Antimicrobial Activity of Fungal Endophytes Associated with Peperomia argyreia (Piperaceae)
Abstract
1. Introduction
2. Materials and Methods
3. Results
3.1. Isolation and Molecular Identification of P. argyreia Fungal Endophytes
3.2. MALDI ToF MS of the Fungal Isolates
3.3. Nuclear Magnetic Resonance
3.4. Antimicrobial Activity
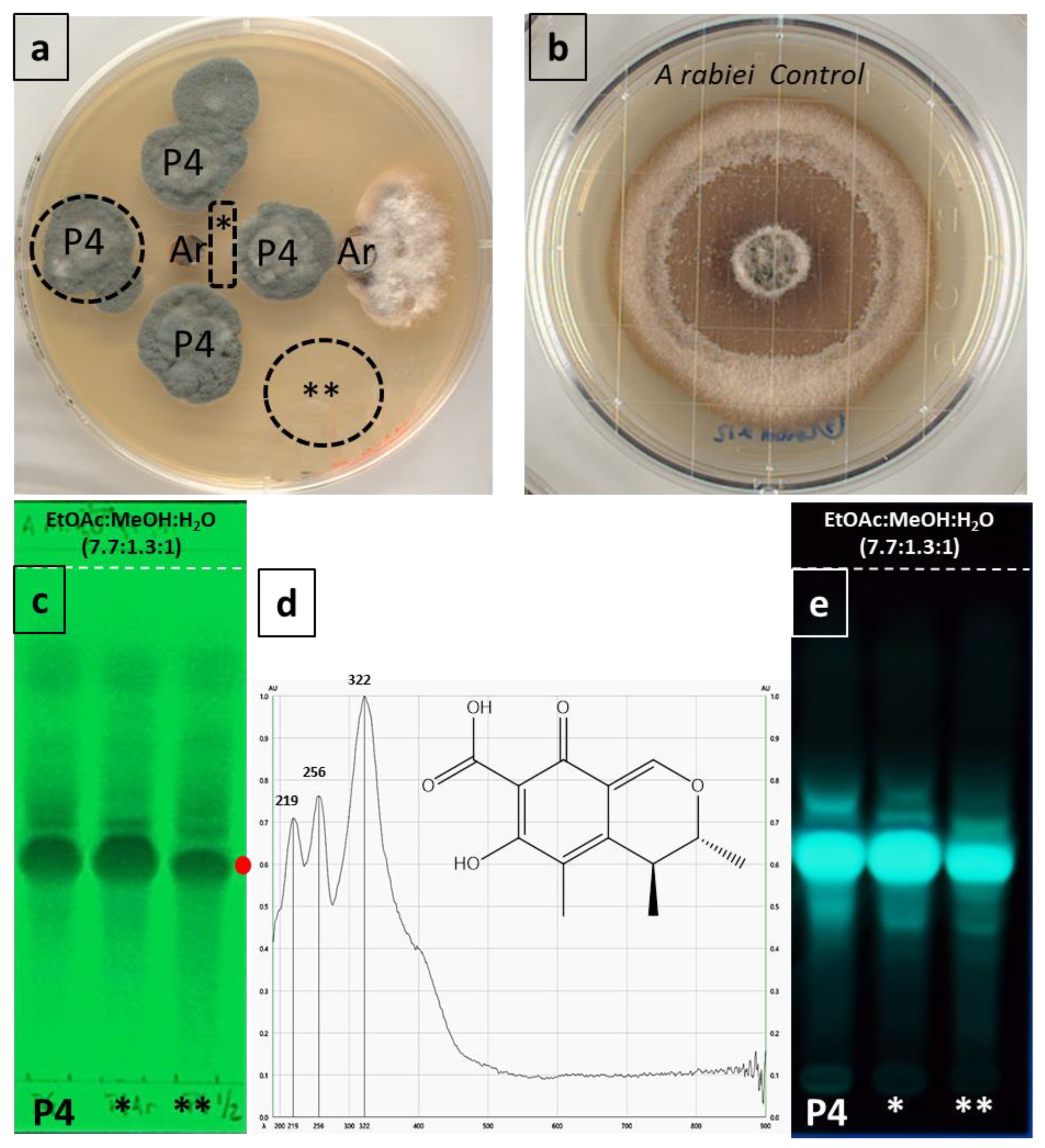
3.5. Inhibitory Effect of P. westlingii P4 on A. rabiei
3.5.1. Chemical Profile and Antifungal Activity
3.5.2. Evaluation of the Interaction of P. westlingii P4 with A. rabiei
3.5.3. Determination of Volatile Compounds in the Inhibition of A. rabiei
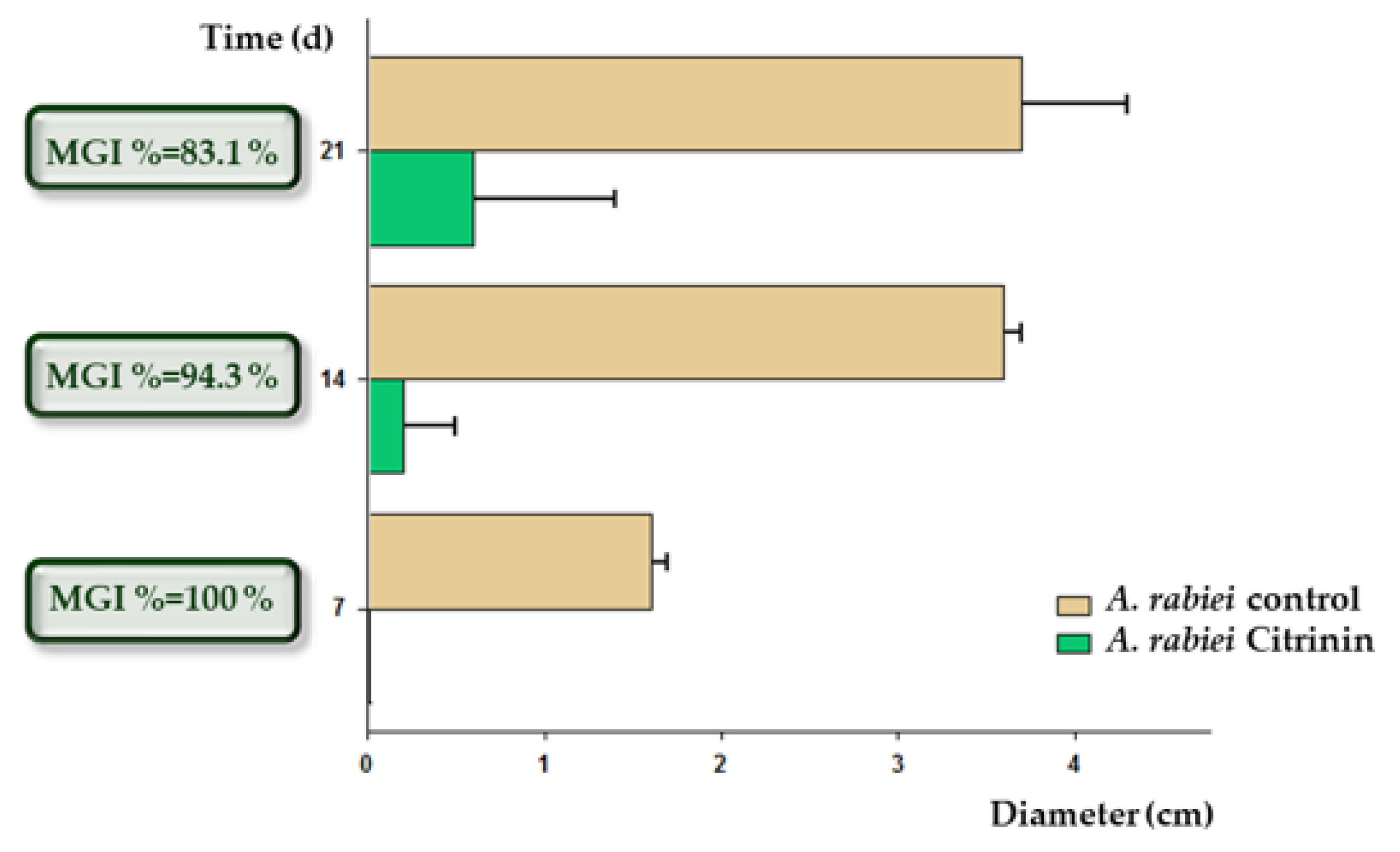
3.5.4. Inhibition of the Mycelial Growth of A. rabiei
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Alam, B.; Lǐ, J.; Gě, Q.; Khan, M.A.; Gōng, J.; Mehmood, S.; Yuán, Y.; Gǒng, W. Endophytic Fungi: From Symbiosis to Secondary Metabolite Communications or Vice Versa? Front. Plant Sci. 2021, 12, 791033. [Google Scholar] [CrossRef]
