Addition of Chicken Litter Compost Changes Bacteriobiome in Fallow Soil
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Site and Conditions
2.2. Experimental Setup
2.3. Soil Sampling and Chemical Analyses
2.4. DNA Extraction, Amplification and Sequencing
2.5. Bioinformatic Analysis
2.6. Statistical Analyses
3. Results
3.1. General Taxonomic Diversity in the CLC Nacteriobiome
3.2. General Taxonomic Diversity in the Soil Bacteriobiome
3.3. Bacterial Taxonomic Diversity as Related to the CLC Addition in Soil
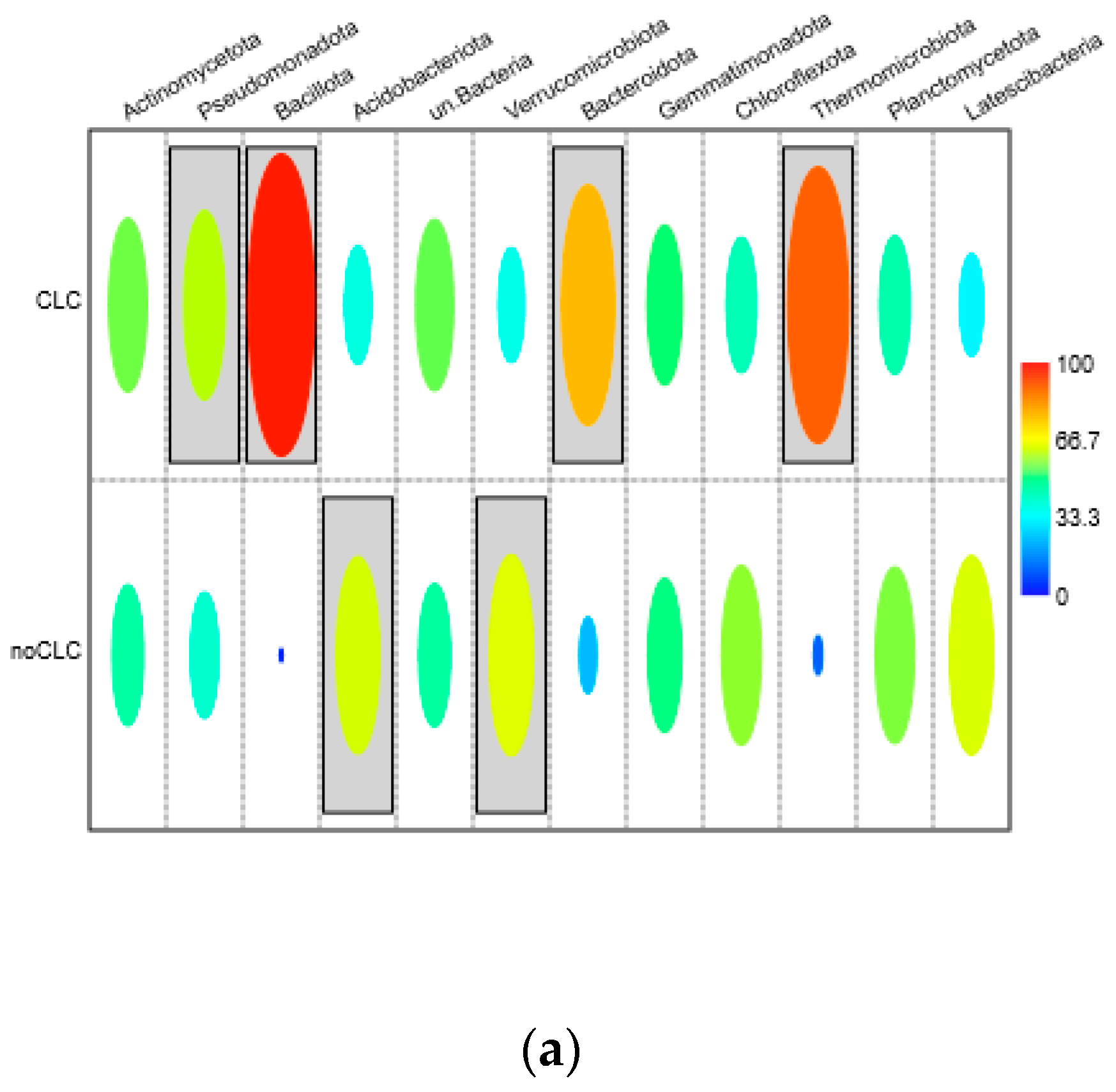
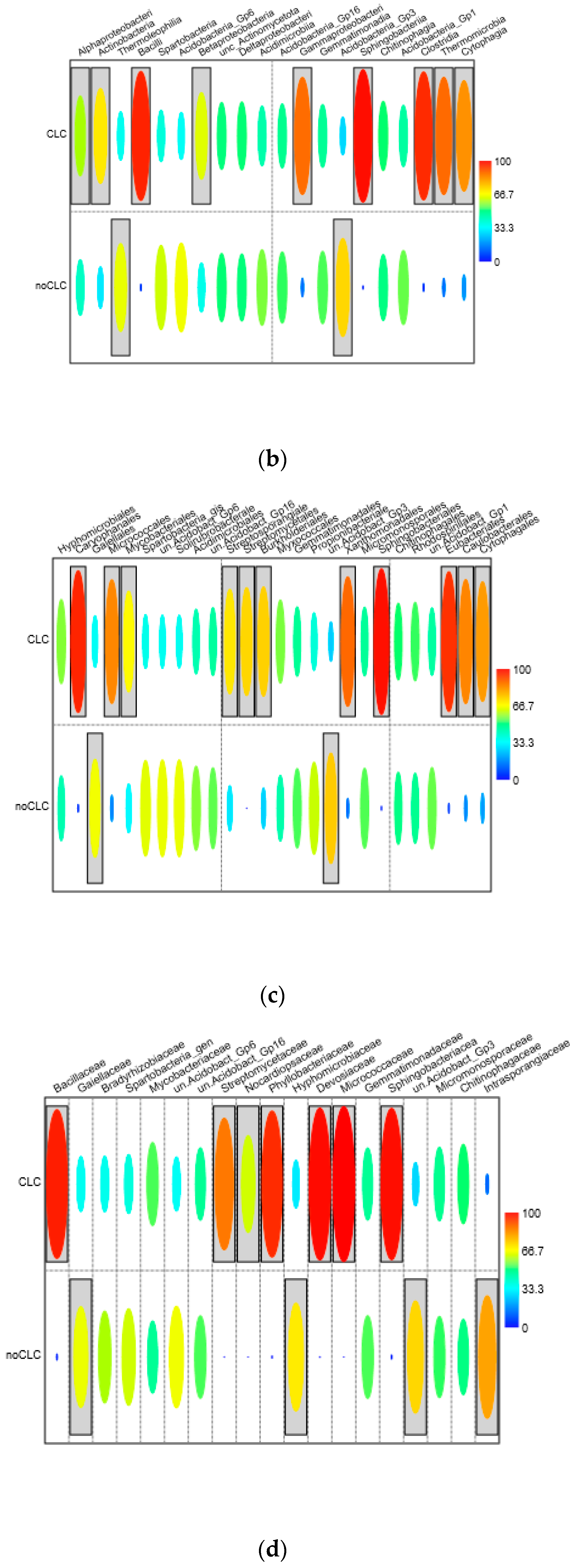
3.3.1. Bacteriobiome Composition and a Comparison of the Relative Abundance
| Taxon | No CLC | CLC | p-Value | ||
|---|---|---|---|---|---|
| Actinomycetota | 38.7 | ±7.0 | 31.2 | ±5.3 | 0.031 |
| Pseudomonadota | 25.4 | ±1.8 | 26.9 | ±4.9 | 0.420 |
| Acidobacteriota | 13.3 | ±2.8 | 6.5 | ±3.6 | 0.000 |
