Abstract
The increasing complexity of food safety concerns, driven by the rising risks of contamination from pathogens, chemical hazards, and environmental influences, has highlighted the need for more effective detection and prevention strategies. Metagenomics, a powerful molecular tool, is transforming the food industry by providing a comprehensive understanding of the microbial communities in fresh produce, poultry, and meat. Advances in microbial detection techniques, such as shotgun sequencing, metabarcoding, and long-read sequencing technologies, have led to faster and more accurate pathogen detection, reducing the risk of outbreaks and ensuring public health protection. Despite its promise, several challenges remain in implementing metagenomics on a broader scale, including the high cost of sequencing technologies, the complexity of analyzing large datasets, and the lack of standardized procedures across platforms. These limitations hinder its adoption, particularly for smaller operations or in regions with fewer resources. This review examines the applications of metagenomics in food safety, focusing on its impact on meat, poultry, and fresh produce, while discussing the obstacles to its widespread use and potential solutions to overcome these barriers.
1. Introduction
Ensuring food safety is becoming increasingly complex as the risk of contamination by foodborne pathogens and chemical hazards continues to grow. These challenges are worsened by extreme weather events and other environmental factors, emphasizing the need for robust strategies to protect public health [1]. At the same time, rising concerns over food fraud, adulteration, and authenticity have driven the demand for greater transparency and stricter regulations. As a result, molecular technologies continue to advance in efforts to establish standardized protocols that ensure the safety and authenticity of food in our society.
Among these innovations, metagenomics, a branch of genomics, has significantly enhanced food analysis by offering a comprehensive understanding of microbial diversity, identifying both beneficial and harmful microorganisms in food products. By examining bacterial consortia, the mycobiome, and the virome, this approach provides critical insights into genes related to virulence, antimicrobial resistance, and spoilage, aiding in the detection of pathogens like Salmonella, Listeria monocytogenes, and Escherichia coli in raw meats, dairy, and fresh produce [2,3,4]. Advancements in metagenomics have also enabled the development of more precise diagnostic tools and improved food safety protocols, revolutionizing the way pathogens and microbial contaminants are monitored in food production systems. The use of techniques, such as shotgun sequencing for the detection of foodborne pathogens in the microbiome of beef production [5], has led to faster and more accurate identification, reducing the risk of outbreaks and improving overall food safety.
Despite significant advancements in metagenomics, several challenges remain in its application to food analysis. One of the primary issues is the high cost of sequencing, which can be a barrier to its widespread use, particularly for smaller companies or in regions with limited funding [6]. Additionally, the vast amount of metadata generated during sequencing complicates the process of data analysis, as it requires advanced computational tools and expertise to process and interpret the results effectively [7]. Furthermore, the lack of standardization in metagenomics techniques across different laboratories and sequencing platforms can result in inconsistent findings, leading to the risk of false positives or false negatives in results [8]. For example, variations in sample preparation, sequencing protocols, and data interpretation methods can cause discrepancies in identifying microbial communities in food products. Addressing these obstacles, including reducing costs, improving data analysis methods, and standardizing procedures, will be crucial for the broader adoption and reliability of metagenomics technologies in the food industry.
This review aims to assess the role of metagenomics in food safety, exploring its application to meat, poultry, and fresh produce, and to identify the limitations in utilizing these technologies for routine food quality monitoring.
2. Basis of Metagenomic Sequencing for Food Analyses
Since the efforts of Hooke and van Leeuwenhoek in the discovery of microorganisms and the development of the first culture method by Koch, significant progress has been made in the study of microorganisms. The utilization of 16S ribosomal RNA (rRNA) sequences has allowed the correct classification of microbes through phylogenetic-based studies [9,10]. Metagenomics has emerged as an alternate molecular tool. Metagenomic analysis, or the phylogenetic analyses from environmental samples of microbial communities, has exponentially grown with the advancement of sequencing techniques. Currently, it is classified as amplicon and shotgun metagenomics. The data used in amplicon metagenomics include amplified sequences of marker genes, such as 16S/18S/26S rRNA and intergenic transcribed spacer (ITS) [11,12]. The processing of this type of information results in the exploration of microbial diversity [13,14], as it is based on the analysis of hypervariable regions of a gene used to identify corresponding taxonomy. On the other side, shotgun metagenomics data includes all DNA sequences in a sample, reveals information about taxonomic diversity, and allows the prediction of major metabolisms in the microbial community [15]. The untargeted sequences obtained from the sample results in information that could be used in the study of all microbes present, such as bacteria, viruses, archaea, and single-cell eukaryotes. The field of metagenomics is further classified into functional and sequencing metagenomics depending on whether it results in the study of the gene function of diversity [16].
Metagenomic studies reveal information regarding community composition at strain-level profiling and above [17,18]. While the framework for metagenomic analysis has been established since the 2010s [19], the development of high-throughput sequencing (HTS) is providing massive microbiome datasets [20,21]. These sequencing platforms have optimized molecular methods for the study of microorganisms, with the integration of functional genomics, transcriptomics, proteomics, and metabolomics [22]. The impact of -omic technologies in the food industry has been great, as they have been used as a tool in food safety, spoilage, authentication, and provenance and to understand microbial composition in probiotics and fermented food [23]. The quick development of next-generation sequencing (NGS) has transformed food microbiology. NGS platforms, such as Illumina, Ion Torrent, PacBio, and Oxford Nanopore, can be used to conduct a wide variety of food microbiological investigations with a focus on amplicon and shotgun sequencing and whole-genome sequencing (WGS) [24]. Its integration into the field of metagenomics (mNGS) has given way to a technology capable of detecting nucleic acids and identifying pathogens, such as bacteria, viruses, fungi, and parasites in a sample.
Metagenomics has been widely used in the food industry, a powerful tool to study microbial communities present in products. Microorganism surveillance is essential to ensure food quality, as microbes affect flavor and texture and cause spoilage. While fermented food microbes can improve organoleptic and nutritive qualities and expand the shelf life of fermented food [25], other bacteria might cause quality problems such as off odors, odd flavors, and gas formation [26]. Metagenomic approaches present a solution to the management of microbial communities, as their applications are wide. Metagenomics has been used as a tool in food safety, spoilage, authentication, and provenance and to understand microbial composition in probiotics and fermented food [23]. NGS has greatly benefited microbiological research by opening the door to the HTS metagenomic analysis of mixed microbial communities from different environments. In the study of food microbes, techniques such as WGS and long-read sequencing (LRS) have transformed microbial profiling by facilitating the identification of pathogens and antibiotic resistance genes (ARGs) and improving the understanding of microbial interactions in food matrixes.
Currently, WGS of microbial pathogens has been used in the determination of the source of infections. WGS provides information regarding the entire genome and is able to describe similarities among strains [24,27]. This information is essential in tracing pathogens along the food chain, as it allows an accurate comparison among bacterial sequences from outbreaks and those present in the production chain. These phylogenetic analyses allow the establishment of the transmission chain of the isolates and the identification of the origin strain if it has evolved. Metabarcoding is one of the preferred methods of studying the progression of microbial populations based on taxon abundances. Shotgun metagenomics permits the identification of individual strains and the prediction of the functional relationships between hosts and bacteria [28,29,30]. Shotgun metagenomic data have been used to understand microbial activity in situ [6,31], to measure population diversity levels in situ [32,33], and to determine family- and habitat-specific genes [34,35]. Among both metagenomic approaches, shotgun has been relatively underutilized up to date mainly due to its high costs and its bioinformatic requirements, which are more demanding [29,36].
3. Current Applications of Metagenomics in Food Safety
Metagenomics has emerged as a transformative tool for analyzing microbial communities in various food products, providing insights into diversity and functionality. In view of the increase in the demand and production of certain types of edible products, such as fresh produce, poultry, and meat, it is necessary to understand how microbial communities affect shelf life and organoleptic and nutritive qualities. Beneficial microbes shape the rheological and organoleptic traits of fermented food, while harmful microbes affect food quality, leading to spoilage and other food safety issues. These microorganisms are influenced by production methods, processing environments, and storage conditions. Although culture-based techniques have historically been used to characterize microorganisms present in food, they have several constraints that are critical for the accurate identification of microorganisms. In addition to being labor-intensive and time-consuming, they can only detect a fraction of microbes present in a sample. Culture-based techniques not only overlook viable but not culturable microorganisms (VBNCs) but also do not evaluate relationships within the microbial community [37].
The application of advanced metagenomic approaches enables the study of complex interactions within the microbiome and a way to identify contamination sources and improve food safety protocols. Metagenomics has been widely employed in environmental, agricultural, and biomedical sciences, proving its versatility and transformative potential. This section delves into the application of metagenomics as a technique for pathogen detection and monitoring, taxonomic research, detecting ARGs, and the understanding of microbial community relationships (Table 1).

Table 1.
Overview of metagenomic techniques for food safety.
3.1. Enhancing Detection Stategies for Food Spoilage and Foodborne Contamination
Microbial food spoilage and contamination occurs when the growth of microorganisms renders the food products unacceptable to the consumer. In the past, the prevention of spoilage and the elimination/exclusion of pathogens from food had been handled separately, within the research and legislative area [84]. However, the consequences of foodborne diseases are matched by the economic loss caused by food spoilage [85]. Metagenomics has been effectively used for bacterial strain profiling [86], essential and foodborne pathogen tracking, and the assessment of spoilage bacteria within various environmental factors [87,88]. Different approaches, such as metabarcoding, shotgun sequencing, and LRS can be used for the detection of foodborne and spoilage pathogens.
Metabarcoding, based on biomarker genes, enables the identification of highly diverse microbial communities [89]. It has become an effective molecular-based biomonitoring and bio-surveillance method for multiple purposes. This is a result of the use of targeted amplicons and HTS, facilitating the identification of microorganisms within complex food matrices regardless of the abundance [58,90]. It is a cost-effective technique that can run on most platforms and sequencers. This approach has several limitations as a quantitative technique, as it yields inaccurate biomass estimates [91]: the requirement of prior information of the targeted microbial group due to the introduction of amplification bias and the restrictive classification of genus level, which encumbers differentiation between pathogenic and nonpathogenic species [77,88,92]. Shotgun sequencing, based on the sequencing of all genomic DNA, enables identification at greater specificity (species and subspecies level) and the detection of organisms from different kingdoms [88,93]. Compared to metabarcoding, there is no amplification bias, as there is no use of a specific target, and it can differentiate between pathogenic and nonpathogenic strains. Its downsides are its high cost, less sensitivity towards less abundant genomes in comparison to metabarcoding, and its inability to differ live bacteria from dead ones.
These approaches have been used to perform pathogen typing from contaminated food samples to foodborne outbreaks. Metabarcoding has been instrumental in profiling microbial communities on food surfaces and in processing environments. For instance, minimally processed fruits, vegetables, and ready-to-eat (RTE) products reveal diverse microbial populations including Pseudomonas spp., Acinetobacter spp., and Psychrobacter spp., which are common contaminants in processing lines [94]. Metabarcoding has been used to trace microbial changes under defined conditions in hopes of determining how these variations can be used to improve shelf life. Similarly, shotgun sequencing has been used to identify foodborne pathogens, such as Proteobacteria and Actinobacteria, and contamination sources in the beef production chain [5]. It has also allowed the detection of functional traits of spoilage organisms and their metabolic role in fermented and processed foods [38,43]. Both techniques provide the high-resolution identification of microbial communities and their contamination sources. Their applications improve food safety by enabling targeted interventions destined to prevent spoilage and foodborne pathogen contamination throughout the food production chain.
3.2. Innovative Approaches for the Management of Antibiotic Resistance in Food Products
Antibiotics remain widespread for both prophylactic and curative purposes in animal breeding and food production. The issue of antibiotic residues in animal products, groundwater, soil, and feed has generated global concern and continues to do so, leading to significant costs associated with combatting antibiotic resistance [39]. Metagenomic technologies have been used to characterize the resistome, virulence, and antimicrobial resistance genes (AMRs) present in food. Targeted (PCR- and/or microarray-based), sequenced-based, and functional metagenomics are the three approaches used in the study of the resistome. In view of the use of known primers needed in PCR- and microarray-based studies, only known resistance genes can be tracked, which is a major limitation [40,95]. The use of NGS enables sequence-based metagenomics. Furthermore, HTS metagenomics is greatly suited to the identification of unknown resistance genes or variations within the abundance of related resistance genes.
Shotgun sequencing has also been used in the detection of ARGs and mobile genetic elements (MGEs) [43,45,96]. With the efforts by the Antibiotic Resistance gene DataBase (ARDB) [44] and the Comprehensive Antibiotic Resistance Database (CARD) [97], ARG annotation and metagenomic analysis have greatly improved. Nevertheless, high sequence depth is needed in order to compensate for the low abundance of bacterial reads and high ration of DNA from the original host [26] persist as restrictions. In view of these obstacles, alternative methods such as host DNA depletion [50,98] have been designed to overcome these issues. Most studies employ HTS shotgun sequencing to characterize the bacterial community and resistome in a wide variety of samples, such as RTE foods [51], pork [55], and beef [60,65,99,100], throughout the manufacturing chain. NGS coupled with functional metagenomics is the only approach able to isolate completely novel ARGs. This approach is able of screening of metagenomic DNA for genes that encode specific molecular functions, and it has been used in the identification of enzymes, bioactive agents, and environmental ARGs [101,102]. Its current limitation is the development of hosts capable of storing metagenomic libraries regardless of the origin and size.