- Wanke, S.; Jaramillo, A.; Borsch, T.; Samain, M.S.; Quandt, D.; Neinhuis, C. Evolution of Piperales-matK gene and trnK intron sequence data reveal lineage specific resolution contrast. Mol. Phylogenet Evol. 2007, 42, 477–497. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.F.; Stevens, A.C.; Tepe, E.J.; Davidson, C.L. Placing the origin of two species-rich genera in the late cretaceous with later species divergence in the tertiary: A phylogenetic, biogeographic and molecular dating analysis of Piper and Peperomia (Piperaceae). Plant Syst. Evol. 2008, 275, 9–30. [Google Scholar] [CrossRef]
- Valarezo, E.; Herrera-García, M.; Astudillo-Dávila, P.; Rosales-Demera, I.; Jaramillo Fierro, X.; Cartuche, L.; Meneses, M.A.; Morocho, V. Study of the Chemical Composition and Biological Activity of the Essential Oil from Congona (Peperomia inaequalifolia Ruiz and Pav.). Plants 2023, 12, 1504. [Google Scholar] [CrossRef]
- Valero Gutierrez, Y.; Yamaguchi, L.F.; Moraes, M.M.; Jeffrey, C.S.; Kato, M.J. Natural products from Peperomia: Occurrence, biogenesis and bioactivity. Phytochem. Rev. 2016, 15, 1009–1033. [Google Scholar] [CrossRef]
- Saepudin, S.; Susilawati, Y. Alpha-glucosidase inhibitor activities and phytochemicals screening of the Peperomia genus cultivated in Indonesia. Int. J. Appl. Pharm. 2022, 14, 117–122. [Google Scholar] [CrossRef]