| Verrucomicrobiota | 6.1 | ±1.7 | 2.1 | ±1.8 | 0.001 |
| Bacteroidota | 2.0 | ±0.8 | 4.2 | ±1.9 | 0.007 |
| Gemmatimonadota | 1.9 | ±0.6 | 1.4 | ±0.7 | 0.076 |
| Chloroflexota | 1.7 | ±0.7 | 1.0 | ±0.4 | 0.030 |
| Bacillota | 1.3 | ±1.0 | 18.6 | ±10.3 | 0.000 |
| Class | |||||
| Alphaproteobacteria | 18.7 | ±1.2 | 18.0 | ±4.2 | 0.661 |
| Thermoleophilia | 18.5 | ±4.7 | 5.9 | ±4.4 | 0.000 |
| Actinobacteria | 13.5 | ±5.3 | 20.5 | ±5.6 | 0.022 |
| Spartobacteria | 5.3 | ±1.6 | 2.0 | ±1.6 | 0.001 |
| Acidobacteria_Gp6 | 4.7 | ±2.5 | 2.0 | ±1.5 | 0.022 |
| Acidimicrobiia | 3.2 | ±0.6 | 1.7 | ±0.7 | 0.001 |
| Betaproteobacteria | 3.0 | ±0.7 | 3.5 | ±1.1 | 0.165 |
| Gammaproteobacteria | 0.8 | ±0.4 | 3.2 | ±1.3 | 0.000 |
| Acidobacteria_Gp16 | 2.9 | ±0.8 | 1.9 | ±0.6 | 0.012 |
| Deltaproteobacteria | 2.8 | ±0.2 | 2.1 | ±1.0 | 0.052 |
| Acidobacteria_Gp3 | 1.9 | ±0.6 | 0.5 | ±0.3 | 0.000 |
| Gemmatimonadia | 1.6 | ±0.5 | 3.3 | ±1.3 | 0.001 |