Various livestock studies have focused on the interactions between AMRs and ARGs in relation to microbial diversity and pathogenicity and its effect in animal production. Over 204 genes associated with AMRs have been identified, and diversity among the bacterial community under different conditions has been studied (such as dietary compositions [74,103,104], the use of feed additives [105,106,107], and the environment [52,108]). Among the ARG classes identified in domesticated animal manure (tetracycline (tet), sulfonamides (sul), β-lactams (bla), macrolide–lincosamide–streptogramin (erm), and fluoroquinolone (fca) [109,110]), tet and sul are the most abundant in domesticated birds [96] and bovine farms [52]. The integrative use of metagenomics in the development of potential regulations and industrial strategies to mitigate AMR risks is a priority. The direct evidence of antimicrobial use in food products is difficult to link to outbreaks of human AMR diseases. Nevertheless, this gap in knowledge can be shrunk with studies analyzing AMR profiles between outbreak clinical isolates and animal isolates [111].
3.3. Unraveling Viral Metagenomics for Stengthening Food Safety
Most emerging diseases have originated from animal viruses capable of interspecies transmission. The intensification of livestock farming has expedited disease transmission between humans and livestock and within herds as a result of greater population and density [112]. Zoonotic pathogens are responsible for 60.3% of emerging infectious diseases (EIDs), with 70% originating from wildlife species [113,114,115]. Food consumption, direct contact, and/or indirect environmental contact are transmission routes of diseases between humans and animals [116]. Food products of animal origin are major vehicles of foodborne diseases, as they are major pathogen reservoirs [117]. Pigs and meats from wild animals and fresh produce are major repositories for known foodborne pathogens, such as norovirus and hepatitis A (HAV) and E virus (HEV) [118].
Two methods have been used in the characterization of viral communities: mNGS and virome sequencing. With mNGS, often coupled with amplicon techniques, evolutionary development, strain identification, and the prediction of the drug resistance of viruses have been studied [119]. Focused on the overall structure and interaction of microbial community present in a sample, mNGS has a reduced sensitivity for detecting low-abundance viruses, which can represent a challenge assembling the viral genome. On the other hand, virome sequencing focuses on enriching virus-like particles (VLP) prior to sequencing to enhance sensitivity for virus detection [120,121,122]. This allows for a more detailed characterization of viral communities, diversity, and dynamics; nevertheless, this type of analysis lacks the context provided with metagenomic sequencing. Despite the novelty of virome and metagenomics integration approaches and the current lack of normalized protocols [123], these procedures hold great potential in providing a holistic understanding of the interaction of a virus within the microbial community and the prediction of zoonotic diseases. Currently, viral metagenomic sequencing has been used due to its cost effectiveness and providing context for bacterial communities present. Such has been the case in the identification of viral families associated with zoonotic diseases in bats [75,124], farm animals [125,126], and non-human primates [127].
3.4. Advancing Authentication Techniques for Mitigating Global Food Fraud
Food transparency, quality, and safety are raising concerns for consumers worldwide. Nevertheless, the determination and quantification of food ingredients is hindered by difficulties in the traceability of trading channels and failures in correct labeling, stocking, and processing procedures [128]. As the global population expands and food demand rises, food adulteration has emerged as a widespread issue, impacting approximately 10–20% of globally consumed food [129]. The cost of global food fraud ranges from USD 10–60 billion, without the potential related losses [70]. The addition or substitution of food ingredients with foreign nonlabelled alternatives represents a significant dietary concern, especially for those with allergies or specific religious beliefs [130]. Several types of identification techniques have developed through the years with DNA-based molecular tools, such as PCR being the most used. However, PCR-based approaches are limited by DNA fragmentation, amplification biases, ‘primer universality’, the effect of DNA amplification inhibitors and food trace, and low yield [8,130,131,132]. It has been shown that deep metagenomic DNA sequencing of WGS is able to overcome these issues. The development of accurate analytical methods is essential for the identification and trace quantity in food products to ensure safe and ethical trade in the diverse worldwide market.
The use of NGS approaches has allowed the development of new techniques focused on the determination of the biological composition of mixed samples, such as NGS-based metabarcoding and mitochondrial metagenomics (MMG). While metabarcoding depends on the PCR amplification of a genetic marker for species identification, MMG uses mitochondrial genomes (mitogenome) as references. Currently, the search for genetic markers for the differentiation of poultry breads and meat origins has received great attention. Numerous markers, such as 12S and 16S rRNA, 18S rRNA from the nuclear genome, and the cytochrome c oxidase I (COX1, CO1 or COI) gene from the mitogenome, have been studied with PCR-related limitations [61,78,133]. The mitogenome has commonly been used in the identification of meat species, and it has been reported that its combined use with NGS has allowed an identification capacity of species down to 1% (w/w) [61,79]. While the integration of HTS into these techniques proposes a powerful for food authentication and fraud detection, its high cost, computation demand, and lack of standardization pose serious limitations. The difficulty of DNA amplification from degraded DNA in processed samples and primer bias results in an amplified DNA mixture that does not reflect the original proportion of the species, which hinders metabarcoding for the effective quantification of food composition [8]. Physical and ecological contamination sources and the need for comprehensive databases demand a standardization in protocol for an efficient metagenomic pipeline.
Food adulteration has targeted processed meat for economic gain, with an increased number of fraudulent practices [41,42,47,61,134]. Mislabeling involves primarily the substitution of declared meats with cheaper-priced protein. For instance, to beef products sheep or chicken meat is added for a cheaper product, and processed products are mixed with unexpected meats and non-declared species [42,61]. In methods targeting conserved genes for mammal barcoding [42], the effective recognition of the taxon relies on the accessibility of sequences in the reference database and their distinctiveness from sequences linked to other taxa. Although research in fresh produce fraud is still awaiting further development, the presence of not-listed and not-expected vegetable and fruit components has been proven in herbal products and fresh foods using metabarcoding techniques [135,136]. While both metagenomics and metabarcoding have restrictions regarding accurate quantification in mixed samples and a lack of regulation, their combined used for food authentication is a promising tool in current development.
4. Metagenomics of Different Types of Edible Products
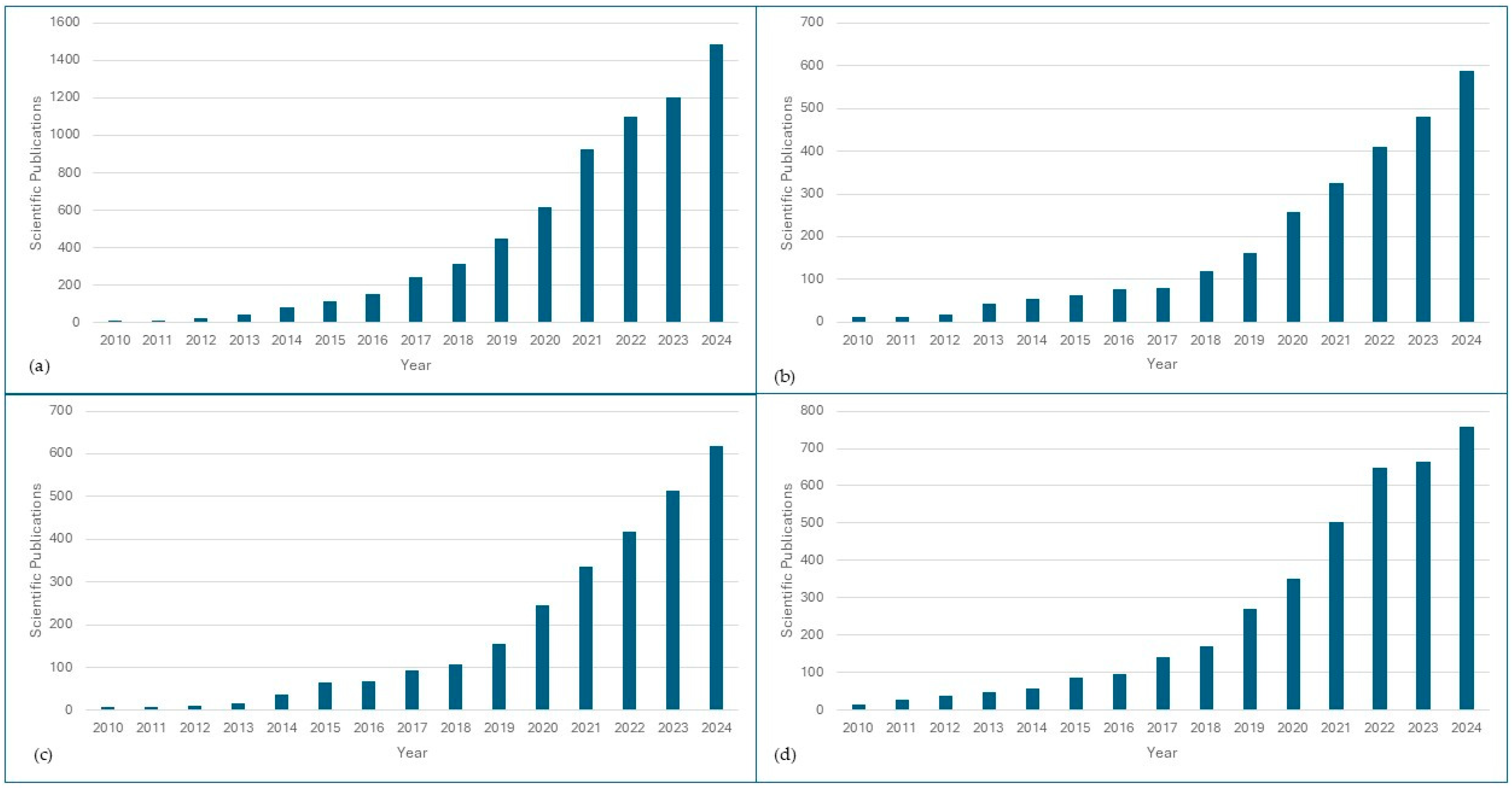
Metagenomic approaches have offered useful insights into the composition, dynamics, and functional traits of microorganisms across various food products. The increasing number of scientific publications on metagenomics and food safety reflects the growing interest in this field (Figure 1). The application of this knowledge has transformed the food industry by providing a powerful way to monitor spoilage, detect contamination, assess antibiotic resistance, and ensure product authentication. Fresh products, poultry, and meat have received significant attention in metagenomics research in view of their high consumption rates and their risks of contamination in the food chain. This section delves into the current state of metagenomics in understanding microbial dynamics in fresh produce, poultry, and meat products.
Figure 1.
Number of scientific publications available in the literature on metagenomics for food safety. (a) Scientific publications retrieved from SCOPUS using the query (metagenomics AND food safety OR microbial contamination OR foodborne pathogens OR food microbiome OR microbial diversity OR food spoilage OR food). (b) Scientific publications retrieved from SCOPUS using the query (metagenomics AND fresh produce OR vegetables OR fruit OR soil-to-fork OR postharvest). (c) Scientific publications retrieved from SCOPUS using the query (metagenomics AND meat OR meat derivatives OR cows OR dairy OR pigs OR beef OR meat products OR meat safety). (d) Scientific publications retrieved from SCOPUS using the query (metagenomics AND poultry OR chicken OR turkey OR goose OR poultry litter OR broiler).
4.1. Fresh Produce
Fresh produce, including fruits and vegetables, is an essential part of the human diet and is known to harbor large bacterial populations [80,137]. This is due to conditions that favor microbial growth, such as high-water activity, nutrient-rich tissue, and neutral pH. The microbiota associated with fresh produce has a dual role in product spoilage and health outcomes. Despite being a significant source of nutrients and a balanced diet, since it is often consumed raw, fresh produce can cause widespread foodborne outbreaks [138]. Some characterized microorganisms are responsible for the infection and microbiological spoilage of fruits and vegetables throughout the food supply chain [46,139], such as the genera Erwinia, Pectobacterium, Xanthomonas, Listeria, and Pseudomonas.
The focus has revolved around microbiome composition, diversity, and functional traits of fresh fruits and vegetables. Studies using 16S rRNA metagenomics have shown the influence of environmental factors (such as season, irrigation, and soil type) on leaf microbes [37,48] and the change of bacterial communities during cold storage (8 °C) of ready-to-eat (RTE) vegetables, with Pseudomonadales (primarily the genus Pseudomonas) being the most abundant high-level taxonomic group across samples [40,53]. Similarly, Ascomycota and Basidiomycota have been consistently identified as the prevailing phyla on grapes, tomatoes, cherries, and wild grasses [54,56], whereas Bacteroidetes and Proteobacteria have been predominant on lettuce [80]. Functional metagenomics have shed light on the influence of management practices on microbial diversity and metabolic pathways. For instance, organic orchards have shown a higher enzymatic activity of the biosynthetic pathways related to plant defense mechanisms, such as the alkaloid and aromatic amino acid pathways, due to the absence of synthetic pesticides [59]. Furthermore, changes within fungal communities have been linked to management [61], with Aureobasidium pullulans demonstrating its potential as a biocontrol agent due to its antagonistic action against several fungal pathogens.
