Abstract
Contamination of coastal-marine environment with multidrug-resistant Escherichia coli has resulted in such bacteria increasingly being detected in the seafood chain. This study aimed to determine the quinolone and colistin resistance genes in extended spectrum-β-lactamase (ESBL)-producing E. coli from seafood. ESBL-producing E. coli isolates (n = 269) were tested for quinolones and colistin resistance phenotypes by disk diffusion and broth microdilution methods, respectively. The isolates were further PCR screened for the plasmid-mediated quinolone resistance (PMQR) genes qnrA, qnrB, and qnrS, genomic mutations in gyrA and parC genes, and the colistin resistance genes mcr-1 and mcr-2. Phylogroup was determined by PCR using the Clermont E. coli phylotyping method. Of 269 isolates tested, 73.60% of E. coli isolates were resistant to moxifloxacin and 8.55% to ofloxacin, the least of all the quinolones tested. Further, 150 (55.76%) E. coli isolates carried at least one of the three PMQR genes tested, where qnrS was the most prevalent gene (53.90%). The colistin resistance gene (mcr-2) was detected in 38 (14.12%) isolates. Twenty-one of these isolates (55.26%) had a colistin minimum inhibitory concentration (MIC) of 16 µg/mL. Based on the Clermont E. coli phylotyping of the isolates harboring at least one of the qnr genes, 66 (44%) belonged to the phylogroup B1, followed by 23 (15.33%) to phylogroup A. Among 38 E. coli isolates carrying colistin resistance gene mcr-2, 27 (71.05%) isolates belonged to phylogroup B1, followed by 4 (10.52%) isolates to phylogroup A. The results suggest that E. coli phylogroups B1 and A harboring plasmid-mediated quinolone and colistin resistance genes are predominant in the seafood supply chain.
1. Introduction
The extended-spectrum β-lactamase (ESBL)-producing Escherichia coli has become a severe challenge to chemotherapy [1,2]. The ESBLs are classified into several groups, the most prominent of them being TEM, SHV, and CTX-M types [3,4]. Pathogenic E. coli are distinct from their commensal counterparts due to the virulence genes they possess, based on which they are classified into at least five pathovars, namely enterotoxigenic E. coli (ETEC), enteropathogenic E. coli (EPEC), enterohaemorrhagic E. coli (EHEC), enteroaggregative/adherent E. coli (EAEC), and enteroinvasive E. coli (EIEC). E. coli causes diverse infections ranging from wound infection to meningitis and is one of the major ESBL-producing Gram-negative bacteria. ESBLs confer resistance to a broad spectrum of cephalosporins, and together with AmpC lactamases, bacteria producing them can resist a majority of cephalosporins and their inhibitor combinations [5,6,7]. The emergence and spread of carbapenemases have further confounded the problem, as carbapenems have been the preferred antibiotics to control ESBL-producing Enterobacteriaceae. The broad-spectrum antibiotics of the fluoroquinolone group are also widely used to treat gastrointestinal and urinary infections caused by E. coli. However, quinolone resistance has dramatically increased in recent years, particularly with the emergence of certain specific lineages of bacteria such as ST131-H30 [8,9]. Quinolone resistance occurs due to amino acid changes in topoisomerase subunits GyrA and ParC, which become recalcitrant to binding by the antibiotic due to structural changes [10]. The plasmid-mediated quinolone resistance (PMQR) genes, collectively called qnr genes and first reported in a Klebsiella pneumoniae isolate, encode pentapeptide proteins that bind with DNA gyrase and topoisomerase IV enzymes, protecting them from the inhibitory effects of quinolones [10,11,12]. The genes qnrA, qnrB, qnrC, qnrD, qnrS, and qnrVC, with over 100 variants, are widely distributed in Gram-negative bacteria [13]. Plasmids harboring qnr may also carry extended-spectrum β-lactamases (ESBLs) [14]. With ESBL- and carbapenemase-resistant Enterobacteriaceae becoming challenging to treat, polymyxins are increasingly being considered as therapeutic options despite their potential nephrotoxicity. Colistin (polymyxin E) is a cationic, multi-component, lipopeptide antibacterial that has been used in veterinary medicine for decades to treat Gram-negative intestinal tract infections. Colistin was initially restricted to ophthalmic and topical use for treating human infections due to its systemic toxicity [15]. The growing evolution of multidrug-resistant (MDR) Gram-negative bacterial infections with limited available therapeutic options has compelled its clinical use as a primary treatment option. The most commonly found mechanism of colistin resistance is associated with the chromosomal mutation in the genes, which leads to modification of the lipid A of LPS, the primary target of colistin, as an adaptative strategy. However, plasmid-mediated polymyxin resistance is attributed to the mcr genes, with at least ten variants, mcr-1 to mcr-10 [16,17]. The horizontal transfer of the plasmid-carried mcr-1 gene encoding PEtN has become an important cause of the spread of colistin resistance among various Gram-negative bacteria [17,18]. The emergence of mcr genes has focused global attention on colistin resistance, one of the last means of treatment. The mcr-gene-containing bacteria (MGCB) are disseminated by horizontal/lateral transfer into diverse ecosystems, including aquatic, soil, botanical, wildlife, animal, and environment ecosystems, and public places.