- Available online: https://www.bcr.com.ar/es/mercados/investigacion-y-desarrollo/informativo-semanal/noticias-informativo-semanal/el-agro-aporto (accessed on 27 December 2023).
- Pandit, M.A.; Kumar, J.; Gulati, S.; Bhandari, N.; Mehta, P.; Katyal, R.; Rawat, C.D.; Mishra, V.; Kaur, J. Major biological control strategies for plant pathogens. Pathogens. 2022, 11, 273. [Google Scholar] [CrossRef] [PubMed]
- Bahr, L.; Castelli, M.V.; Barolo, M.I.; Ruiz Mostacero, N.; Tosello, M.E.; López, S.N. Ascochyta blight: Isolation, characterization, and development of a rapid method to detect inhibitors of the chickpea fungal pathogen Ascochyta rabiei. Fungal Biol. 2016, 120, 424–432. [Google Scholar] [CrossRef] [PubMed]
- Ruiz Mostacero, N.; Castelli, M.V.; Barolo, M.I.; Amigot, S.L.; Fulgueira, C.L.; López, S.N. Fungal endophytes in Peperomia obtusifolia and their potential as inhibitors of chickpea fungal pathogens. World J. Microbiol. Biotechnol. 2021, 37, 14. [Google Scholar] [CrossRef]
- Available online: https://fcagr.unr.edu.ar/herbario/ (accessed on 27 December 2023).