Recent research highlights the potential of metagenomics for tracking contaminated sources, managing toxins in food supply chains, and investigating foodborne outbreaks. HTS and metagenomic tools can assist in the development of storage practices to trigger specific microbial changes that will influence fruit and vegetable quality and storability [62,63]. With these advances, potential mitigation strategies for spoilage and contamination should be at the forefront of research. Current studies could be used to refine cold storage practices and sustainable agricultural methods according to natural resistance and biocontrol. Additionally, future research should prioritize the integration of microbial functional analysis with practical applications, such as pre-/post-harvest protocols and the development of microbial inoculants that enhance shelf life.
Researchers in recent years have suggested that the microbiota and mycobiota of fresh produce are characterized by high biodiversity, and their composition is susceptible to the product. While the type of edible fraction and leaf morphology have been determined as critical factors in the growth of specific microbial communities, more information about the specific handling, washing, transport, and storage process is needed to ensure a complete assessment of microbial contamination. It is crucial to comprehend the intricate microbial ecosystem unique to each product to implement targeted control measures during harvesting, handling, and distribution to the end consumer. By exploring microbiota dynamics alongside practical solutions, a more holistic understanding of spoilage and contamination mitigation can be achieved. Therefore, researchers and the food industry must collaboratively invest more effort in exploring product biodiversity and specific microbial communities to develop and validate these control measures.
4.2. Poultry and Meat Products
The safety of meat and poultry has become a significant concern in the commercial food industry. The increase in apprehensiveness is due to several factors, including alterations in animal production, processing, and distribution methods; expanded international trade; a global increase in meat consumption; evolving consumer preferences and consumption habits, such as a preference for minimally processed foods; a higher number of consumers susceptible to infection; and increased consumer interest, awareness, and scrutiny [62,64]. Metagenomic research on meat and poultry has focused on detecting and reducing pathogenic organisms and developing safety applications for public health [63]. This analysis proves beneficial in managing pathogens and spoilage organisms, as it identifies organisms whose abundance correlates positively or negatively with the pathogens or spoilage organisms of interest. Many researchers have directed their efforts towards utilizing metagenomics to identify potential targets for influencing the microbiome of food animals before slaughter. The objectives of such studies include reducing pathogen levels within animals, modifying growth status, mitigating undesired outcomes like liver abscesses in cattle, and manipulating the nutrient profile of the produced meat.
4.2.1. Poultry
The demand for poultry products in the last years has resulted in an intensification of poultry production. Current poultry biomass represents approximately 70% of the total biomass of birds worldwide [64,71]. The multiple steps involved in poultry meat production and the commercial egg industry, from breeder and hatcheries to commercial layer and broiler flocks, increase the risk of microbial contamination. In poultry farms, the combination of bedding material, chicken excrement, and feathers plays a crucial role in pathogen development, posing a threat to the food chain. Both 16S amplicon and HTS shotgun metagenomics have been used to characterize the microbiome from the gastrointestinal tracts of poultry [72,73,140]. Lactobacillus (Firmicutes), Lachnoclostridium (Firmicutes), Clostridium (Firmicutes), and Bacteroides (Bacteroidetes) have been reported as the most dominant genera in chicken [76,104,141], with phyla diversity (Firmicutes, Bacteroidetes, and Proteobacteria) variating according to feeding patterns and environmental factors [81,82,83,141]. Foodborne pathogens, such as Staphylococcus, Clostridium, Bacillus, Listeria, and Salmonella, have also been detected in poultry [4,140,142]. Monitoring the diversity of microbiomes is essential for public health, as chicken gastrointestinal tracts (GITs) are significant reservoirs for various foodborne pathogens [143], and several spoilage species have proven resistant in cold poultry packed under aerobic conditions [144,145].
Metagenomic analysis has also been extensively applied to study the chicken gut microbiota to improve metabolism and health. In addition to identifying pathogenic microbes, metagenomic analysis is able to assess poultry performance and health through a functional approach. From metagenomic assembled genomes (MAGs) from GITs, different enzymes, genes, and six phyla (Firmicutes, Proteobacteria, Bacteroidota, Verrucomicrobiota, Actinobacteriota, and Cyanobacteria) have been identified [146,147,148,149] in an effort to understand the metabolic processes involved in the gut microbiota development of chickens. Metagenomic sequencing has proven invaluable in the understanding of a chicken’s resistome and how ARGs vary across different conditions. mNGS and deep shotgun sequencing have shown breed-specific differences in ARG prevalence [108,150,151,152], with over 50 ARGs alongside virulent genes associated with MGEs [82,151,152] and major resistance to specific antibiotics [153,154]. The virome of these domestic birds, chicken [64,151,155,156,157], turkeys [158,159], geese, and ducks [160,161], has also been an area of study, with the focus being community-based analysis of the gut microbiome.
The increase in poultry production and consumption has highlighted the importance of understanding the role of the poultry microbiome in human health, its productivity, and food safety. The application of techniques such as 16S amplicon and deep shotgun sequencing has been instrumental in understanding the functionality of bacterial diversity when integrated with metatranscriptomics. The characterization of the resistome and identifying how horizontal gene transfer (HGT) affects different domesticated birds highlights the potential these procedures have in the development of regulations. Despite the progress made, critical gaps remain, especially in the study of other poultry products. Future research integrating ‘big data’ obtained from metagenomics will be crucial in enhancing poultry health and performance, mitigating the risks of spoilage and foodborne pathogens and addressing antimicrobial resistance.
4.2.2. Meat and Derivatives
The growing population, projected to reach 9 billion by 2025 [162], and the economic development of developing countries [163,164] are the driving forces in the global demand for meat. With the rise of meat production and consumption, contamination and adulteration have become global issues. Contamination along the food production process represents a risk to humans due to recontaminated RTE meats: Taenia saginata in meat of domestic cattle; Trichinella spp. in meat of Suidae; and nontyphoidal Salmonella spp. in beef and pork meat [3,165]. On the other hand, meat is often mixed with cheaper chicken, duck, pork, and other animal meat [166], which disrupts market order and harms consumers’ rights and interests. Therefore, identifying meat and its derivatives and viral and bacterial genomes in these products is essential in improving food quality and safety and avoiding food fraud.
The bacterial community in the GITs of cattle has been studied by various techniques, such as 16S RNAs and MAGs. Around 542 genera belonging to 23 phyla have been identified, with Firmicutes, Bacteroides, and Proteobacteria predominating [100,142,167,168,169]. Common spoilage bacteria like Lactococcus, Leuconostoc, and Lactobacillus have also been identified. However, this bacterial composition drastically varies according to the feeding patterns (high/low density), cattle breed, and breeding location, with Ruminococcus, Butyrivibrio, Coprococcus, Shuttleworthia, Prevotella, and Treponema being prominent microbiota as well [65,168,170,171,172]. Nevertheless, the understanding of meat and milk production, host health, and metabolic pathways is still limited.
Metagenomics has also been a tool used to identify foodborne pathogen and contamination throughout the production pipeline. This process can involve more than 50 different operations with multiple contamination sources, such as slaughtering, leakage of gut content, and animal hides [173,174]. Furthermore, pathogens might undergo transfer from cross-contamination, fecal origin (from rearing environments such as soil, bedding, and water), and human origin. The sources of contamination have been studied, with species from the genera Pseudomonas, Staphylococcus, Brochothrix, Acinetobacter, and Psychrobacter being prominent members of the core microbiota for all facilities, with some with greater relative abundance in raw materials, food processing environments, and/or the final product [3,175,176,177]. Given the high likelihood of cross-contamination through the food supply chain, meat and its derivatives are a source of enteric infections and other zoonotic diseases originating from viruses [60,117,178]. This has caused an increase in the efforts to characterize these viruses and meat viromes. Metagenomic-derived viral sequences of parvovirus, polyomaviruses (BPyV2-SF being novel), anelloviruses, and circoviruses have been detected, and hokovirus has been identified in both pork and beef [60,157].
The resistome of cattle has been largely studied through 16S amplicon approaches to study its changes under various environments and hosts. In dairy cows, 80 distinct ARGs were identified, with approximately 60% of the deduced protein sequences to known sequences [99,167,179]. Research analyzing the impact of conventional and organic production systems identified a varied composition of the resistome in the colon but not on meat trimmings [107,180,181]. Interestingly, studies comparing AMR levels among cattle raised without antibiotics were generally similar to conventionally raised cattle [103,106]. With 43 ARGs detected, only those conferring resistance to macrolide–lincosamide–streptogramin B (MLS) and tetracyclines were more abundant in conventional beef. These results highlight the utility of metagenomic approaches in uncovering the diversity and distribution of bacterial communities and ARGs in different agricultural settings.
5. Limitations of Metagenomics for Food Safety
Metagenomics has significantly improved food safety protocols through the detection of foodborne pathogens, spoilage microorganisms, and antibiotic resistance genes. However, despite its advancements, several limitations hinder its widespread implementation. These barriers, which stem from technological constraints, computational challenges, and standardization issues, must be addressed before it can be considered as a gold standard.
One of the primary impediments to fully integrating metagenomics into food safety protocols is the high cost and computational capacity required for sequencing and bioinformatics analysis. While HTS has reduced costs, shotgun sequencing remains expensive, particularly when deep sequencing is required to detect low-abundance pathogens [6,102]. Additionally, the large volume of sequencing data represents a complication for the storage of this information and limits accessibility in routine food testing. Limited availability of public data, mainly due to the privatization of the metadata as well as the lack of bioinformatics expertise in the food industry due to the lack of user-friendly graphical interfaces [182], hinders microorganism identification efforts. Furthermore, the development of bioinformatic and big data procedures is slowed down by the increasing number of metadata [7], despite the publication of approaches such as alignment-based AFS pipeline [183]. Additionally, the use of techniques that produce massive amounts of data requires the establishment of datasets that help determine what ‘normal’ diversity is in humans, wildlife, and farm animals [64,75,126,184] in order to use approaches such as mNGS for food surveillance.
Unlike qPCR, metagenomics struggles with the accurate quantification of mixed samples and still faces sensitivity limitations. Current techniques offer relative abundance estimates, which complicates the establishment of contamination thresholds for food safety risk assessments. The amplification bias of metabarcoding and shotgun sequencing often leads to false negatives or the overrepresentation of mixed samples [8,102,106,109]. Along with the bioinformatics limitations, the lack of normalization, especially for small sample preparation, might interfere with the quality and accuracy of the metagenomic data. Different factors such as freezing time, manipulation, and sampling time can alter the microbial community, as can the contamination of rapidly growing species during the enrichment method [185].
This current situation calls for a diverse set of advancement and standardization efforts. The development of techniques focused on the improvement of pathogen detection in complex food matrices, such as host DNA depletion [98], is key to increasing sensitivity in metagenomics. Integrating functional and quantitative metagenomics will greatly improve foodborne pathogen tracking and processing the data generated by the sequencing. Functional metagenomics will allow maintaining the genetic context, such as adjacent markers of horizontal gene transfer and indicators of host bacterium or origin [186,187]. The training of machine learning as a prediction tool is an emerging area within bioinformatics and a highly interesting proposition for taxonomic studies. PICRUST (Phylogenetic Investigation of Communities by Reconstruction of Unobserved States) [188] and FASER (Functional Annotation of Sequencing Reads) [189] have recently been developed for the functional prediction of bacterial composition from the use of biomarkers and the identification of gene molecular functions corresponding to mapped reads.
While metagenomics has revolutionized food safety, numerous barriers persist, particularly in prediction and data interpretation and processing. The integration of advanced sequencing technologies, to reduce costs according to the main objective, and the implementation of bioinformatics tools promise a way of overcoming these challenges. Nevertheless, achieving standardization for the monitoring of different concerns, such as contamination, antibiotic resistance, viruses, and food authentication, and the development of user-friendly pipelines are essential for closing the gap between research and practical applications. The main limitations of the current metagenomic techniques used in the research area of the food microbiome, resistome, and virulome and possible solutions can be seen in Table 2.

Table 2.
Barriers and future prospectives for metagenomic techniques in food safety applications.
Contrasting Metabolomics and Metagenomics in Food Analysis
Metagenomic has improved the surveillance of foodborne diseases, yet these genomic approaches primarily focus on microbial identification rather than detecting harmful chemical compounds such as toxins, which have an important role in the food industry. Toxins in food can originate from microbial metabolism, fungal contamination, and environmental pollutants. Some examples include Staphylococcus aureus enterotoxins in dairy and meat [190], the botulinum toxin from Clostridium botulinum in improperly processed canned foods [191], aflatoxins from Aspergillus in grains and nuts [192], and marine toxins like saxitoxin in shellfish [193]. Since these toxins pose severe health risks and are not directly detectable through genomic techniques, molecular approaches like metabolomics are employed to analyze small molecules in food, ensuring the early detection and mitigation of contamination risks.