| Chitinophagia | 1.3 | ±0.6 | 0.9 | ±0.9 | 0.002 |
| Acidobacteria_Gp1 | 1.3 | ±0.8 | 0.6 | ±0.5 | 0.065 |
| Bacilli | 1.2 | ±0.9 | 16.9 | ±10.6 | 0.001 |
| Sphingobacteriia | 0.2 | ±0.2 | 2.0 | ±1.4 | 0.002 |
| Clostridia | 0.1 | ±0.1 | 1.4 | ±1.2 | 0.012 |
| Thermomicrobia | 0.2 | ±0.2 | 1.0 | ±0.8 | 0.011 |
3.3.2. The Indicator Taxa
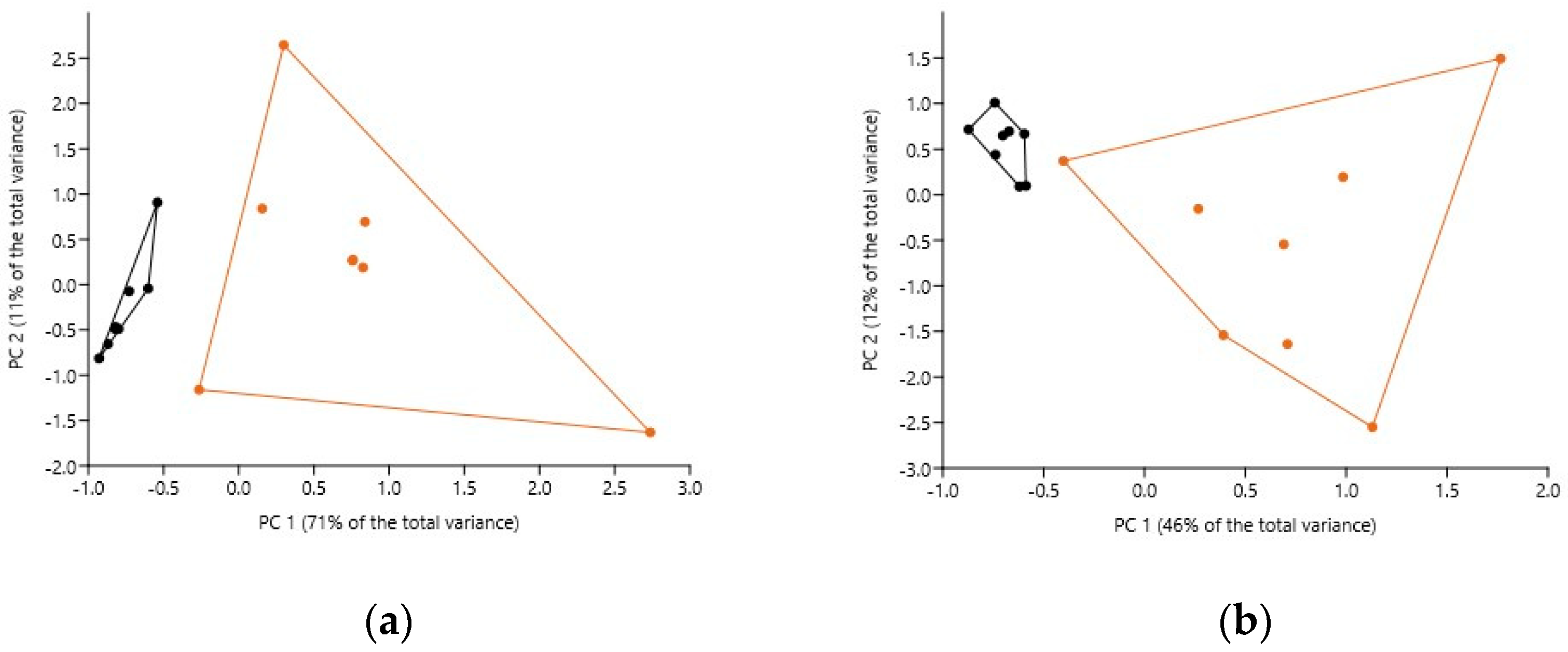
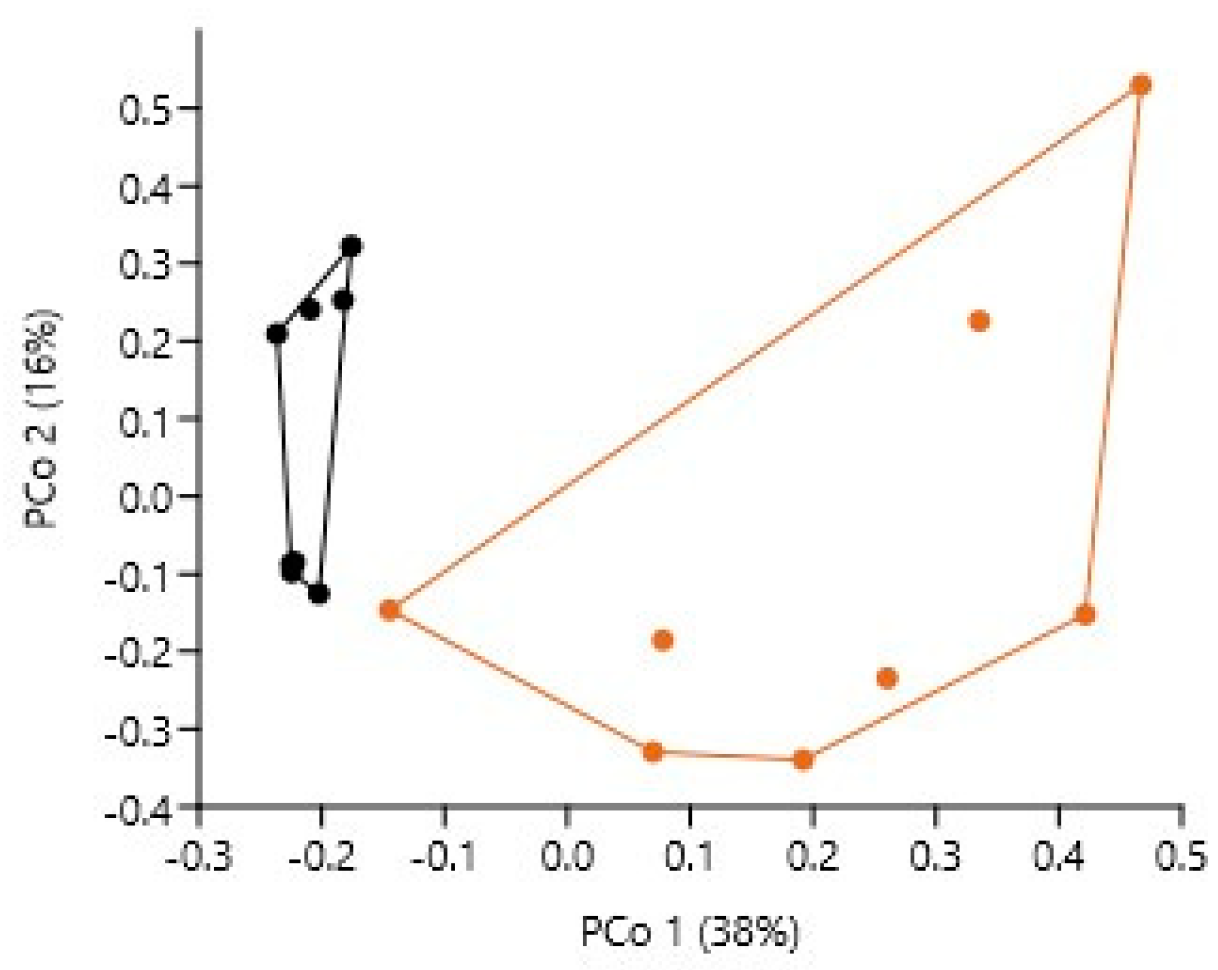
3.3.3. Bacteriome Similarity
3.3.4. Bacteriobiome α- and β-Biodiversity
4. Discussion
4.1. The Indicator Taxa for the CLC Soil
4.2. Bacteriobiome Composition and the Indicator Taxa in the no-CLC Soil
4.3. Bacteriobiome α- and β-Biodiversity
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Food and Agriculture Organization of the United Nations. Gateway to Poultry Production and Products. Available online: https://www.fao.org/poultry-production-products/en/ (accessed on 5 February 2024).