- Schulz, B.; Boyle, C.; Draeger, S.; Rommert, A.K.; Krohn, K. Endophytic fungi: A source of novel biologically active secondary metabolites. Mycol. Res. 2002, 106, 996–1004. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplifcation and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- Villa-Carvajal, M.R.; Coque, J.J.; Álvarez-Rodríguez, M.L.; Uruburu, F.; Belloch, C. Polyphasic identification of yeasts isolated from bark of cork oak during the manufacturing process of cork stoppers. FEMS Yeast Res. 2004, 4, 745–750. [Google Scholar] [CrossRef] [PubMed]
- Glass, N.L.; Donaldson, G.C. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomicetes. Appl. Environ. Microbiol. 1995, 61, 1323–1330. [Google Scholar] [CrossRef] [PubMed]
- Scholtz, I.; Korsten, L. Profile of Penicillium species in the pear supply chain. Plant Pathol. 2016, 65, 1126–1132. [Google Scholar] [CrossRef]
- Raja, H.A.; Miller, A.N.; Pearce, C.J.; Oberlies, N.H. Fungal identification using molecular tools: A primer for the natural products research community. J. Nat. Prod. 2017, 80, 756–770. [Google Scholar] [CrossRef]
- Nilsson, R.H.; Anslan, S.; Bahram, M.; Wurzbacher, C.; Baldrian, P.; Tedersoo, L. Mycobiome diversity: High-throughput sequencing and identification of fungi. Nat. Rev. Microbiol. 2019, 17, 95–109. [Google Scholar] [CrossRef] [PubMed]
- Reich, E.; Schibli, A. High Performance Thin-Layer Chromatography for the Analysis of Medicinal Plants; Thieme Medical Publishers Inc.: New York, NY, USA, 2007; p. 237. [Google Scholar]
- Barolo, M.I.; Castelli, M.V.; López, S.N. Antimicrobial properties and biotransforming ability of fungal endophytes from Ficus carica L. (Moraceae). Mycology 2023, 14, 108–132. [Google Scholar] [CrossRef] [PubMed]
- Soylu, E.M.; Soylu, S.; Kurt, S. Antimicrobial activities of the essential oils of various plants against tomato late blight disease agent Phytophthora infestans. Mycopathologia 2006, 161, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Fleitas Centurión, A.; Grabowski Ocampos, C.J. Biological control of the fungi complex that cause leaf spot on sweet corn (Zea mays var. saccharata) with beneficial bacteria. Investig. Agrar. 2014, 16, 83–92. [Google Scholar]
- Hetherington, A.C.; Raistrick, H. Studies in biochemistry of microorganism XIV. On the production and chemical constitution of a new yellow colouring matter, citrinin, produced from glucose by Penicillium citrinum Thom. Philos. Trans. R. Soc. Lond. Ser. B—Biol. Sci. 1931, 220, 269–295. [Google Scholar] [CrossRef]
- He, Y.; Cox, R.J. The molecular steps of citrinin biosynthesis in fungi. Chem. Sci. 2016, 7, 2119–2127. [Google Scholar] [CrossRef]
- Ei-Banna, A.A.; Pitt, J.I.; Leistner, L. Production of mycotoxins by Penicillium species. Syst. Appl. Microbiol. 1987, 10, 42–46. [Google Scholar] [CrossRef]
- Kurata, H. Mycotoxins and mycotoxicoses. In Microbial Toxins in Foods and Feeds; Pohland, A.E., Dowell, V.R., Richards, J.L., Eds.; Plenum Press: New York, NY, USA, 1990; pp. 249–259. [Google Scholar]
- Blanc, P.J.; Loret, M.O.; Goma, G. Production of citrinin by various species of Monascus. Biotechnol. Lett. 1995, 17, 291. [Google Scholar] [CrossRef]
- Blanc, P.J.; Laussac, J.P.; Le, J.B.; Le, B.P.; Loret, M.O.; Pareilleux, A.; Prome, D.; Prome, J.C.; Santerre, A.L.; Goma, G. Characterisation of monascidin A from Monascus as citrinin. Int. J. Food Microbiol. 1995, 27, 201–213. [Google Scholar] [CrossRef] [PubMed]
- Samsudin, N.I.; Abdullah, N. A preliminary survey on the occurrence of mycotoxigenic fungi and mycotoxins contaminating red rice at consumer level in Selangor, Malaysia. Mycotoxin Res. 2013, 29, 89–96. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.; Cartwright, N.; Robertson, A.; Whalley, W.B. Structure of Citrinin. Nature 1948, 162, 72–73. [Google Scholar] [CrossRef] [PubMed]
- Hill, R.K.; Gardella, L.A. The Absolute Configuration of Citrinin 1. JOC 1964, 29, 766–767. [Google Scholar] [CrossRef]
- Krogh, P.; Hasselager, E.; Friis, P. Studies on fungal nephrotoxicity. APMIS 1970, 78, 401–413. [Google Scholar] [CrossRef]
- Marcon, E.L. Fungos Endofíticos de Piper Peltatum e Peperomia Pellucida: Caracterização Metabólitos Secundários e Atividades Biológicas. Doctoral Thesis, Federal University of Amazonas, Manaus, Brazil, 2013. Available online: http://tede.ufam.edu.br/handle/tede/4442 (accessed on 22 January 2024).