Metabolomics has been widely applied to qualitatively and quantitatively assess metabolites that reflect the quality, safety, and authenticity of foods. It is also used for detecting spoilage-associated microbial communities, foodborne pathogens, and chemical contaminants [194,195]. Metabolomic studies in food science encompass analyses conducted during different periods of food development processes as well as throughout the shelf life of products. The primary analytical methods in metabolomics are classified into fingerprinting and profiling. Fingerprinting focuses on determining the overall pattern of metabolites in a sample without requiring detailed identification or quantification. Profiling, on the other hand, emphasizes the identification and quantification of specific compounds present in the sample [196]. Metabolomics has proven to be a powerful tool in food analysis, as it enables the simultaneous determination of multiple components. However, several limitations restrict its broader application. These include the lack of comprehensive food-related databases and advanced data processing technologies capable of handling the large volumes of data generated. Another significant challenge lies in the limited coverage of metabolites due to their sensitivity and stability, which often necessitates specific environmental conditions for detection by the analytical equipment used [195,197,198].
Both metagenomics and metabolomics are powerful tools in food analysis, each offering unique insights into different aspects of food safety, quality, and authenticity. While metagenomics focuses on detecting and characterizing microorganisms, including pathogens and beneficial microbes, metabolomics provides a detailed chemical profile of food composition and contaminants. Their combined use may allow for a more comprehensive and holistic approach, integrating microbial community analysis with metabolic profiling to enhance food safety, improve quality control, and optimize both food and crop production processes.
6. Conclusions
Metagenomics has emerged as a groundbreaking tool in food analysis, offering insights into various products’ microbial communities, including fruits, vegetables, poultry, and meat. The evolution of sequencing technologies, from the use of amplicon-based methods to metabarcoding, has enhanced the characterization of food-associated microbiomes, tracks pathogens, and assesses the presence of antibiotic resistance genes. The integration of HTS and NGS platforms has transformed food microbiology, offering real-time surveillance and rapid pathogen identification. Nonetheless, metagenomics still faces several obstacles that hinder its application in standardized food safety protocols. The high cost of deep sequencing and the complexity of bioinformatics pipelines pose significant limitations to its widespread implementation, which encumbers the normalization of protocols needed for food analysis regulation. Despite the identification of microbial taxa and functional genes, metagenomic approaches remain limited in the quantification of viable pathogens and contaminants. Tackling these obstacles requires more efficient computational tools, cost-effective sequencing methods, and a standard analytical pipeline.
As food demand increases, reliable and comprehensive food safety measures are greatly required. The continued integration of this approach with other -omic technologies will enhance its potential as a food safety standard, ensuring product authenticity and optimizing the food supply chain with reduced contaminants and economic loss. Refining sequencing approaches and enhancing bioinformatics capabilities are the key points of the next phase in the evolution of metagenomics. Future research will be able to accurately identify microbial communities, ARGs, and viruses and provide a precise risk assessment. By overcoming these barriers, metagenomics can become a keystone approach for ensuring global food safety and public health.
Author Contributions
Conceptualization, L.O.M.-M. and E.F.F.D.L.S.; methodology, T.I.M.-M., B.R.-H., Y.F., C.R., M.R.-M., J.A., R.T.R., E.F.F.D.L.S. and L.O.M.-M.; validation, L.O.M.-M. and E.F.F.D.L.S.; formal analysis, T.I.M.-M., Y.F., C.R., B.R.-H., M.R.-M. and J.A.; investigation, T.I.M.-M., B.R.-H., M.R.-M. and J.A.; resources, R.T.R., L.O.M.-M. and E.F.F.D.L.S.; data curation, T.I.M.-M., B.R.-H., M.R.-M., Y.F., C.R. and J.A.; writing—original draft preparation, T.I.M.-M., B.R.-H., M.R.-M., Y.F., C.R. and J.A.; writing—review and editing, R.T.R., L.O.M.-M. and E.F.F.D.L.S.; supervision, R.T.R., L.O.M.-M. and E.F.F.D.L.S.; project administration, L.O.M.-M. and E.F.F.D.L.S.; funding acquisition, R.T.R., L.O.M.-M. and E.F.F.D.L.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Ministerio de Educación Superior Ciencia y Tecnología (MESCYT) through Fondo Nacional de Innovación y Desarrollo Científico y Tecnológico (FONDOCYT), grant number 2022-2A4-204: Una Sola Salud-Inocuidad Alimentaria. Evaluación De Riesgos Microbiológicos Patogénicos En Carnes De Cerdo Y Pollo, Y Subproductos.
Data Availability Statement
Not applicable.
Acknowledgments
This research project was successfully conducted thanks to the support provided by the Research Vice-Rectory and the Deanship of Basic and Environmental Sciences at the Instituto Tecnologico de Santo Domingo (INTEC).
Conflicts of Interest
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Duchenne-Moutien, R.A.; Neetoo, H. Climate Change and Emerging Food Safety Issues: A Review. J. Food Prot. 2021, 84, 1884–1897. [Google Scholar] [CrossRef] [PubMed]
- Sequino, G.; Valentino, V.; Torrieri, E.; De Filippis, F. Specific Microbial Communities Are Selected in Minimally-Processed Fruit and Vegetables according to the Type of Product. Foods 2022, 11, 2164. [Google Scholar] [CrossRef] [PubMed]
- Bonaldo, F.; Avot, B.J.P.; De Cesare, A.; Aarestrup, F.M.; Otani, S. Foodborne Pathogen Dynamics in Meat and Meat Analogues Analysed Using Traditional Microbiology and Metagenomic Sequencing. Antibiotics 2024, 13, 16. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.I.; Kim, S.A.; Park, S.H.; Ricke, S.C. Molecular and New-Generation Techniques for Rapid Detection of Foodborne Pathogens and Characterization of Microbial Communities in Poultry Meat. In Food Safety in Poultry Meat Production; Springer: Berlin/Heidelberg, Germany, 2019; pp. 235–260. [Google Scholar] [CrossRef]
- Yang, X.; Noyes, N.R.; Doster, E.; Martin, J.N.; Linke, L.M.; Magnuson, R.J.; Yang, H.; Geornaras, I.; Woerner, D.R.; Jones, K.L.; et al. Use of Metagenomic Shotgun Sequencing Technology to Detect Foodborne Pathogens within the Microbiome of the Beef Production Chain. Appl. Environ. Microbiol. 2016, 82, 2433. [Google Scholar] [CrossRef] [PubMed]
- Emiola, A.; Oh, J. High throughput in situ metagenomic measurement of bacterial replication at ultra-low sequencing coverage. Nat. Commun. 2018, 9, 4956. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Liu, M.; Yang, J. Recovering metagenome-assembled genomes from shotgun metagenomic se-quencing data: Methods, applications, challenges, and opportunities. Microbiol. Res. 2022, 260, 127023. [Google Scholar] [CrossRef]
- Bruno, A.; Sandionigi, A.; Agostinetto, G.; Bernabovi, L.; Frigerio, J.; Casiraghi, M.; Labra, M. Food Tracking Perspective: DNA Metabarcoding to Identify Plant Composition in Complex and Processed Food Products. Genes 2019, 10, 248. [Google Scholar] [CrossRef]
- Johnson, J.S.; Spakowicz, D.J.; Hong, B.Y.; Petersen, L.M.; Demkowicz, P.; Chen, L.; Leopold, S.R.; Hanson, B.M.; Agresta, H.O.; Gerstein, M. Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nat. Commun. 2019, 10, 5029. [Google Scholar] [CrossRef]
- Woese, C.R.; Fox, G.E. Phylogenetic structure of the prokaryotic domain: The primary kingdoms. Proc. Natl. Acad. Sci. USA 1977, 74, 5088–5090. [Google Scholar] [CrossRef]
- Liu, Y.X.; Qin, Y.; Chen, T.; Lu, M.; Qian, X.; Guo, X.; Bai, Y. A practical guide to amplicon and metagenomic analysis of microbiome data. Protein Cell 2021, 12, 315–330. [Google Scholar] [CrossRef]
- Kinoshita, Y.; Niwa, H.; Uchida-Fujii, E.; Nukada, T. Establishment and assessment of an amplicon sequencing method targeting the 16S-ITS-23S rRNA operon for analysis of the equine gut microbiome. Sci. Rep. 2021, 11, 11884. [Google Scholar] [CrossRef] [PubMed]
- Franzosa, E.A.; Hsu, T.; Sirota-Madi, A.; Shafquat, A.; Abu-Ali, G.; Morgan, X.C.; Huttenhower, C. Sequencing and beyond: Integrating molecular “omics” for microbial community profiling. Nat. Rev. Microbiol. 2015, 13, 360–372. [Google Scholar] [CrossRef]
- Kamble, A.; Sawant, S.; Singh, H. 16S ribosomal RNA gene-based metagenomics: A review. Biomed. Res. J. 2020, 7, 5. [Google Scholar] [CrossRef]
- Quince, C.; Walker, A.W.; Simpson, J.T.; Loman, N.J.; Segata, N. Shotgun metagenomics, from sampling to analysis. Nat. Biotechnol. 2017, 35, 833–844. [Google Scholar] [CrossRef]
- Zhang, L.; Chen, F.X.; Zeng, Z.; Xu, M.; Sun, F.; Yang, L.; Bi, X.; Lin, Y.; Gao, Y.J.; Hao, H.X.; et al. Advances in Metagenomics and Its Application in Environmental Microorganisms. Front. Microbiol. 2021, 12, 766364. [Google Scholar] [CrossRef] [PubMed]
- Milani, C.; Alessandri, G.; Mangifesta, M.; Mancabelli, L.; Lugli, G.A.; Fontana, F.; Longhi, G.; Anzalone, R.; Viappiani, A.; Duranti, S. Untangling Species-Level Composition of Complex Bacterial Communities through a Novel Metagenomic Approach. mSystems 2020, 5, e00404-20. [Google Scholar] [CrossRef] [PubMed]
- Buytaers, F.E.; Verhaegen, B.; Van Nieuwenhuysen, T.; Roosens, N.H.C.; Vanneste, K.; Marchal, K.; De Keersmaecker, S.C.J. Strain-level characterization of foodborne pathogens without culture enrichment for outbreak investigation using shotgun metagenomics facilitated with nanopore adaptive sampling. Front. Microbiol. 2024, 15, 1330814. [Google Scholar] [CrossRef]
- Qin, J.; Li, R.; Raes, J.; Arumugam, M.; Burgdorf, K.S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; Yamada, T.; et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010, 464, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Ning, K.; Tong, Y. The Fast Track for Microbiome Research. Genom. Proteom. Bioinform. 2019, 17, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Li, X.; Yang, L.; Liu, C.; Wang, Q.; Chi, W.; Zhu, H. How Microbes Shape Their Communities? A Microbial Community Model Based on Functional Genes. Genom. Proteom. Bioinform. 2019, 17, 91–105. [Google Scholar] [CrossRef] [PubMed]
- O’Flaherty, S.; Klaenhammer, T.R. The impact of omic technologies on the study of food microbes. Annu. Rev. Food Sci. Technol. 2011, 2, 353–371. [Google Scholar] [CrossRef] [PubMed]
- Juszczuk-Kubiak, E.; Greguła-Kania, M.; Sokołowska, B. “Food-Omics” Applications in the Food Metagenom Profiling. Postępy Mikrobiol. Adv. Microbiol. 2021, 60, 59–75. [Google Scholar] [CrossRef]