- FAOSTAT. Crops and Livestock Products. 2024. Available online: https://www.fao.org/faostat/en/#data/QCL (accessed on 6 February 2024).
- Dróżdż, D.; Wystalska, K.; Malińska, K.; Grosser, A.; Grobelak, A.; Kacprzak, M. Management of poultry manure in Poland—Current state and future perspectives. J. Environ. Manag. 2020, 264, 110327. [Google Scholar] [CrossRef] [PubMed]
- Wan, L.; Wang, X.; Cong, C.; Li, J.; Xu, Y.; Li, X.; Hou, F.; Wu, Y.; Wang, L. Effect of inoculating microorganisms in chicken manure composting with maize straw. Bioresour. Technol. 2020, 301, 122730. [Google Scholar] [CrossRef]
- Bolan, N.S.; Szogi, A.A.; Chuasavathi, T.; Seshadri, B.; Rothrock, M.J., Jr.; Panneerselvam, P. Uses and management of poultry litter. World’s Poult. Sci. J. 2010, 66, 673–698. [Google Scholar] [CrossRef]
- FAOSTAT. Livestock Manure, Livestock Manure Applied to Soils (N Content, Chicken Broilers and Layers). Available online: https://www.fao.org/faostat/en/#data/EMN (accessed on 6 February 2024).
- López-Mosquera, M.E.; Cabaleiro, F.; Sainz, M.J.; López-Fabal, A.; Carral, E. Fertilizing value of broiler litter: Effects of drying and pelletizing. Bioresour. Technol. 2008, 99, 5626–5633. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Huang, Y.; Duan, W.; Qiao, C.; Shen, Q.; Li, R. Microbial community composition turnover and function in the mesophilic phase predetermine chicken manure composting efficiency. Bioresour. Technol. 2020, 313, 123658. [Google Scholar] [CrossRef]
- Zhang, D.; Sun, J.; Wang, D.; Peng, S.; Wang, Y.; Lin, X.; Yang, X.; Hua, Q.; Wu, P. Comparison of bacterial and fungal communities’ structure and dynamics during chicken manure and pig manure composting. Environ. Sci. Pollut. Res. 2023, 30, 94347–94360. [Google Scholar] [CrossRef] [PubMed]
- Martins, G.L.; de Souza, A.J.; Mendes, L.W.; Gontijo, J.B.; Rodrigues, M.M.; Coscione, A.R.; Oliveira, F.C.; Regitano, J.B. Physicochemical and bacterial changes during composting of vegetable and animal-derived agro-industrial wastes. Bioresour. Technol. 2023, 376, 128842. [Google Scholar] [CrossRef]
- Gupta, C.L.; Avidov, R.; Kattusamy, K.; Saadi, I.; Varma, V.S.; Blum, S.E.; Zhu, Y.; Zhou, X.; Su, J.; Laor, Y.; et al. Spatial and temporal dynamics of microbiomes and resistomes in broiler litter stockpiles. Comput. Struct. Biotechnol. J. 2021, 19, 6201–6211. [Google Scholar] [CrossRef]
- Zhan, Y.; Chang, Y.; Tao, Y.; Zhang, H.; Lin, Y.; Deng, J.; Ma, T.; Ding, G.; Wei, Y.; Li, J. Insight into the dynamic microbial community and core bacteria in composting from different sources by advanced bioinformatics methods. Environ. Sci. Pollut. Res. 2023, 30, 8956–8966. [Google Scholar] [CrossRef]
- Wang, F.; Xie, L.; Gao, W.; Wu, D.; Chen, X.; Wei, Z. The role of microbiota during chicken manure and pig manure co-composting. Bioresour. Technol. 2023, 384, 129360. [Google Scholar] [CrossRef]
- Steiner, C.; Teixeira, W.G.; Lehmann, J.; Nehls, T.; de Macêdo, J.L.V.; Blum, W.E.H.; Zech, W. Long term effects of manure, charcoal and mineral fertilization on crop production and fertility on a highly weathered Central Amazonian upland soil. Plant Soil 2007, 291, 275–290. [Google Scholar] [CrossRef]
- Miyao, G.; Davis, R.M.; Leveau, J.; Maharaj, N. Evaluation of composted poultry manure on plant health of processing tomatoes in the lower Sacramento Valley of California. In Proceedings of the XIII International Symposium on Processing Tomato, Sirmione, Italy, 8–11 June 2014; ISHS Acta Horticulturae 1081. pp. 121–125. [Google Scholar] [CrossRef]