- Gutiérrez, V.; Yasmin, L. Molecular Phylogeny and Secondary Metabolism from Peperomia Species. Doctoral Thesis, Instituto de Química, Universidade de Sao Paulo, São Paulo, Brazil, 2015. [Google Scholar] [CrossRef]
- Chlebicki, A. Some endophytes of Juncus trifidus from Tatra Mts. in Poland. Acta Mycol. 2009, 44, 11–17. [Google Scholar] [CrossRef]
- Panda, A.; Ghosh, A.K.; Mirdha, B.R.; Xess, I.; Paul, S.; Samantaray, J.C.; Srinivasan, A.; Khalil, S.; Rastogi, N.; Dabas, Y. MALDI-TOF mass spectrometry for rapid identification of clinical fungal isolates based on ribosomal protein biomarkers. J. Microbiol. Methods 2015, 109, 93–105. [Google Scholar] [CrossRef]
- Newman, D.J.; Cragg, G.M. Natural Products as Sources of New Drugs over the Nearly Four Decades from 01/1981 to 09/2019. J. Nat. Prod. 2020, 83, 770–803. [Google Scholar] [CrossRef]
- Sadrati, N.; Zerroug, A.; Demirel, R.; Harzallah, D. Anti-multidrug-resistant Staphylococcus aureus and anti-dermatophyte activities of secondary metabolites of the endophytic fungus Penicillium brevicompactum ANT13 associated with the Algerian endemic plant Abies numidica. Arch. Microbiol. 2023, 205, 110. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.W.; Bai, F.Y.; Bensch, K.; Meijer, M.; Sun, B.D.; Han, Y.F.; Crous, P.W.; Samson, R.A.; Yang, F.Y.; Houbraken, J. Phylogenetic re-evaluation of Thielavia with the introduction of a new family Podosporaceae. Stud. Mycol. 2019, 93, 155–252. [Google Scholar] [CrossRef] [PubMed]
- Fleming, A. On the antibacterial action of cultures of a Penicillium, with special reference to their use in the isolation of B. influenzae. Br. J. Exp. Pathol. 1929, 10, 226–236. [Google Scholar] [CrossRef]
- Chain, E.; Florey, H.; Gardner, A.; Heatley, N.G.; Jennings, M.A.; Orr-Erwing, J.; Sanders, A.G. Penicillin as a chemotherapeutic agent. Lancet 1940, 236, 226–228. [Google Scholar] [CrossRef]
- Gao, N.; Shang, Z.C.; Yu, P.; Luo, J.; Jian, K.L.; Kong, L.Y.; Yang, M.H. Alkaloids from the endophytic fungus Penicillium brefeldianum and their cytotoxic activities. Chin. Chem. Lett. 2017, 28, 1194–1199. [Google Scholar] [CrossRef]
- Yang, M.H.; Li, T.X.; Wang, Y.; Liu, R.H.; Luo, J.; Kong, L.Y. Antimicrobial metabolites from the plant edophytic fungus Penicillium sp. Fitoterapia 2017, 116, 72–76. [Google Scholar] [CrossRef] [PubMed]
- Marinho, A.M.; Marinho, P.S.; Filho, E.R. Esteroides produzidos por Penicillium herquei, um fungo endofítico isolado dos frutos de Melia azedarach (Meliaceae). Quim. Nova 2009, 32, 1710–1712. [Google Scholar] [CrossRef][Green Version]
- Koul, M.; Meena, S.; Kumar, A.; Sharma, P.R.; Singamaneni, V.; Hassan, S.R.; Hamid, A.; Chaubey, A.; Prabhakar, A.; Gupta, P.; et al. Secondary metabolites from endophytic fungus Penicillium pinophilum induce ROS-mediated apoptosis through mitochondrial pathway in pancreatic cancer cells. Planta Med. 2016, 82, 344–355. [Google Scholar] [CrossRef] [PubMed]
