271. https://doi.org/10.3390/applmicrobiol2010019
Chicago/Turabian StyleKamlungmak, Sukanjana, Monwadee Wonglapsuwan, Komwit Surachat, Panchalika Deachamag, and Teerapol Srichana. 2022. "Bioactivity of Mupirocin Nanoparticle-Loaded Hydrogel against Methicillin-Resistant Staphylococcus aureus (MRSA)" Applied Microbiology 2, no. 1: 260-271. https://doi.org/10.3390/applmicrobiol2010019
APA StyleKamlungmak, S., Wonglapsuwan, M., Surachat, K., Deachamag, P., & Srichana, T. (2022). Bioactivity of Mupirocin Nanoparticle-Loaded Hydrogel against Methicillin-Resistant Staphylococcus aureus (MRSA). Applied Microbiology, 2(1), 260-271. https://doi.org/10.3390/applmicrobiol2010019