- Nguyen, V.T.; Wang, C.-H. Effects of Organic Materials on Growth, Yield, and Fruit Quality of Honeydew Melon. Commun. Soil Sci. Plant Anal. 2016, 47, 495–504. [Google Scholar] [CrossRef]
- Jiang, L.; Lin, J.L.; Jia, L.X.; Liu, Y.; Pan, B.; Yang, Y.; Lin, Y. Effects of two different organic amendments addition to soil on sorption–desorption, leaching, bioavailability of penconazole and the growth of wheat (Triticum aestivum L.). J. Environ. Manag. 2016, 167, 130–138. [Google Scholar] [CrossRef] [PubMed]
- Ortega, R.; Domene, M.A.; Soriano, M.; Sánchez-Marañón, M.; Asensio, C.; Miralles, I. Improving the fertility of degraded soils from a limestone quarry with organic and inorganic amendments to support vegetation restoration with semiarid Mediterranean plants. Soil Tillage Res. 2020, 204, 104718. [Google Scholar] [CrossRef]
- Semenov, M.V.; Zhelezova, A.D.; Ksenofontova, N.A.; Ivanova, E.A.; Nikitin, D.A. Chicken manure as an organic fertilizer: Composting technologies and impact on soil properties (a review). Dokuchaev Soil Bull. 2023, 115, 160–198. [Google Scholar] [CrossRef]
- Oakley, B.B.; Lillehoj, H.S.; Kogut, M.H.; Kim, W.K.; Maurer, J.J.; Pedroso, A.; Lee, M.D.; Collett, S.R.; Johnson, T.J.; Cox, N.A. The chicken gastrointestinal microbiome. FEMS Microbiol. Lett. 2014, 360, 100–112. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Xiang, Y.; Zhou, W.; Chen, J.; Li, K.; Yang, H. Microbial community mapping in intestinal tract of broiler chicken. Poult. Sci. 2016, 96, 1387–1393. [Google Scholar] [CrossRef]
- Huang, P.; Zhang, Y.; Xiao, K.; Jiang, F.; Wang, H.; Tang, D.; Liu, D.; Liu, B.; Liu, Y.; He, X.; et al. The chicken gut metagenome and the modulatory effects of plant-derived Benzylisoquinoline alkaloids. Microbiome 2018, 6, 211. [Google Scholar] [CrossRef]
- Wang, Y.; Gong, J.; Li, J.; Xin, Y.; Hao, Z.; Chen, C.; Li, H.; Wang, B.; Ding, M.; Li, W.; et al. Insights into bacterial diversity in compost: Core microbiome and prevalence of potential pathogenic bacteria. Sci. Total Environ. 2020, 718, 137304. [Google Scholar] [CrossRef]
- Sharaf, H.; Thompson, A.A.; Williams, M.A.; Peck, G.M. Compost applications increase bacterial community diversity in the apple rhizosphere. Soil. Sci. Soc. Am. J. 2021, 85, 1105–1121. [Google Scholar] [CrossRef]
- Kyakuwaire, M.; Olupot, G.; Amoding, A.; Nkedi-Kizza, P.; Basamba, T.A. How Safe is Chicken Litter for Land Application as an Organic Fertilizer? A Review. Int. J. Environ. Res. Public Health 2019, 16, 3521. [Google Scholar] [CrossRef]
- Błażejewska, A.; Zalewska, M.; Grudniak, A.; Popowska, M. A Comprehensive Study of the Microbiome, Resistome, and Physical and Chemical Characteristics of Chicken Waste from Intensive Farms. Biomolecules 2022, 12, 1132. [Google Scholar] [CrossRef] [PubMed]
- Subirats, J.; Murray, R.; Scott, A.; Lau, C.H.; Topp, E. Composting of chicken litter from commercial broiler farms reduces the abundance of viable enteric bacteria, Firmicutes, and selected antibiotic resistance genes. Sci. Total Environ. 2020, 746, 141113. [Google Scholar] [CrossRef]
- Qiu, T.; Wu, D.; Zhang, L.; Zou, D.; Sun, Y.; Gao, M.; Wang, X. A comparison of antibiotics, antibiotic resistance genes, and bacterial community in broiler and layer manure following composting. Environ. Sci. Pollut. Res. 2021, 28, 14707–14719. [Google Scholar] [CrossRef]
- Hydrometcenter of Russia. Available online: https://meteoinfo.ru/en/climate/monthly-climate-means-for-towns-of-russia-temperature-and-precipitation (accessed on 26 August 2024).