- Jagadeesan, B.; Gerner-Smidt, P.; Allard, M.W.; Leuillet, S.; Winkler, A.; Xiao, Y.; Chaffron, S.; Van Der Vossen, J.; Tang, S.; Katase, M.; et al. The use of next generation sequencing for improving food safety: Translation into practice. Food Microbiol. 2019, 79, 96–115. [Google Scholar] [CrossRef] [PubMed]
- Hatti-Kaul, R.; Chen, L.; Dishisha, T.; Enshasy, H. El: Lactic acid bacteria: From starter cultures to producers of chemicals. FEMS Microbiol. Lett. 2018, 365, 213. [Google Scholar] [CrossRef] [PubMed]
- Yap, M.; Ercolini, D.; Álvarez-Ordóñez, A.; O’Toole, P.W.; O’Sullivan, O.; Cotter, P.D. Next-Generation Food Research: Use of Meta-Omic Approaches for Characterizing Microbial Communities Along the Food Chain. Annu. Rev. Food Sci. Technol. 2022, 13, 361–384. [Google Scholar] [CrossRef]
- Gerner-Smidt, P.; Hyytia-Trees, E.; Barrett, T.J. Molecular Source Tracking and Molecular Subtyping. In Food Microbiology: Fundamentals and Frontiers; Wiley: Hoboken, NJ, USA, 2014; pp. 1059–1077. [Google Scholar] [CrossRef]
- Huttenhower, C.; Gevers, D.; Knight, R.; Abubucker, S.; Badger, J.H.; Chinwalla, A.T.; Creasy, H.H.; Earl, A.M.; Fitzgerald, M.G.; Fulton, R.S.; et al. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [Google Scholar] [CrossRef]
- Walsh, A.M.; Crispie, F.; O’Sullivan, O.; Finnegan, L.; Claesson, M.J.; Cotter, P.D. Species classifier choice is a key consideration when analysing low-complexity food microbiome data. Microbiome 2018, 6, 50. [Google Scholar] [CrossRef] [PubMed]
- Jovel, J.; Patterson, J.; Wang, W.; Hotte, N.; O’Keefe, S.; Mitchel, T.; Perry, T.; Kao, D.; Mason, A.L.; Madsen, K.L.; et al. Characterization of the gut microbiome using 16S or shotgun metagenomics. Front. Microbiol. 2016, 7, 180723. [Google Scholar] [CrossRef]
- Emiola, A.; Zhou, W.; Oh, J. Metagenomic growth rate inferences of strains in situ. Sci. Adv. 2020, 6, eaaz2299. [Google Scholar] [CrossRef]
- Venter, J.C.; Remington, K.; Heidelberg, J.F.; Halpern, A.L.; Rusch, D.; Eisen, J.A.; Wu, D.; Paulsen, I.; Nelson, K.E.; Nelson, W.; et al. Environmental Genome Shotgun Sequencing of the Sargasso Sea. Science 2004, 304, 66–74. [Google Scholar] [CrossRef] [PubMed]
- Baker, B.J.; Tyson, G.W.; Webb, R.I.; Flanagan, J.; Hugenholtz, P.; Allen, E.E.; Banfield, J.F. Lineages of acidophilic archaea revealed by community genomic analysis. Science 2006, 314, 1933–1935. [Google Scholar] [CrossRef] [PubMed]
- Riesenfeld, S.J.; Pollard, K.S. Beyond classification: Gene-family phylogenies from shotgun metagenomic reads enable accurate community analysis. BMC Genom. 2013, 14, 419. [Google Scholar] [CrossRef]
- Khan, A.S.; Afrin, S.; Ahmed, F.; Rahman, S.R. Shotgun metagenomic analysis reveals the emergence of plasmid-encoded mcr-5.1 gene in hospital wastewater in Bangladesh. J. Glob. Antimicrob. Resist. 2024, 39, 22–26. [Google Scholar] [CrossRef]
- Knight, R.; Jansson, J.; Field, D.; Fierer, N.; Desai, N.; Fuhrman, J.A.; Hugenholtz, P.; Van Der Lelie, D.; Meyer, F.; Stevens, R.; et al. Unlocking the potential of metagenomics through replicated experimental design. Nat. Biotechnol. 2012, 30, 513–520. [Google Scholar] [CrossRef]
- Fanning, S.; Proos, S.; Jordan, K.; Srikumar, S. A Review on the Applications of Next Generation Sequencing Technologies as Applied to Food-Related Microbiome Studies. Front. Microbiol. 2017, 8, 1829. [Google Scholar] [CrossRef]
- Verce, M.; De Vuyst, L.; Weckx, S. Shotgun Metagenomics of a Water Kefir Fermentation Ecosystem Reveals a Novel Oenococcus Species. Front. Microbiol. 2019, 10, 479. [Google Scholar] [CrossRef] [PubMed]
- Treiber, F.M.; Beranek-Knauer, H. Antimicrobial Residues in Food from Animal Origin—A Review of the Literature Focusing on Products Collected in Stores and Markets Worldwide. Antibiotics 2021, 10, 534. [Google Scholar] [CrossRef]
- Mullany, P. Functional metagenomics for the investigation of antibiotic resistance. Virulence 2014, 5, 443–447. [Google Scholar] [CrossRef] [PubMed]
- Pazartzi, T.; Siaperopoulou, S.; Gubili, C.; Maradidou, S.; Loukovitis, D.; Chatzispyrou, A.; Griffiths, A.M.; Minos, G.; Imsiridou, A. High levels of mislabeling in shark meat—Investigating patterns of species utilization with DNA barcoding in Greek retailers. Food Control 2019, 98, 179–186. [Google Scholar] [CrossRef]
- Xing, R.R.; Wang, N.; Hu, R.R.; Zhang, J.K.; Han, J.X.; Chen, Y. Application of next generation sequencing for species identification in meat and poultry products: A DNA metabarcoding approach. Food Control 2019, 101, 173–179. [Google Scholar] [CrossRef]
- Yasir, M.; Al-Zahrani, I.A.; Bibi, F.; Abd El Ghany, M.; Azhar, E.I. New insights of bacterial com-munities in fermented vegetables from shotgun metagenomics and identification of antibiotic resistance genes and probiotic bacteria. Food Res. Int. 2022, 157, 111190. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Pop, M. ARDB—Antibiotic Resistance Genes Database. Nucleic Acids Res. 2009, 37, D443–D447. [Google Scholar] [CrossRef]
- Bengtsson-Palme, J.; Kristiansson, E.; Larsson, D.G.J. Environmental factors influencing the devel-opment and spread of antibiotic resistance. FEMS Microbiol. Rev. 2018, 42, 68–80. [Google Scholar] [CrossRef] [PubMed]
- Alegbeleye, O.; Odeyemi, O.A.; Strateva, M.; Stratev, D. Microbial spoilage of vegetables, fruits and cereals. Appl. Food Res. 2022, 2, 100122. [Google Scholar] [CrossRef]
- Almerón-Souza, F.; Sperb, C.; Castilho, C.L.; Figueiredo, P.I.C.C.; Gonçalves, L.T.; Machado, R.; Oliveira, L.R.; Valiati, V.H.; Fagundes, N.J.R. Molecular identification of shark meat from local markets in Southern Brazil based on DNA barcoding: Evidence for mislabeling and trade of endangered species. Front. Genet. 2018, 9, 326887. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.S.; Bae, J.W.; Park, E.J. Geographic and Host-Associated Variations in Bacterial Communities on the Floret Surfaces of Field-Grown Broccoli. Appl. Environ. Microbiol. 2018, 84, e02837-17. [Google Scholar] [CrossRef]
- Söderqvist, K.; Ahmed Osman, O.; Wolff, C.; Bertilsson, S.; Vågsholm, I.; Boqvist, S. Emerging microbiota during cold storage and temperature abuse of ready-to-eat salad. Infect. Ecol. Epidemiol. 2017, 7, 1328963. [Google Scholar] [CrossRef]
- Charalampous, T.; Kay, G.L.; Richardson, H.; Aydin, A.; Baldan, R.; Jeanes, C.; Rae, D.; Grundy, S.; Turner, D.J.; Wain, J.; et al. Nanopore metagenomics enables rapid clinical diagnosis of bacterial lower respiratory infection. Nat. Biotechnol. 2019, 37, 783–792. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.M.; Cao, W.W.; Liang, S.L.; Yamasaki, S.; Chen, X.; Shi, L.; Ye, L. Metagenomic characterization of bacterial community and antibiotic resistance genes in representative ready-to-eat food in southern China. Sci. Rep. 2020, 10, 15175. [Google Scholar] [CrossRef]
- Cadena, M.; Durso, L.M.; Miller, D.N.; Waldrip, H.M.; Castleberry, B.L.; Drijber, R.A.; Wortmann, C. Tetracycline and sulfonamide antibiotic resistance genes in soils from nebraska organic farming operations. Front. Microbiol. 2018, 9, 371278. [Google Scholar] [CrossRef] [PubMed]
- Tatsika, S.; Karamanoli, K.; Karayanni, H.; Genitsaris, S. Metagenomic Characterization of Bacterial Communities on Ready-to-Eat Vegetables and Effects of Household Washing on their Diversity and Composition. Pathogens 2019, 8, 37. [Google Scholar] [CrossRef] [PubMed]
- Kokaeva, L.; Chudinova, E.; Berezov, A.; Yarmeeva, M.; Balabko, P.; Belosokhov, A.; Elansky, S. Fungal diversity in tomato (Solanum lycopersicum) leaves and fruits in Russia. J. Cent. Eur. Agric. 2020, 21, 809–816. [Google Scholar] [CrossRef]
- Li, L.; Xiao, Y.; Wang, C.; Olsen, R.H.; Meng, H.; Shi, L. Exploring the resistome, virulome, mobilome and microbiome along pork production chain using metagenomics. Int. J. Food Microbiol. 2022, 371, 109674. [Google Scholar] [CrossRef] [PubMed]
- Stanevičienė, R.; Lukša, J.; Strazdaitė-žielienė, Ž.; Ravoitytė, B.; Losinska-sičiūnienė, R.; Mozūraitis, R.; Servi-enė, E. Mycobiota in the carposphere of sour and sweet cherries and antagonistic features of potential bio-control yeasts. Microorganisms 2021, 9, 1423. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Santoni, S.; Weber, A.; This, P.; Péros, J.P. Understanding the phyllosphere microbiome assemblage in grape species (Vitaceae) with amplicon sequence data structures. Sci. Rep. 2019, 9, 14294. [Google Scholar] [CrossRef]
- Milián-García, Y.; Young, R.; Madden, M.; Bullas-Appleton, E.; Hanner, R.H. Optimization and validation of a cost-effective protocol for biosurveillance of invasive alien species. Ecol. Evol. 2021, 11, 1999. [Google Scholar] [CrossRef] [PubMed]
- Bartuv, R.; Berihu, M.; Medina, S.; Salim, S.; Feygenberg, O.; Faigenboim-Doron, A.; Zhimo, V.Y.; Abdelfattah, A.; Piombo, E.; Wisniewski, M.; et al. Functional analysis of the apple fruit microbiome based on shotgun metagenomic sequencing of conventional and organic orchard samples. Environ. Microbiol. 2023, 25, 1728–1746. [Google Scholar] [CrossRef]
- Cibulski, S.; Alves de Lima, D.; Fernandes dos Santos, H.; Teixeira, T.F.; Tochetto, C.; Mayer, F.Q.; Roehe, P.M. A plate of viruses: Viral metagenomics of supermarket chicken, pork and beef from Brazil. Virology 2021, 552, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Angeli, D.; Sare, A.R.; Jijakli, M.H.; Pertot, I.; Massart, S. Insights gained from metagenomic shotgun sequencing of apple fruit epiphytic microbiota. Postharvest Biol. Technol. 2019, 153, 96–106. [Google Scholar] [CrossRef]
- Padmaperuma, G.; Butler, T.O.; Shuhaili, F.A.B.A.; Almalki, W.J.; Vaidyanathan, S. Microbial consortia: Concept and application in fruit crop management. In Fruit Crops Diagnosis and Management of Nutrient Constraints; Elsevier: Amsterdam, The Netherlands, 2019; pp. 353–366. [Google Scholar] [CrossRef]
- Kusstatscher, P.; Cernava, T.; Abdelfattah, A.; Gokul, J.; Korsten, L.; Berg, G. Microbiome approaches provide the key to biologically control postharvest pathogens and storability of fruits and vegetables. FEMS Microbiol. Ecol. 2020, 96, 119. [Google Scholar] [CrossRef]
- Sofos, J.N. Challenges to meat safety in the 21st century. Meat. Sci. 2008, 78, 3–13. [Google Scholar] [CrossRef]
- Xie, Y.; Sun, H.; Xue, M.; Liu, J. Metagenomics reveals differences in microbial composition and metabolic functions in the rumen of dairy cows with different residual feed intake. Anim. Microbiome 2022, 4, 19. [Google Scholar] [CrossRef] [PubMed]
- Chaora, N.S.; Khanyile, K.S.; Magwedere, K.; Pierneef, R.; Tabit, F.T.; Muchadeyi, F.C. A 16S Next Generation Sequencing Based Molecular and Bioinformatics Pipeline to Identify Processed Meat Products Contamination and Mislabelling. Animals 2022, 12, 416. [Google Scholar] [CrossRef]