- Challinor, V.L.; Bode, H.B. Bioactive natural products from novel microbial sources. Ann. N. Y. Acad. Sci. 2015, 1354, 82–97. [Google Scholar] [CrossRef]
- Chen, M.; Shen, N.X.; Chen, Z.Q.; Zhang, F.M.; Chen, Y. Penicilones A-D, Anti-MRSA Azaphilones from the Marine-Derived Fungus Penicillium janthinellum HK 1-6. J. Nat. Prod. 2017, 80, 1081–1086. [Google Scholar] [CrossRef]
- Nicoletti, R.; Gresa, L.; Pilar, M.; Manzo, E.; Carella, A.; Ciavatta, M.L. Production and fungitoxic activity of Sch 642305, a secondary metabolite of Penicillium canescens. Mycopathologia 2007, 163, 295–301. [Google Scholar] [CrossRef] [PubMed]
- Flajs, D.; Peraica, M. Toxicological properties of citrinin. Arh. Hig. Rada Toksikol. 2009, 60, 457–464. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.H.; Yu, F.Y.; Wang, L.T.; Lin, Y.S.; Liu, B.H. Activation of ERK and JNK signaling pathways by mycotoxin citrinin in human cells. Toxicol. Appl. Pharmacol. 2009, 237, 281–287. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, Y.; Iguchi, H.; Kamisuki, S.; Sugawara, F.; Furuichi, T.; Shinoda, Y. Low doses of the mycotoxin citrinin protect cortical neurons against glutamate-induced. J. Toxicol. Sci. 2016, 41, 311–319. [Google Scholar] [CrossRef] [PubMed]
- Hiroyuki, H.; Kensuke, H.; Kozo, S. Mechanism of antifungal action of citrinin. Agric. Biol. Chem. 1987, 51, 1373–1378. [Google Scholar] [CrossRef]
- Mazumder, P.M.; Mazumder, R.; Mazumder, A.; Sasmal, D.S. Antimicrobial activity of the mycotoxin citrinin obtained from the fungus Penicillium citrinum. Anc. Sci. Life 2002, XXI, 191–197. [Google Scholar]
- Wakana, D.; Hosoe, T.; Itabashi, T.; Okada, K.; de Campos Takaki, G.M.; Yaguchi, T.; Fukushima, K.; Kawai, K.I. New citrinin derivatives isolated from Penicillium citrinum. J. Nat. Med. 2006, 60, 279–284. [Google Scholar] [CrossRef]
- Xu, B.J.; Jia, X.Q.; Gu, L.J.; Sung, C.K. Review on the qualitative and quantitative analysis of the mycotoxin citrinin. Food Control 2006, 17, 271–285. [Google Scholar] [CrossRef]



| Strain | Isolate ID (GenBank Accession) | GenBank Sequence with Major % of Identity (GenBank Accession) | Match Identity (%) | Query Cover (%) | Primers |
|---|---|---|---|---|---|
| P1 | Arcopilus sp. (MH165192.1) | Arcopilus globulus (NG_070473.1) | 95 | 100 | NL1-NL4 |
| P2 | Sporothrix stylites (MH165193.1) | Sporothrix stylites (MH874594.1) | 98 | 100 | NL1-NL4 |
| P3 | Cladosporium devikae (MH165229.1) | Cladosporium devikae (MZ303808.1) | 100 | 100 | ITS1-ITS4 |