- IUSS Working Group WRB. World Reference Base for Soil Resources. In International Soil Classification System for Naming Soils and Creating Legends for Soil Maps, 4th ed.; International Union of Soil Sciences (IUSS): Vienna, Austria, 2022; 236p. [Google Scholar]
- Patterson, G.T.; Carter, M.R. Soil Sampling and Methods of Analysis, 2nd ed.; Carter, M.R., Gregorich, E.G., Eds.; CRC Press: Boca Raton, FL, USA, 2008; Volume 44, ISBN 978-0-8493-3586-0. [Google Scholar]
- Naumova, N.; Barsukov, P.; Baturina, O.; Rusalimova, O.; Kabilov, M. West-Siberian Chernozem: How Vegetation and Tillage Shape Its Bacteriobiome. Microorganisms 2023, 11, 2431. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naïve Bayesian Classifier for Rapid Assignment of rRNA Sequences into the New Bacterial Taxonomy. App Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [PubMed]
- Hammer, O.; Harper, D.A.T.; Ryan, P.D. PAST: Paleontological Statistics Software Package for Education and Data Analysis. Palaeontol. Electron. 2001, 4, 1. [Google Scholar]
- Hughes, J.B.; Hellmann, J.J. The Application of Rarefaction Techniques to Molecular Inventories of Microbial Diversity. Methods Enzymol. 2005, 397, 292–308. [Google Scholar] [CrossRef]
- Qiu, C.; Bao, Y.; Petropoulos, E.; Wang, Y.; Zhong, Z.; Jiang, Y.; Ye, X.; Lin, X.; Feng, Y. Organic and Inorganic Amendments Shape Bacterial Indicator Communities That Can, In Turn, Promote Rice Yield. Microorganisms 2022, 10, 482. [Google Scholar] [CrossRef]
- Naumova, N.B.; Alikina, T.Y.; Zolotova, N.S.; Konev, A.V.; Pleshakova, V.I.; Lescheva, N.A.; Kabilov, M.R. Bacillus-Based Probiotic Treatment Modified Bacteriobiome Diversity in Duck Feces. Agriculture 2021, 11, 406. [Google Scholar] [CrossRef]
- Lysko, S.B.; Baturina, O.A.; Naumova, N.B.; Lescheva, N.A.; Pleshakova, V.I.; Kabilov, M.R. No-Antibiotic-Pectin-Based Treatment Differently Modified Cloaca Bacteriobiome of Male and Female Broiler Chickens. Agriculture 2022, 12, 24. [Google Scholar] [CrossRef]
- Shi, T.; Wang, Y.F.; Wang, H.; Wang, B. Genus Nocardiopsis: A Prolific Producer of Natural Products. Mar. Drugs 2022, 20, 374. [Google Scholar] [CrossRef] [PubMed]
- Chanama, M.; Suriyachadkun, C.; Chanama, S. Streptomyces antimicrobicus sp. nov., a novel clay soil-derived actinobacterium producing antimicrobials against drug-resistant bacteria. PLoS ONE 2023, 18, e0286365. [Google Scholar] [CrossRef] [PubMed]
- Cuervo, L.; Álvarez-García, S.; Salas, J.A.; Méndez, C.; Olano, C.; Malmierca, M.G. The Volatile Organic Compounds of Streptomyces spp.: An In-Depth Analysis of Their Antifungal Properties. Microorganisms 2023, 11, 1820. [Google Scholar] [CrossRef]
- El-Sayed, M.H.; Kobisi, A.E.A.; Elsehemy, I.A.; El-Sakhawy, M.A. Rhizospheric-Derived Nocardiopsis alba BH35 as an Effective Biocontrol Agent Actinobacterium with Antifungal and Plant Growth-Promoting Effects: In Vitro Studies. J. Microbiol. Biotechnol. 2023, 33, 607–620. [Google Scholar] [CrossRef]
- Chhetri, G.; Kim, I.; Kang, M.; Kim, J.; So, Y.; Seo, T. Devosia rhizoryzae sp. nov., and Devosia oryziradicis sp. nov., novel plant growth promoting members of the genus Devosia, isolated from the rhizosphere of rice plants. J. Microbiol. 2022, 60, 1–10. [Google Scholar] [CrossRef]
- Zhang, C.; van der Heijden, M.G.A.; Dodds, B.K.; Nguyen, T.B.; Spooren, J.; Valzano-Held, A.; Cosme, M.; Berendsen, R.L. A tripartite bacterial-fungal-plant symbiosis in the mycorrhiza-shaped microbiome drives plant growth and mycorrhization. Microbiome 2024, 12, 13. [Google Scholar] [CrossRef]
- Mghazli, N.; Bruneel, O.; Zouagui, R.; Hakkou, R.; Sbabou, L. Characterization of plant growth promoting activities of indigenous bacteria of phosphate mine wastes, a first step toward revegetation. Front. Microbiol. 2022, 13, 1026991. [Google Scholar] [CrossRef]
- Manetsberger, J.; Caballero Gómez, N.; Soria-Rodríguez, C.; Benomar, N.; Abriouel, H. Simply Versatile: The Use of Peribacillus simplex in Sustainable Agriculture. Microorganisms 2023, 11, 2540. [Google Scholar] [CrossRef]