- Flint, A.; Cooper, A.; Rao, M.; Weedmark, K.; Carrillo, C.; Tamber, S. Targeted metagenomics using bait-capture to detect antibiotic resistance genes in retail meat and seafood. Front. Microbiol. 2023, 14, 1188872. [Google Scholar] [CrossRef] [PubMed]
- Weinroth, M.D.; Noyes, N.R.; Morley, P.M.; Belk, K.E. Metagenomics of Meat and Poultry. In Food Microbiology Fundamentals and Frontiers; Wiley: Hoboken, NJ, USA, 2019; pp. 939–962. [Google Scholar] [CrossRef]
- François, S.; Pybus, O.G. Towards an understanding of the avian virome. J. Gen. Virol. 2020, 101, 785–790. [Google Scholar] [CrossRef] [PubMed]
- Robson, K.; Dean, M.; Haughey, S.; Elliott, C. A comprehensive review of food fraud terminologies and food fraud mitigation guides. Food Control. 2021, 120, 107516. [Google Scholar] [CrossRef]
- Bar-On, Y.M.; Phillips, R.; Milo, R. The biomass distribution on Earth. Proc. Natl. Acad. Sci. USA 2018, 115, 6506–6511. [Google Scholar] [CrossRef]
- Wilkinson, T.J.; Cowan, A.A.; Vallin, H.E.; Onime, L.A.; Oyama, L.B.; Cameron, S.J.; Gonot, C.; Moorby, J.M.; Waddams, K.; Theobald, V.J.; et al. Charac-terization of the microbiome along the gastrointestinal tract of growing Turkeys. Front. Microbiol. 2017, 8, 259475. [Google Scholar] [CrossRef]
- Pan, D.; Yu, Z. Intestinal microbiome of poultry and its interaction with host and diet. Gut Microbes 2014, 5, 108–119. [Google Scholar] [CrossRef] [PubMed]
- Min, B.R.; Wright, C.; Ho, P.; Eun, J.-S.; Gurung, N.; Shange, R. The Effect of Phytochemical Tannins-Containing Diet on Rumen Fermentation Characteristics and Microbial Diversity Dynamics in Goats Using 16S rDNA Amplicon Pyrosequencing. Agric. Food Anal. Bacteriol. 2014, 4, 141909. [Google Scholar]
- Van Brussel, K.; Holmes, E.C. Zoonotic disease and virome diversity in bats. Curr. Opin. Virol. 2021, 52, 192. [Google Scholar] [CrossRef]
- Lou, C.; Chen, Z.; Bai, Y.; Chai, T.; Guan, Y.; Wu, B. Exploring the Microbial Community Structure in the Chicken House Environment by Metagenomic Analysis. Animals 2024, 14, 55. [Google Scholar] [CrossRef]
- Gohl, D.M.; Vangay, P.; Garbe, J.; MacLean, A.; Hauge, A.; Becker, A.; Gould, T.J.; Clayton, J.B.; Johnson, T.J.; Hunter, R.; et al. Systematic improvement of amplicon marker gene methods for increased accuracy in microbiome studies. Nat. Biotechnol. 2016, 34, 942–949. [Google Scholar] [CrossRef] [PubMed]
- Rennstam Rubbmark, O.; Sint, D.; Horngacher, N.; Traugott, M. A broadly applicable COI primer pair and an efficient single-tube amplicon library preparation protocol for metabarcoding. Ecol. Evol. 2018, 8, 12335–12350. [Google Scholar] [CrossRef]
- Cottenet, G.; Blancpain, C.; Chuah, P.F.; Cavin, C. Evaluation and application of a next generation sequencing approach for meat species identification. Food Control. 2020, 110, 107003. [Google Scholar] [CrossRef]
- Rastogi, G.; Sbodio, A.; Tech, J.J.; Suslow, T.V.; Coaker, G.L.; Leveau, J.H.J. Leaf microbiota in an agroecosystem: Spatiotemporal variation in bacterial community composition on field-grown lettuce. ISME J. 2012, 6, 1812. [Google Scholar] [CrossRef] [PubMed]
- Andreani, N.A.; Donaldson, C.J.; Goddard, M. A reasonable correlation between cloacal and cecal microbiomes in broiler chickens. Poult. Sci. 2020, 99, 6062–6070. [Google Scholar] [CrossRef]
- Eckstrom, K.; Barlow, J.W. Resistome metagenomics from plate to farm: The resistome and microbial composition during food waste feeding and composting on a Vermont poultry farm. PLoS ONE 2019, 14, e0219807. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Lv, H.; Song, Y.; Sun, C.; Zhang, Z.; Chen, S. Community composition of cecal microbiota in commercial yellow broilers with high and low feed efficiencies. Poult. Sci. 2021, 100, 100996. [Google Scholar] [CrossRef] [PubMed]
- Petruzzi, L.; Corbo, M.R.; Sinigaglia, M.; Bevilacqua, A. Microbial Spoilage of Foods: Fundamentals. In The Microbiological Quality of Food Foodborne Spoilers; Elsevier: Amsterdam, The Netherlands, 2017; pp. 1–21. [Google Scholar] [CrossRef]
- Di Renzo, L.; Colica, C.; Carraro, A.; Cenci Goga, B.; Marsella, L.T.; Botta, R.; Colombo, M.L.; Gratteri, S.; Chang, T.F.M.; Droli, M.; et al. Food safety and nutritional quality for the prevention of non communicable diseases: The Nutrient, hazard Analysis and Critical Control Point process (NACCP). J. Transl. Med. 2015, 13, 1–13. [Google Scholar] [CrossRef]
- Buytaers, F.E.; Saltykova, A.; Denayer, S.; Verhaegen, B.; Vanneste, K.; Roosens, N.H.C.; Piérard, D.; Marchal, K.; De Keersmaecker, S.C.J. A Practical Method to Implement Strain-Level Metagenomics-Based Foodborne Outbreak In-vestigation and Source Tracking in Routine. Microorganisms 2020, 8, 1191. [Google Scholar] [CrossRef]
- Odeyemi, O.A.; Alegbeleye, O.O.; Strateva, M.; Stratev, D. Understanding spoilage microbial community and spoilage mechanisms in foods of animal origin. Compr. Rev. Food Sci. Food Saf. 2020, 19, 311–331. [Google Scholar] [CrossRef]
- Siraj, K.; Kathireshan, A.; Gayathri, G. Amzad Basha: Metagenomic Analysis of Bacterial Communities in Food Spoilage. Eur. Chem. Bull. 2023, 12, 7693–7707. [Google Scholar]
- Leech, J.; Cabrera-Rubio, R.; Walsh, A.M.; Macori, G.; Walsh, C.J.; Barton, W.; Finnegan, L.; Crispie, F.; O’Sullivan, O.; Claesson, M.J.; et al. Fermented-Food Metagenomics Reveals Substrate-Associated Differences in Taxonomy and Health-Associated and Antibiotic Resistance Determinants. mSystems 2020, 5, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- de Kerdrel, G.A.; Andersen, J.C.; Kennedy, S.R.; Gillespie, R.; Krehenwinkel, H. Rapid and cost-effective generation of single specimen multilocus barcoding data from whole arthropod commu-nities by multiple levels of multiplexing. Sci. Rep. 2020, 10, 78. [Google Scholar] [CrossRef]
- Lamb, P.D.; Hunter, E.; Pinnegar, J.K.; Creer, S.; Davies, R.G.; Taylor, M.I. How quantitative is metabarcoding: A meta-analytical approach. Mol. Ecol. 2019, 28, 420–430. [Google Scholar] [CrossRef]
- Rausch, P.; Rühlemann, M.; Hermes, B.M.; Doms, S.; Dagan, T.; Dierking, K.; Domin, H.; Fraune, S.; Von Frieling, J.; Hentschel, U.; et al. Comparative analysis of amplicon and metagenomic sequencing methods reveals key features in the evolution of animal metaorganisms. Microbiome 2019, 7, 133. [Google Scholar] [CrossRef]
- Ranjan, R.; Rani, A.; Metwally, A.; McGee, H.S.; Perkins, D.L. Analysis of the microbiome: Advantages of whole genome shotgun versus 16S amplicon sequencing. Biochem. Biophys. Res. Commun. 2016, 469, 967–977. [Google Scholar] [CrossRef] [PubMed]
- Van Dyk, B.N.; De Bruin, W.; Du Plessis, E.M.; Korsten, L. Microbiological Food Safety Status of Commercially Produced Tomatoes from Production to Marketing. J. Food Prot. 2016, 79, 392–406. [Google Scholar] [CrossRef] [PubMed]
- Card, R.M.; Warburton, P.J.; MacLaren, N.; Mullany, P.; Allan, E.; Anjum, M.F. Application of Mi-croarray and Functional-Based Screening Methods for the Detection of Antimicrobial Resistance Genes in the Microbiomes of Healthy Humans. PLoS ONE 2014, 9, e86428. [Google Scholar] [CrossRef]
- He, Y.; Yuan, Q.; Mathieu, J.; Stadler, L.; Senehi, N.; Sun, R.; Alvarez, P.J.J. Antibiotic resistance genes from livestock waste: Occurrence, dissemination, and treatment. npj Clean Water 2020, 3, 4. [Google Scholar] [CrossRef]
- McArthur, A.G.; Waglechner, N.; Nizam, F.; Yan, A.; Azad, M.A.; Baylay, A.J.; Bhullar, K.; Canova, M.J.; De Pascale, G.; Ejim, L.; et al. The comprehensive antibiotic resistance database. Antimicrob. Agents Chemother. 2013, 57, 3348–3357. [Google Scholar] [CrossRef] [PubMed]
- Bloomfield, S.J.; Zomer, A.L.; O’Grady, J.; Kay, G.L.; Wain, J.; Janecko, N.; Palau, R.; Mather, A.E. Determination and quantification of microbial communities and antimicrobial resistance on food through host DNA-depleted metagenomics. Food Microbiol. 2023, 110, 104162. [Google Scholar] [CrossRef] [PubMed]
- Pitta, D.W.; Dou, Z.; Kumar, S.; Indugu, N.; Toth, J.D.; Vecchiarelli, B.; Bhukya, B. Metagenomic Evidence of the Prevalence and Distribution Patterns of Antimicrobial Resistance Genes in Dairy Agroecosystems. Foodborne Pathog. Dis. 2016, 13, 296–302. [Google Scholar] [CrossRef]
- Doster, E.; Thomas, K.M.; Weinroth, M.D.; Parker, J.K.; Crone, K.K.; Arthur, T.M.; Schmidt, J.W.; Wheeler, T.L.; Belk, K.E.; Morley, P.S. Metagenomic Characterization of the Microbiome and Resistome of Retail Ground Beef Products. Front. Microbiol. 2020, 11, 541972. [Google Scholar] [CrossRef]
- Apjok, G.; Számel, M.; Christodoulou, C.; Seregi, V.; Vásárhelyi, B.M.; Stirling, T.; Eszenyi, B.; Sári, T.; Vidovics, F.; Nagrand, E.; et al. Characterization of antibiotic resistomes by repro-grammed bacteriophage-enabled functional metagenomics in clinical strains. Nat. Microbiol. 2023, 8, 410–423. [Google Scholar] [CrossRef] [PubMed]
- Mathieu, E.; Léjard, V.; Ezzine, C.; Govindin, P.; Morat, A.; Giat, M.; Lapaque, N.; Doré, J.; Blottière, H.M. An Insight into Functional Metagenomics: A High-Throughput Approach to Decipher Food–Microbiota–Host Interactions in the Human Gut. Int. J. Mol. Sci. 2023, 24, 17630. [Google Scholar] [CrossRef] [PubMed]
- Auffret, M.D.; Dewhurst, R.J.; Duthie, C.A.; Rooke, J.A.; John Wallace, R.; Freeman, T.C.; Stewart, R.; Watson, M.; Roehe, R. The rumen microbiome as a reservoir of antimicrobial resistance and patho-genicity genes is directly affected by diet in beef cattle. Microbiome 2017, 5, 159. [Google Scholar] [CrossRef]
- Panyako, P.M.; Ommeh, S.C.; Kuria, S.N.; Lichoti, J.K.; Musina, J.; Nair, V.; Nene, V.; Munir, M.; Oyola, S.O. Metagenomic Characterization of Poultry Cloacal and Oropharyngeal Swabs in Kenya Reveals Bacterial Pathogens and Their Antimicrobial Resistance Genes. Int. J. Microbiol. 2024, 2024, 8054338. [Google Scholar] [CrossRef] [PubMed]
- Vikram, A.; Rovira, P.; Agga, G.E.; Arthur, T.M.; Bosilevac, J.M.; Wheeler, T.L.; Morley, P.S.; Belk, K.E.; Schmidt, J.W. Impact of “raised without antibiotics” beef cattle production practices on occurrences of antimicrobial resistance. Appl. Environ. Microbiol. 2017, 83, e01682-17. [Google Scholar] [CrossRef]
- Vikram, A.; Miller, E.; Arthur, T.M.; Bosilevac, J.M.; Wheeler, T.L.; Schmidt, J.W. Similar Levels of Antimicrobial Resistance in U.S. Food Service Ground Beef Products with and without a “Raised without Antibiotics” Claim. J. Food Prot. 2018, 81, 2007–2018. [Google Scholar] [CrossRef] [PubMed]
- Weinroth, M.D.; Scott, H.M.; Norby, B.; Loneragan, G.H.; Noyes, N.R.; Rovira, P.; Doster, E.; Yang, X.; Woerner, D.R.; Morley, P.S.; et al. Effects of ceftiofur and chlortetracycline on the resistomes of feedlot cattle. Appl. Environ. Microbiol. 2018, 84, e00610-18. [Google Scholar] [CrossRef] [PubMed]
- Rothrock, M.J.; Min, B.R.; Castleberry, L.; Waldrip, H.; Parker, D.; Brauer, D.; Pitta, D.; Indugu, N. Antibiotic resistance, antimicrobial residues, and bacterial community diversity in pasture-raised poultry, swine, and beef cattle manures. J. Anim. Sci. 2021, 99, skab144. [Google Scholar] [CrossRef] [PubMed]