| P4 | Penicillium sp. | P. miczynskii (NR_077156.1) P. aurantiacobrunneum (NR_121509.1) CBS 126228, P. quebencense (NR_121507.1) CBS 101623, P. ubiquetum (KX011020.1), P. neomiczynskii (KP714288.1), P. cairnsense (NR_121508.1) CBS 124325, P. westlingii (JN617668.1) CBS 124313, P. decaturense (HM469399.1) | 91 | 97 | ITS1-ITS4 |
| Penicillium westlingii (MH165230.1) | Penicillium westlingii (JN606717.1) CBS 127008 | 99 | 90 | Bt2a-Bt2b | |
| P5 | Chaetomium cupreum (MH165194.1) | Chaetomium cupreum (KJ439108.1) | 99 | 99 | NL1-NL4 |
| P6 | Trichoderma koningiopsis (MH165231.1) | Trichoderma koningiopsis (NR_131281.1) | 100 | 100 | ITS1-ITS4 |
| P7 | Trichoderma hamatum (MH165232.1) | Trichoderma hamatum (KT827285.1) | 100 | 99 | ITS1-ITS4 |
| P8 | Trichoderma koningiopsis (MH165233.1) | Trichoderma koningiopsis (NR_131281.1) | 100 | 100 | ITS1-ITS4 |
| P9 | Thermothielavioides sp. (MH165195.1) | Thermothielavioides maryleeae (OR731504.1) | 92 | 93 | NL1-NL4 |
| P10 | Thermothielavioides maryleeae (MH165196.1) | Thermothielavioides maryleeae (OR731504.1) | 100 | 98 | NL1-NL4 |
| P11 | Thermothielavioides terrestris (MH165197.1) | Thermothielavioides terrestris (OR731504.1) | 100 | 98 | NL1-NL4 |
| P12 | Thermothielavioides maryleeae (MH165198.1) | Thermothielavioides maryleeae (OR731504.1) | 98 | 100 | NL1-NL4 |
| P13 | Thermothielavioides sp. (MH165199.1) | Thermothielavioides terrestris (MK926837.1) | 91 | 99 | NL1-NL4 |
| P14 | Thermothielavioides maryleeae (MH165200.1) | Thermothielavioides maryleeae (OR731504.1) | 97 | 100 | NL1-NL4 |
| P15 | Thermothielavioides maryleeae (MH165201.1) | Thermothielavioides maryleeae (OR731504.1) | 98 | 100 | NL1-NL4 |
| P16 | Thermothielavioides sp. (MH165202.1) | Thermothielavioides maryleeae (OR731504.1) | 86 | 99 | NL1-NL4 |
| P17 | Thermothielavioides sp. (MH165203.1) | Thermothielavioides terrestris (MK926837.1) | 93 | 99 | NL1-NL4 |
| P18 | Thermothielavioides maryleeae (MH165204.1) | Thermothielavioides maryleeae (OR731504.1) | 98 | 100 | NL1-NL4 |
| P19 | Plectosphaerella cucumerina (MH165234.1) | Plectosphaerella cucumerina (LR026809.1) | 100 | 100 | ITS1-ITS4 |
| P20 | Cyphellophora sp. (MH165235.1) | Cyphellophora goniomatis (NR_166332.1) | 91 | 84 | ITS1-ITS4 |
| P21 | Thermothielavioides maryleeae (MH165205.1) | Thermothielavioides maryleeae (OR731504.1) | 98 | 100 | NL1-NL4 |
| P22 | Thermothielavioides maryleeae (MH165206.1) | Thermothielavioides maryleeae (OR731504.1) | 98 | 100 | NL1-NL4 |
| P23 | Thermothielavioides maryleeae (MH165207.1) | Thermothielavioides maryleeae (OR731504.1) | 97 | 100 | NL1-NL4 |
| P24 | Thermothielavioides maryleeae (MH165208.1) | Thermothielavioides maryleeae (OR731504.1) | 98 | 100 | NL1-NL4 |
| P25 | Thermothielavioides sp. (MH165209.1) | Thermothielavioides maryleeae (OR731504.1) | 95 | 100 | NL1-NL4 |
| P26 | Thermothielavioides sp. (MH165210.1) | Thermothielavioides maryleeae (OR731504.1) | 95 | 99 | NL1-NL4 |