- Senchenkov, V.Y.; Lyakhovchenko, N.S.; Nikishin, I.A.; Myagkov, D.A.; Chepurina, A.A.; Polivtseva, V.N.; Abashina, T.N.; Delegan, Y.A.; Nikulicheva, T.B.; Nikulin, I.S.; et al. Whole-Genome Sequencing and Biotechnological Potential Assessment of Two Bacterial Strains Isolated from Poultry Farms in Belgorod, Russia. Microorganisms 2023, 11, 2235. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Yang, G.; Yu, Z.; Zhuang, L.; Jin, Y.; Zhou, S. Oceanobacillus luteolus sp. nov., isolated from soil. Int. J. Syst. Evol. Microbiol. 2014, 64, 1495–1500. [Google Scholar] [CrossRef]
- Guevara-Luna, J.; Arroyo-Herrera, I.; Tapia-García, E.Y.; Santos, P.E.-D.L.; Ortega-Nava, A.J.; Vásquez-Murrieta, M.S. Diversity and Biotechnological Potential of Cultivable Halophilic and Halotolerant Bacteria from the “Los Negritos” Geothermal Area. Microorganisms 2024, 12, 482. [Google Scholar] [CrossRef]
- Teles, E.A.P.; Xavier, J.F.; Arcênio, F.S.; Amaya, R.L.; Gonçalves, J.V.S.; Rouwns, L.F.M.; Zonta, E.; Coelho, I.S. Characterization and evaluation of potential halotolerant phosphate solubilizing bacteria from Salicornia fruticosa rhizosphere. Front. Plant Sci. 2024, 14, 1324056. [Google Scholar] [CrossRef]
- Naumova, N.B.; Belanov, I.P.; Alikina, T.Y.; Kabilov, M.R. Undisturbed Soil Pedon under Birch Forest: Characterization of Microbiome in Genetic Horizons. Soil Syst. 2021, 5, 14. [Google Scholar] [CrossRef]
- Evdokimova, E.V.; Gladkov, G.V.; Kuzina, N.I.; Ivanova, E.A.; Kimeklis, A.K.; Zverev, A.O.; Kichko, A.A.; Aksenova, T.S.; Pinaev, A.G.; Andronov, E.E. The difference between cellulolytic ‘culturomes’ and microbiomes inhabiting two contrasting soil types. PLoS ONE 2020, 15, e0242060. [Google Scholar] [CrossRef] [PubMed]
- Ivanova, E.; Abakumov, E.; Vasilyeva, N.; Zverev, A.; Vladimirov, A.; Ksenofontova, N.; Andronov, E.; Kostenko, I. The shifts in the structure of the prokaryotic community of mountain-grassland soil under the influence of artificial larch plantations. PLoS ONE 2022, 17, e0263135. [Google Scholar] [CrossRef] [PubMed]
- Hou, M.; Zhao, X.; Wang, Y.; Lv, X.; Chen, Y.; Jiao, X.; Sui, Y. Pedogenesis of typical zonal soil drives belowground bacterial communities of arable land in the Northeast China Plain. Sci. Rep. 2023, 13, 14555. [Google Scholar] [CrossRef]
- Zhang, Z.; Han, X.; Yan, J.; Zou, W.; Wang, E.; Lu, X.; Chen, X. Keystone Microbiomes Revealed by 14 Years of Field Restoration of the Degraded Agricultural Soil Under Distinct Vegetation Scenarios. Front. Microbiol. 2020, 11, 1915. [Google Scholar] [CrossRef]
- Zhang, S.; Han, S.; Gao, J.; Yu, X.; Hu, S. Low-temperature corn straw-degrading bacterial agent and moisture effects on indigenous microbes. Appl. Microbiol. Biotechnol. 2023, 107, 5241–5255. [Google Scholar] [CrossRef]
- Borowik, A.; Wyszkowska, J.; Kucharski, J. Impact of Various Grass Species on Soil Bacteriobiome. Diversity 2020, 12, 212. [Google Scholar] [CrossRef]
- Pershina, E.V.; Ivanova, E.A.; Korvigo, I.O.; Chirak, E.L.; Sergaliev, N.H.; Abakumov, E.V.; Provorov, N.A.; Andronov, E.E. Investigation of the core microbiome in main soil types from the East European plain. Sci. Total Environ. 2018, 631–632, 1421–1430. [Google Scholar] [CrossRef]
- Li, X.; Jin, Z.; Xiong, L.; Tong, L.; Zhu, H.; Zhang, X.; Qin, G. Effects of Land Reclamation on Soil Bacterial Community and Potential Functions in Bauxite Mining Area. Int. J. Environ. Res. Public Health 2022, 19, 16921. [Google Scholar] [CrossRef] [PubMed]
- Minkina, T.; Sushkova, S.; Delegan, Y.; Bren, A.; Mazanko, M.; Kocharovskaya, Y.; Filonov, A.; Rajput, V.D.; Mandzhieva, S.; Rudoy, D.; et al. Effect of chicken manure on soil microbial community diversity in poultry keeping areas. Environ. Geochem. Health 2023, 45, 9303–9319. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Zou, J.; Chen, D.; He, X.; Zhang, C.; Li, J.; Lan, S.; Liu, Z.J.; Zou, S.; Qian, X. Chicken manure application alters microbial community structure and the distribution of antibiotic-resistance genes in rhizosphere soil of Cinnamomum camphora forests. FEMS Microbiol. Ecol. 2023, 99, fiad155. [Google Scholar] [CrossRef]
- Bilal, M.; Diarra, M.S.; Islam, M.R.; Lepp, D.; Mastin-Wood, R.E.; Topp, E.; Bittman, S.; Zhao, X. Effects of litter from antimicrobial-fed broiler chickens on soil bacterial community structure and diversity. Can. J. Microbiol. 2022, 68, 643–653. [Google Scholar] [CrossRef]