- Qian, X.; Gu, J.; Sun, W.; Wang, X.J.; Su, J.Q.; Stedfeld, R. Diversity, abundance, and persistence of antibiotic resistance genes in various types of animal manure following industrial composting. J. Hazard. Mater. 2018, 344, 716–722. [Google Scholar] [CrossRef] [PubMed]
- Hurst, J.J.; Oliver, J.P.; Schueler, J.; Gooch, C.; Lansing, S.; Crossette, E.; Wigginton, K.; Raskin, L.; Aga, D.S.; Sassoubre, L.M. Trends in Antimicrobial Resistance Genes in Manure Blend Pits and Long-Term Storage Across Dairy Farms with Comparisons to Antimicrobial Usage and Residual Concentrations. Environ. Sci. Technol. 2019, 53, 2405–2415. [Google Scholar] [CrossRef]
- Cameron, A.; McAllister, T.A. Antimicrobial usage and resistance in beef production. J. Anim. Sci. Biotechnol. 2016, 7, 68. [Google Scholar] [CrossRef] [PubMed]
- Jones, B.A.; Grace, D.; Kock, R.; Alonso, S.; Rushton, J.; Said, M.Y.; McKeever, D.; Mutua, F.; Young, J.; McDermott, J.; et al. Zoonosis emergence linked to agricultural intensification and environ-mental change. Proc. Natl. Acad. Sci. USA 2013, 110, 8399–8404. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.T.; Sobur, M.A.; Islam, M.S.; Ievy, S.; Hossain, M.J.; Zowalaty, M.E.E.; Rahman, A.M.M.T.; Ashour, H.M. Zoonotic Diseases: Etiology, Impact, and Control. Microorganisms 2020, 8, 1405. [Google Scholar] [CrossRef] [PubMed]
- European Food Safety Authority (EFSA); European Centre for Disease Prevention and Control (ECDC). The European Union One Health 2023 Zoonoses report. EFSA J. 2024, 22, e9106. [Google Scholar] [CrossRef]
- Jones, K.E.; Patel, N.G.; Levy, M.A.; Storeygard, A.; Balk, D.; Gittleman, J.L.; Daszak, P. Global trends in emerging infectious diseases. Nature 2008, 451, 990–993. [Google Scholar] [CrossRef]
- Chlebicz, A.; Śliżewska, K. Campylobacteriosis, Salmonellosis, Yersiniosis, and Listeriosis as Zoonotic Foodborne Diseases: A Review. Int. J. Environ. Res. Public Health 2018, 15, 863. [Google Scholar] [CrossRef] [PubMed]
- Heredia, N.; García, S. Animals as sources of food-borne pathogens: A review. Anim. Nutr. 2018, 4, 250–255. [Google Scholar] [CrossRef]
- Velebit, B.; Djordjevic, V.; Milojevic, L.; Babic, M.; Grkovic, N.; Jankovic, V.; Yushina, Y. The common foodborne viruses: A review. IOP Conf. Ser. Earth Environ. Sci. 2019, 333, 012110. [Google Scholar] [CrossRef]
- Gu, W.; Miller, S.; Chiu, C.Y. Clinical Metagenomic Next-Generation Sequencing for Pathogen Detection. Annu. Rev. Pathol. Mech. Dis. 2019, 14, 319–338. [Google Scholar] [CrossRef] [PubMed]
- Garin-Fernandez, A.; Pereira-Flores, E.; Glöckner, F.O.; Wichels, A. The North Sea goes viral: Oc-currence and distribution of North Sea bacteriophages. Mar. Genom. 2018, 41, 31–41. [Google Scholar] [CrossRef] [PubMed]
- Holmfeldt, K.; Solonenko, N.; Shah, M.; Corrier, K.; Riemann, L.; VerBerkmoes, N.C.; Sullivan, M.B. Twelve previously unknown phage genera are ubiquitous in global oceans. Proc. Natl. Acad. Sci. USA 2013, 110, 12798–12803. [Google Scholar] [CrossRef] [PubMed]
- Kosmopoulos, J.C.; Klier, K.M.; Langwig, M.V.; Tran, P.Q.; Anantharaman, K. Viromes vs. mixed community metagenomes: Choice of method dictates interpretation of viral community ecology. Microbiome 2024, 12, 195. [Google Scholar] [CrossRef] [PubMed]
- Rose, R.; Constantinides, B.; Tapinos, A.; Robertson, D.L.; Prosperi, M. Challenges in the analysis of viral metagenomes. Virus Evol. 2016, 2, vew022. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, P.; Han, Y.; Zhao, W.; Zhao, L.; Li, R.; Zhang, J.; Zhang, S.; Lu, J.; Daszak, P.; et al. Unveiling bat-borne viruses: A comprehensive classification and analysis of virome evolution. Microbiome 2024, 12, 235. [Google Scholar] [CrossRef]
- Kwok, K.T.T.; Nieuwenhuijse, D.F.; Phan, M.V.T.; Koopmans, M.P.G. Virus Metagenomics in Farm Animals: A Systematic Review. Viruses 2020, 12, 107. [Google Scholar] [CrossRef] [PubMed]
- Shan, T.; Yang, S.; Wang, H.; Wang, H.; Zhang, J.; Gong, G.; Xiao, Y.; Yang, J.; Wang, X.; Lu, J.; et al. Virome in the cloaca of wild and breeding birds revealed a diversity of significant viruses. Microbiome 2022, 10, 60. [Google Scholar] [CrossRef]
- dos Santos Miranda, T.; Schiffler, F.B.; D’arc, M.; Moreira, F.R.R.; Cosentino, M.A.C.; Coimbra, A.; Mouta, R.; Medeiros, G.; Girardi, D.L.; Wanderkoke, V.; et al. Metagenomic analysis reveals novel dietary-related viruses in the gut virome of marmosets hybrids (Callithrix jacchus x Callithrix penicillata), Brazil. Virus Res. 2022, 325, 199017. [Google Scholar] [CrossRef] [PubMed]
- Hensel, A. Food Safety Assessment in the European and Global Context; German Federal Office for Risk Assessment: Berlin, Germany, 2015.
- Sajali, N.; Wong, S.C.; Abu Bakar, S.; Khairil Mokhtar, N.F.; Manaf, Y.N.; Yuswan, M.H.; Mohd Desa, M.N. Analytical approaches of meat authentication in food. Int. J. Food Sci. Technol. 2021, 56, 1535–1543. [Google Scholar] [CrossRef]
- Bansal, S.; Singh, A.; Mangal, M.; Mangal, A.K.; Kumar, S. Food adulteration: Sources, health risks, and detection methods. Crit. Rev. Food Sci. Nutr. 2017, 57, 1174–1189. [Google Scholar] [CrossRef] [PubMed]
- Tedersoo, L.; Anslan, S.; Bahram, M.; Põlme, S.; Riit, T.; Liiv, I.; Kõljalg, U.; Kisand, V.; Nilsson, R.H.; Hildebrand, F.; et al. Shotgun metagenomes and multiple primer pair-barcode combinations of amplicons reveal biases in metabarcoding analyses of fungi. MycoKeys 2015, 10, 1–43. [Google Scholar] [CrossRef]
- Hellmann, S.L.; Ripp, F.; Bikar, S.E.; Schmidt, B.; Köppel, R.; Hankeln, T. Identification and quanti-fication of meat product ingredients by whole-genome metagenomics (All-Food-Seq). Eur. Food Res. Technol. 2020, 246, 193–200. [Google Scholar] [CrossRef]
- Dirong, G.; Nematbakhsh, S.; Selamat, J.; Chong, P.P.; Idris, L.H.; Nordin, N.; Fatchiyah, F.; Razis, A.F.A. Omics-Based Analytical Approaches for Assessing Chicken Species and Breeds in Food Au-thentication. Molecules 2021, 26, 6502. [Google Scholar] [CrossRef] [PubMed]
- Vaithiyanathan, S.; Vishnuraj, M.R.; Reddy, G.N.; Kulkarni, V.V. Application of DNA technology to check misrepresentation of animal species in illegally sold meat. Biocatal. Agric. Biotechnol. 2018, 16, 564–568. [Google Scholar] [CrossRef]
- Anthoons, B.; Karamichali, I.; Schrøder-Nielsen, A.; Drouzas, A.D.; de Boer, H.; Madesis, P. Metabarcoding reveals low fidelity and presence of toxic species in short chain-of-commercialization of herbal products. J. Food Compos. Anal. 2021, 97, 103767. [Google Scholar] [CrossRef]
- Dobrovolny, S.; Blaschitz, M.; Weinmaier, T.; Pechatschek, J.; Cichna-Markl, M.; Indra, A.; Hufnagl, P.; Hochegger, R. Development of a DNA metabarcoding method for the identification of fifteen mam-malian and six poultry species in food. Food Chem. 2019, 272, 354–361. [Google Scholar] [CrossRef] [PubMed]
- Nguyen-the, C.; Carlin, F. The microbiology of minimally processed fresh fruits and vegetables. Crit. Rev. Food Sci. Nutr. 1994, 34, 371–401. [Google Scholar] [CrossRef] [PubMed]
- Critzer, F.J.; Doyle, M.P. Microbial ecology of foodborne pathogens associated with produce. Curr. Opin. Biotechnol. 2010, 21, 125–130. [Google Scholar] [CrossRef] [PubMed]
- Siroli, L.; Patrignani, F.; Serrazanetti, D.I.; Chiavari, C.; Benevelli, M.; Grazia, L.; Lanciotti, R. Survival of spoilage and pathogenic microorganisms on cardboard and plastic packaging materials. Front. Microbiol. 2017, 8, 313909. [Google Scholar] [CrossRef] [PubMed]
- Deblais, L.; Kathayat, D.; Helmy, Y.A.; Closs, G.; Rajashekara, G. Translating ‘big data’: Better un-derstanding of host-pathogen interactions to control bacterial foodborne pathogens in poultry. Anim. Health Res. Rev. 2020, 21, 15–35. [Google Scholar] [CrossRef] [PubMed]
- Bhogoju, S.; Nahashon, S.; Wang, X.; Darris, C.; Kilonzo-Nthenge, A. A comparative analysis of mi-crobial profile of Guinea fowl and chicken using metagenomic approach. PLoS ONE 2018, 13, e0191029. [Google Scholar] [CrossRef]
- Kim, H.; Cho, J.H.; Song, M.; Cho, J.H.; Kim, S.; Kim, E.S.; Keum, G.B.; Kim, H.B.; Lee, J.H. Evaluating the Prevalence of Foodborne Pathogens in Livestock Using Metagenomics Approach. J. Microbiol. Biotechnol. 2021, 31, 1701. [Google Scholar] [CrossRef]
- Ricke, S.C.; Lee, S.I.; Kim, S.A.; Park, S.H.; Shi, Z. Prebiotics and the poultry gastrointestinal tract microbiome. Poult. Sci. 2020, 99, 670–677. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.-Y.; Wang, H.-H.; Han, Y.-W.; Xing, T.; Ye, K.-P.; Xu, X.-L.; Zhou, G.-H. Evaluation of the spoilage potential of bacteria isolated from chilled chicken in vitro and in situ. Food Microbiol. 2017, 63, 139–146. [Google Scholar] [CrossRef]
- Gomes, B.; Pena, P.; Cervantes, R.; Dias, M.; Viegas, C. Microbial Contamination of Bedding Material: One Health in Poultry Production. Int. J. Environ. Res. Public Health 2022, 19, 16508. [Google Scholar] [CrossRef] [PubMed]
- Segura-Wang, M.; Grabner, N.; Koestelbauer, A.; Klose, V.; Ghanbari, M. Genome-Resolved Metagenomics of the Chicken Gut Microbiome. Front. Microbiol. 2021, 12, 726923. [Google Scholar] [CrossRef] [PubMed]
- Glendinning, L.; Stewart, R.D.; Pallen, M.J.; Watson, K.A.; Watson, M. Assembly of hundreds of novel bacterial genomes from the chicken caecum. Genome Biol. 2020, 21, 34. [Google Scholar] [CrossRef] [PubMed]
- Medvecky, M.; Cejkova, D.; Polansky, O.; Karasova, D.; Kubasova, T.; Cizek, A.; Rychlik, I. Whole genome sequencing and function prediction of 133 gut anaerobes isolated from chicken caecum in pure cultures. BMC Genom. 2018, 19, 561. [Google Scholar] [CrossRef] [PubMed]
- Beauclercq, S.; Nadal-Desbarats, L.; Hennequet-Antier, C.; Gabriel, I.; Tesseraud, S.; Calenge, F.; Le Bihan-Duval, E.; Mignon-Grasteau, S. Relationships between digestive efficiency and metabolomic profiles of serum and intestinal contents in chickens. Sci. Rep. 2018, 8, 6678. [Google Scholar] [CrossRef]
- Xu, Y.; Huang, Y.; Guo, L.; Zhang, S.; Wu, R.; Fang, X.; Xu, H.; Nie, Q. Metagenomic analysis reveals the microbiome and antibiotic resistance genes in indigenous Chinese yellow-feathered chickens. Front. Microbiol. 2022, 13, 930289. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Tong, C.; Xiao, D.; Xie, L.; Zhao, R.; Huo, Z.; Tang, Z.; Hao, J.; Zeng, Z.; Xiong, W. Met-agenomic Insights into Chicken Gut Antibiotic Resistomes and Microbiomes. Microbiol. Spectr. 2022, 10, e01907-21. [Google Scholar] [CrossRef] [PubMed]
- Jochum, J.M.; Redweik, G.A.J.; Ott, L.C.; Mellata, M. Bacteria Broadly-Resistant to Last Resort Antibiotics Detected in Commercial Chicken Farms. Microorganisms 2021, 9, 141. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Hu, Y.; Liu, F.; Cao, J.; Lv, N.; Zhu, B.; Zhang, G.; Gao, G.F. Integrated metagenomic and meta-transcriptomic profiling reveals differentially expressed resistomes in human, chicken, and pig gut microbiomes. Environ. Int. 2020, 138, 105649. [Google Scholar] [CrossRef] [PubMed]