| P27 | Alboefibula sp. (MH165236.1) | Alboefibula bambusicola (NR_175177.1) | 93 | 100 | ITS1-ITS4 |
| P28 | Thermothielavioides sp. (MH165211.1) | Thermothielavioides maryleeae (OR731504.1) | 92 | 99 | NL1-NL4 |
| P29 | Thermothielavioides maryleeae (MH165212.1) | Thermothielavioides maryleeae (OR731504.1) | 98 | 100 | NL1-NL4 |
| P30 | Thermothielavioides maryleeae (MH165213.1) | Thermothielavioides maryleeae (OR731504.1) | 98 | 100 | NL1-NL4 |
| P31 | Cyphellophora sp. (MH165237.1) | Cyphellophora goniomatis (NR_166332.1) | 91 | 81 | ITS1-ITS4 |
| Microorganism Tested | |||
|---|---|---|---|
| E. coli ATCC 25922 | S. aureus ATCC 25923 | A. rabiei AR2 | |
| P1—Arcopilus sp. | 100 | 100 | -* |
| P2—Sporothrix stylites | - | 100 | - |
| P4—Penicillium westlingii | - | 25 | 25 |
| P5—Chaetomium cupreum | 25 | 25 | - |
| P9—Thermothielavioides sp. | - | 25 | - |
| P10—Thermothielavioides maryleeae | - | 50 | - |
| P11—Thermothielavioides terrestris | - | 100 | - |
| P12—Thermothielavioides maryleeae | - | 25 | - |
| P13—Thermothielavioides sp. | - | 25 | - |
| P14—Thermothielavioides maryleeae | - | 25 | - |
| P15—Thermothielavioides maryleeae | - | 100 | - |
| P17—Thermothielavioides sp. | - | 100 | - |
| P18—Thermothielavioides maryleeae | - | 100 | - |
| P21—Thermothielavioides maryleeae | - | 100 | - |
| P22—Thermothielavioides maryleeae | - | 25 | - |
| P23—Thermothielavioides maryleeae | - | 25 | - |
| P24—Thermothielavioides maryleeae | - | 100 | - |
| P25—Thermothielavioides sp. | - | 25 | - |
| P26—Thermothielavioides sp. | - | 100 | - |
| P28—Thermothielavioides sp. | - | 100 | - |
| P29—Thermothielavioides maryleeae | - | 25 | - |
| P30—Thermothielavioides maryleeae | - | 100 | - |
| Chlorotalonyl | - | - | 0.5 |
| Vancomycin | 0.3 | 0.3 | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barolo, M.I.; Castelli, M.V.; López, S.N. Antimicrobial Activity of Fungal Endophytes Associated with Peperomia argyreia (Piperaceae). Appl. Microbiol. 2024, 4, 753-770. https://doi.org/10.3390/applmicrobiol4020052
Barolo MI, Castelli MV, López SN. Antimicrobial Activity of Fungal Endophytes Associated with Peperomia argyreia (Piperaceae). Applied Microbiology. 2024; 4(2):753-770. https://doi.org/10.3390/applmicrobiol4020052
Chicago/Turabian StyleBarolo, Melisa Isabel, María Victoria Castelli, and Silvia Noelí López. 2024. "Antimicrobial Activity of Fungal Endophytes Associated with Peperomia argyreia (Piperaceae)" Applied Microbiology 4, no. 2: 753-770. https://doi.org/10.3390/applmicrobiol4020052
APA StyleBarolo, M. I., Castelli, M. V., & López, S. N. (2024). Antimicrobial Activity of Fungal Endophytes Associated with Peperomia argyreia (Piperaceae). Applied Microbiology, 4(2), 753-770. https://doi.org/10.3390/applmicrobiol4020052