- Barbosa, L.M.P.; Santos, T.P.S.; Rocha, S.M.B.; Santos, T.O.d.S.; Oliveira, L.M.d.S.; Neto, F.d.A.; de Araújo, A.S.F.; de Souza, H.A.; Nunes, L.A.P.L.; de Sousa, R.S. Composts from bovine rumen and chicken litter improve soil fertility and promote the growth of pepper (Capsicum chinense Jacq.). Res. Sq. 2023. [Google Scholar] [CrossRef]
- Naumova, N.B.; Kabilov, M.R. About the Biodiversity of the Air Microbiome. Acta Naturae 2022, 14, 50–56. [Google Scholar] [CrossRef]

| Property | ||
|---|---|---|
| Initial Water Content, % | 24.5 | ±0.7 |
| pHH2O | 7.14 | ±0.05 |
| EC, mS/cm | 8.98 | ±0.20 |
| Organic Matter, % | 85.0 | ±0.6 |
| N, % | 4.48 | ±0.39 |
| P, % | 0.95 | ±0.09 |
| K, % | 1.75 | ±0.19 |
| Soil Property | No CLC | CLC | ||
|---|---|---|---|---|
| SOM 1, % | 7.69 | ±0.04 b 2 | 7.58 | ±0.07 a |
| SOC 1, % | 3.84 | ±0.03 b | 3.79 | ±0.04 a |
| pHH2O | 6.03 | ±0.01 a | 6.36 | ±0.01 b |
| Nitrates, mg N/kg | 12.2 | ±5.5 a | 63.8 | ±7.4 b |
| Readily Available P, mg P/kg | 0.61 | ±0.10 a | 3.14 | ±0.33 b |
| Available P, mg P/kg | 10.7 | ±3.7 a | 100.7 | ±9.6 b |
| Exchangeable K, mg K/kg | 171 | ±26 a | 687 | ±52 b |
| Sand, % | 6.6 | ±3.3 | 7.5 | ±3.0 |
| Silt, % | 68.0 | ±3.0 | 66.2 | ±2.2 |
| Clay, % | 25.4 | ±4.3 | 26.3 | ±3.8 |
| Dominant Phyla | Bacillota | Actinomycetota |
|---|---|---|
| 80.9 ± 0.7 | 18.9 ± 0.6 | |
| Dominant Classes | ||
| Bacilli | 80.8 ± 0.7 | |
| Actinobacteria | 18.9 ± 0.6 | |
| Gammaproteobacteria | 0.2 ± 0.1 | |
| Dominant Genera | ||
| Unclassified Bacillaceae | 50.1± 1.2 | |
| Staphylococcus | 13.0 ± 1.6 | |
| Brevibacterium | 6.2 ± 0.7 | |
| Nosocomiicoccus | 5.5 ± 1.1 | |
| Corynebacterium | 5.4 ± 0.4 | |
| Oceanobacillus | 5.3 ± 1.7 | |
| Mammaliicoccus | 4.1 ± 0.5 | |
| Yaniella | 3.2 ± 0.3 | |
| Enteractinococcus | 2.7 ± 0.7 | |
| Brachybacterium | 1.0 ± 0.2 | |
| Sum | 78.1 | 18.4 |
| Genus | No CLC | CLC | p-Value | ||
|---|---|---|---|---|---|
| Gaiella | 12.1 | ±2.9 | 3.8 | ±3.4 | 0.000 |
| Bradyrhizobium | 5.8 | ±0.7 | 2.0 | ±1.9 | 0.000 |
| Spartobacteria_gis * | 5.3 | ±1.6 | 1.7 | ±1.6 | 0.001 |
| un. # Acidobacteria_Gp6 | 4.7 | ±2.5 | 1.8 | ±1.7 | 0.022 |
| Mycobacterium | 3.7 | ±1.0 | 2.5 | ±1.3 | 0.069 |
| un. Acidobacteria_Gp16 | 2.7 | ±0.7 | 1.5 | ±0.8 | 0.006 |
| un.Acidobacteria_Gp3 | 1.9 | ±0.6 | 0.4 | ±0.3 | 0.000 |
| un.Acidobacteria_Gp1 | 1.3 | ±0.8 | 0.6 | ±0.5 | 0.065 |
| Streptomyces | 0.1 | ±0.4 | 3.6 | ±3.2 | 0.001 |
| Oceanobacillus | 0.0 | ±0.0 | 3.1 | ±4.3 | 0.034 |
| Nocardiopsis | 0.0 | ±0.0 | 3.0 | ±6.9 | 0.040 |
| Devosia | 0.1 | ±0.2 | 2.3 | ±1.7 | 0.001 |
| Peribacillus | 0.2 | ±0.4 | 1.8 | ±1.9 | 0.016 |
| Neobacillus | 0.2 | ±0.4 | 1.4 | ±1.2 | 0.002 |
| Cerasibacillus | 0.0 | ±0.0 | 1.0 | ±1.9 | 0.080 |
| Taxon | No CLC | CLC | p-Value | ||
|---|---|---|---|---|---|
| OTU Richness | 937 | ±128 | 900 | ±336 | 0.769 |
| Chao-1 | 1321 | ±142 | 1158 | ±414 | 0.310 |
| Simpson (S) | 0.987 | ±0.002 | 0.982 | ±0.017 | 0.387 |
| Shannon’s | 5.4 | ±0.2 | 5.2 | ±0.6 | 0.354 |
| Equitability | 0.80 | ±0.01 | 0.78 | ±0.04 | 0.222 |
| Berger–Parker | 0.06 | ±0.01 | 0.07 | ±0.06 | 0.546 |
| Dominance (1-S) | 0.013 | ±0.002 | 0.018 | ±0.017 | 0.396 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Naumova, N.; Barsukov, P.; Baturina, O.; Rusalimova, O.; Kabilov, M. Addition of Chicken Litter Compost Changes Bacteriobiome in Fallow Soil. Appl. Microbiol. 2024, 4, 1268-1282. https://doi.org/10.3390/applmicrobiol4030087
Naumova N, Barsukov P, Baturina O, Rusalimova O, Kabilov M. Addition of Chicken Litter Compost Changes Bacteriobiome in Fallow Soil. Applied Microbiology. 2024; 4(3):1268-1282. https://doi.org/10.3390/applmicrobiol4030087
Chicago/Turabian StyleNaumova, Natalia, Pavel Barsukov, Olga Baturina, Olga Rusalimova, and Marsel Kabilov. 2024. "Addition of Chicken Litter Compost Changes Bacteriobiome in Fallow Soil" Applied Microbiology 4, no. 3: 1268-1282. https://doi.org/10.3390/applmicrobiol4030087
APA StyleNaumova, N., Barsukov, P., Baturina, O., Rusalimova, O., & Kabilov, M. (2024). Addition of Chicken Litter Compost Changes Bacteriobiome in Fallow Soil. Applied Microbiology, 4(3), 1268-1282. https://doi.org/10.3390/applmicrobiol4030087