- Huang, P.; Zhang, Y.; Xiao, K.; Jiang, F.; Wang, H.; Tang, D.; Liu, D.; Liu, B.; Liu, Y.; He, X.; et al. The chicken gut metagenome and the modulatory effects of plant-derived benzylisoquinoline alkaloids. Microbiome 2018, 6, 211. [Google Scholar] [CrossRef] [PubMed]
- Day, J.M.; Oakley, B.B.; Seal, B.S.; Zsak, L. Comparative Analysis of the Intestinal Bacterial and RNA Viral Communities from Sentinel Birds Placed on Selected Broiler Chicken Farms. PLoS ONE 2015, 10, e0117210. [Google Scholar] [CrossRef]
- Devaney, R.; Trudgett, J.; Trudgett, A.; Meharg, C.; Smyth, V. A metagenomic comparison of endemic viruses from broiler chickens with runting-stunting syndrome and from normal birds. Avian Pathol. 2016, 45, 616–629. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Li, L.; Deng, X.; Kapusinszky, B.; Delwart, E. What is for dinner? Viral metagenomics of US store bought beef, pork, and chicken. Virology 2014, 468–470, 303–310. [Google Scholar] [CrossRef] [PubMed]
- de Wit, J.J.; ten Dam, G.B.; vande Laar, J.M.A.M.; Biermann, Y.; Verstegen, I.; Edens, F.; Schrier, C.C. Detection and characterization of a new astrovirus in chicken and turkeys with enteric and locomotion disorders. Avian Pathol. 2011, 40, 453–461. [Google Scholar] [CrossRef]
- Aruwa, C.E.; Pillay, C.; Nyaga, M.M.; Sabiu, S. Poultry gut health—Microbiome functions, environmental impacts, microbiome engineering and advancements in characterization technologies. J. Anim. Sci. Biotechnol. 2021, 12, 119. [Google Scholar] [CrossRef]
- Vibin, J.; Chamings, A.; Collier, F.; Klaassen, M.; Nelson, T.M.; Alexandersen, S. Metagenomics detection and characterisation of viruses in faecal samples from Australian wild birds. Sci. Rep. 2018, 8, 8686. [Google Scholar] [CrossRef]
- Ayginin, A.A.; Pimkina, E.V.; Matsvay, A.D.; Speranskaya, A.S.; Safonova, M.V.; Blinova, E.A.; Artyushin, I.V.; Dedkov, V.G.; Shipulin, G.A.; Khafizov, K. The Study of Viral RNA Diversity in Bird Samples Using De Novo Designed Multiplex Genus-Specific Primer Panels. Adv. Virol. 2018, 2018, 3248285. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.J.; Yong, H.I.; Kim, M.; Choi, Y.S.; Jo, C. Status of meat alternatives and their potential role in the future meat market—A review. Asian Australas J. Anim. Sci. 2020, 33, 1533. [Google Scholar] [CrossRef]
- Nam, K.C.; Jo, C.; Lee, M. Meat products and consumption culture in the East. Meat Sci. 2010, 86, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Bryant, C.; Barnett, J. Consumer acceptance of cultured meat: A systematic review. Meat Sci. 2018, 143, 8–17. [Google Scholar] [CrossRef] [PubMed]
- Thomas, J.; Govender, N.; McCarthy, K.M.; Erasmus, L.K.; Doyle, T.J.; Allam, M.; Ismail, A.; Ramalwa, N.; Sekwadi, P.; Ntshoe, G.; et al. Outbreak of Listeriosis in South Africa Associated with Processed Meat. N. Engl. J. Med. 2020, 382, 632–643. [Google Scholar] [CrossRef]
- Hassoun, A.; Måge, I.; Schmidt, W.F.; Temiz, H.T.; Li, L.; Kim, H.Y.; Nilsen, H.; Biancolillo, A.; Aït-Kaddour, A.; Sikorski, M.; et al. Fraud in Animal Origin Food Products: Advances in Emerging Spectroscopic Detection Methods over the Past Five Years. Foods 2020, 9, 1069. [Google Scholar] [CrossRef] [PubMed]
- Mao, S.; Zhang, M.; Liu, J.; Zhu, W. Characterising the bacterial microbiota across the gastrointestinal tracts of dairy cattle: Membership and potential function. Sci. Rep. 2015, 5, 16116. [Google Scholar] [CrossRef] [PubMed]
- Matthews, C.; Crispie, F.; Lewis, E.; Reid, M.; O’Toole, P.W.; Cotter, P.D. The rumen microbiome: A crucial consideration when optimising milk and meat production and nitrogen utilisation efficiency. Gut Microbes 2018, 10, 115. [Google Scholar] [CrossRef] [PubMed]
- Xie, F.; Jin, W.; Si, H.; Yuan, Y.; Tao, Y.; Liu, J.; Wang, X.; Yang, C.; Li, Q.; Yan, X.; et al. An integrated gene catalog and over 10,000 metagenome-assembled genomes from the gastrointestinal microbiome of ruminants. Microbiome 2021, 9, 137. [Google Scholar] [CrossRef]
- Xue, M.; Sun, H.; Wu, X.; Guan, L.L.; Liu, J. Assessment of rumen microbiota from a large dairy cattle cohort reveals the pan and core bacteriomes contributing to varied phenotypes. Appl. Environ. Microbiol. 2018, 84, 970–988. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Chen, J.; Wang, X.; Han, L.; Yang, Y.; Wang, Q.; Yu, Q. Metagenomic and Transcriptomic Analyses Reveal the Differences and Associations Between the Gut Microbiome and Muscular Genes in Angus and Chinese Simmental Cattle. Front. Microbiol. 2022, 13, 815915. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Lai, Z.; Zhang, J.; Zhu, W.; Mao, S. The gastrointestinal microbiome in dairy cattle is constrained by the deterministic driver of the region and the modified effect of diet. Microbiome 2023, 11, 10. [Google Scholar] [CrossRef] [PubMed]
- Nastasijevic, I.; Schmidt, J.W.; Boskovic, M.; Glisic, M.; Kalchayanand, N.; Shackelford, S.D.; Wheeler, T.L.; Koohmaraie, M.; Bosilevac, J.M. Seasonal Prevalence of Shiga Toxin-Producing Escherichia colion Pork Carcasses for Three Steps of the Harvest Process atTwo Commercial Processing Plants in the United States. Appl. Environ. Microbiol. 2020, 87, e01711-20. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Stanford, K.; Bie, X.M.; Niu, Y.D.; McAllister, T.A. Effects of Beef Juice on Biofilm Formation by Shiga Toxin–Producing Escherichia coli on Stainless Steel. Foodborne Pathog. Dis. 2020, 17, 235–242. [Google Scholar] [CrossRef]
- Barcenilla, C.; Cobo-Díaz, J.F.; Puente, A.; Valentino, V.; De Filippis, F.; Ercolini, D.; Carlino, N.; Pinto, F.; Segata, N.; Prieto, M.; et al. In-depth characterization of food and en-vironmental microbiomes across different meat processing plants. Microbiome 2024, 12, 199. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; He, A.; Yang, X. Dynamics of microflora on conveyor belts in a beef fabrication facility during sanitation. Food Control 2018, 85, 42–47. [Google Scholar] [CrossRef]
- Yang, X.; Narvaez-Bravo, C.; Zhang, P. Driving forces shaping the microbial ecology in meat packing plants. Front. Microbiol. 2023, 14, 1333696. [Google Scholar] [CrossRef]
- Santiago-Rodriguez, T.M.; Hollister, E.B. Viral Metagenomics as a Tool to Track Sources of Fecal Contamination: A One Health Approach. Viruses 2023, 15, 236. [Google Scholar] [CrossRef]
- Andrews, T.; Neher, D.A.; Weicht, T.R.; Barlow, J.W. Mammary microbiome of lactating organic dairy cows varies by time, tissue site, and infection status. PLoS ONE 2019, 14, e0225001. [Google Scholar] [CrossRef] [PubMed]
- Weinroth, M.D.; Thomas, K.M.; Doster, E.; Vikram, A.; Schmidt, J.W.; Arthur, T.M.; Wheeler, T.L.; Parker, J.K.; Hanes, A.S.; Alekoza, N.; et al. Resistomes and microbiome of meat trimmings and colon content from culled cows raised in conventional and organic production systems. Anim. Microbiome 2022, 4, 21. [Google Scholar] [CrossRef] [PubMed]
- Noyes, N.R.; Yang, X.; Linke, L.M.; Magnuson, R.J.; Cook, S.R.; Zaheer, R.; Yang, H.; Woerner, D.R.; Geornaras, I.; McArt, J.A.; et al. Characterization of the resistome in manure, soil and wastewater from dairy and beef production systems. Sci. Rep. 2016, 6, 24645. [Google Scholar] [CrossRef] [PubMed]
- Sequino, G.; Valentino, V.; Villani, F.; De Filippis, F. Omics-based monitoring of microbial dynamics across the food chain for the improvement of food safety and quality. Food Res. Int. 2022, 157, 111242. [Google Scholar] [CrossRef] [PubMed]
- Kobus, R.; Abuín, J.M.; Müller, A.; Hellmann, S.L.; Pichel, J.C.; Pena, T.F.; Hildebrandt, A.; Hankeln, T.; Schmidt, B. A big data approach to metagenomics for all-food-sequencing. BMC Bioinform. 2020, 21, 102. [Google Scholar] [CrossRef] [PubMed]
- Yan, A.; Butcher, J.; Mack, D.; Stintzi, A. Virome Sequencing of the Human Intestinal Mucosal–Luminal Interface. Front. Cell. Infect. Microbiol. 2020, 10, 582187. [Google Scholar] [CrossRef]
- Billington, C.; Kingsbury, J.M.; Rivas, L. Metagenomics Approaches for Improving Food Safety: A Review. J. Food Prot. 2022, 85, 448–464. [Google Scholar] [CrossRef]
- Ngara, T.R.; Zhang, H. Recent Advances in Function-Based Metagenomic Screening. Genom. Proteom. Bioinform. 2018, 16, 405–415. [Google Scholar] [CrossRef]
- Allen, H.K. Antibiotic resistance gene discovery in food-producing animals. Curr. Opin. Microbiol. 2014, 19, 25–29. [Google Scholar] [CrossRef]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 2020, 38, 685–688. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Miller, M.; Marpaka, S.; Vaysberg, P.; Rühlemann, M.C.; Wu, G.; Heinsen, F.A.; Tempel, M.; Zhao, L.; Lieb, W.; et al. Functional sequencing read annotation for high precision microbiome analysis. Nucleic Acids Res. 2018, 46, e23. [Google Scholar] [CrossRef] [PubMed]
- Shalaby, M.; Reboud, J.; Forde, T.; Zadoks, R.N.; Busin, V. Distribution and prevalence of enterotoxigenic Staphylococcus aureus and staphylococcal enterotoxins in raw ruminants’ milk: A systematic review. Food Microbiol. 2024, 118, 104405. [Google Scholar] [CrossRef] [PubMed]
- Munir, M.T.; Mtimet, N.; Guillier, L.; Meurens, F.; Fravalo, P.; Federighi, M.; Kooh, P. Physical Treatments to Control Clostridium botulinum Hazards in Food. Foods 2023, 12, 1580. [Google Scholar] [CrossRef] [PubMed]
- Navale, V.; Vamkudoth, K.R.; Ajmera, S.; Dhuri, V. Aspergillus derived mycotoxins in food and the environment: Prevalence, detection, and toxicity. Toxicol. Rep. 2021, 8, 1008–1030. [Google Scholar] [CrossRef]
- Vilariño, N.; Louzao, M.C.; Abal, P.; Cagide, E.; Carrera, C.; Vieytes, M.R.; Botana, L.M. Human Poisoning from Marine Toxins: Unknowns for Optimal Consumer Protection. Toxins 2018, 10, 324. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Chen, J.; Gao, F.; Su, W.; Li, T.; Wang, Y. Foodomics as a Tool for Evaluating Food Authenticity and Safety from Field to Table: A Review. Foods 2024, 14, 15. [Google Scholar] [CrossRef] [PubMed]
- Selamat, J.; Rozani, N.A.A.; Murugesu, S. Application of the Metabolomics Approach in Food Authentication. Molecules 2021, 26, 7565. [Google Scholar] [CrossRef] [PubMed]
- Pedrosa, M.C.; Lima, L.; Heleno, S.; Carocho, M.; Ferreira, I.C.F.R.; Barros, L. Food Metabolites as Tools for Authentication, Processing, and Nutritive Value Assessment. Foods 2021, 10, 2213. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Zhang, L.; Zheng, X.; Huang, Q.; Farag, M.A.; Zhu, R.; Zhao, C. Emerging applications of metabolomics in food science and future trends. Food Chem. X 2022, 16, 100500. [Google Scholar] [CrossRef] [PubMed]
- Winder, C.L.; Witting, M.; Tugizimana, F.; Dunn, W.B.; Reinke, S.N. Providing metabolomics edu-cation and training: Pedagogy and considerations. Metabolomics 2022, 18, 106. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).