The anthropogenic contamination of the coastal-marine environment can introduce multidrug-resistant (MDR) E. coli, eventually leading to their presence in seafood. E. coli from seafood may show different levels of quinolone and colistin resistance due to mutations in QRDR genes and the acquisition of plasmids containing quinolone and colistin resistance genes by horizontal gene transfer. The plasmid-mediated quinolone and colistin resistance genes can be mobilized into other Enterobacterales members, leading to wider dissemination of antibiotic resistance phenotypes and genotypes. Phylogenetic evaluation of E. coli can help understand the distribution and origin of MDR E. coli in seafood and the aquatic environment. The present study aimed to understand the quinolone and colistin resistance in ESBL-producing E. coli from seafood in Mumbai, India and their phylogenetic diversity.
2. Materials and Methods
2.1. Detection of ESBL Phenotype in Escherichia coli Isolates
Escherichia coli isolates used in this study were from our previous study [19], in which we reported the prevalence of ESBL-producing E. coli isolates from seafood samples from markets of Mumbai, India. Briefly, for the phenotypic detection of ESBL production, the double-disk synergy test was followed. The isolate was grown to 0.5 McFarland units in Mueller–Hinton (MH) broth and 100 µL of the culture was spread plated evenly on a Mueller–Hinton (MH) agar plate. An amoxicillin/clavulanic acid (30/10 µg) disk was placed at the center, and cefpodoxime (10 µg), cefotaxime (30 µg), and ceftazidime (30 µg) disks were positioned at 20–30 mm away from the central disk. An extension in the zone of inhibition around the peripheral disk towards the centrally placed disk by at least 5 mm was considered positive for ESBL production. This was further confirmed by using the Triple ESBL detection Ezy MICTM Strip (Hi-Media, Mumbai, India) according to the manufacturer’s instructions. The test strain was spread plated evenly on a Mueller–Hinton (MH) agar plate and then a Triple ESBL detection Ezy MICTM Strip was placed and minimum inhibitory concentrations (MICs) were recorded. A total of 269 isolates of ESBL-positive E. coli were included in this study.
2.2. Quinolone Susceptibility Testing Using Disk Diffusion Assay
The isolates were retrieved from −80 °C glycerol stock on Mueller–Hinton agar (HiMedia, Mumbai, India) by streak plating. A single colony from the plate was inoculated into Mueller–Hinton (MH) broth and was grown for 12 h at 37 °C. The broth culture was evenly spread on the MH agar using a sterile swab and allowed to dry for 5 min before placing antibiotic disks. The isolates were screened for quinolone resistance by the standard disk diffusion method using the following quinolone antibiotics: ciprofloxacin (CIP)—5 µg, nalidixic acid (NA)—30 µg, levofloxacin (LE)—5 µg, ofloxacin (OF)—5 µg, norfloxacin (NX)—10 µg, and moxifloxacin (MO)—5 µg. The zones of inhibition were measured and results were interpreted according to the Clinical Laboratory Standards Institute guidelines [20].
2.3. Screening of Colistin-Resistant Isolates Using Chromogenic Agar Medium
A fresh single pure colony was streaked on HiCrome™ colistin-resistant agar (HiMedia, India) and incubated for 18–24 h at 37 °C. Luxuriant purple colonies indicate the presence of presumptive colistin-resistant E. coli.
2.4. PCR Detection of Quinolone and Polymyxin Resistance Genes
Bacterial DNA was extracted using the Wizard DNA kit (Promega, Madison, WI, USA) according to the instructions therein. The quinolone resistance genes qnrA, qnrB, and qnrS, and the colistin resistance gene mcr-1 and mcr-2 were tested by PCR using previously described oligonucleotide primers and protocols [21,22,23,24]. Briefly, each PCR reaction mixture of 30 µL consisted of 15 µL of 2X EmeraldAmp® GT master mix (TaKaRa Bio Inc., Tokyo, Japan), 10 µM of each of forward and reverse primers, and 3 µL of template DNA. The genomic mutations in gyrA (Ser83 and Asp87) and parC (Ser80 and Glu84) genes were tested by multiplex allele-specific PCR [25]. All the PCRs were performed in a SimpliAmp thermocycler (Thermo Fisher Scientific, Agawam, MA 01001, USA). The PCR products were separated on 1.5% agarose gel, stained with 0.5 µg/mL ethidium bromide, and photographed using the Gel Doc XR+ gel documentation system (Bio-Rad, Hercules, CA 94547, USA).
2.5. Determination of Colistin Minimum Inhibitory Concentration (MIC) by Broth-Microdilution
The colistin minimum inhibitory concentrations (MICs) of the isolates were determined by the broth microdilution method according to CLSI protocol using cation-adjusted Mueller–Hinton II broth (CA-MHBII) [26]. From a stock solution of 64 µg/mL, colistin was serially diluted in CA-MHBII up to a concentration of 0.125 µg/mL. The test isolate was grown to 0.5 McFarland units in CA-MHBII and inoculated into microtiter plates to obtain 5 × 104 CFU/well. The plates were incubated at 35 ± 2 °C for 16 to 20 h. The lowest concentration inhibiting the visible growth of test isolates was recorded, and the streak plating method confirmed the absence of growth. According to the guidelines of the European Committee on Antimicrobial Susceptibility Testing (EUCAST), isolates with a MIC of ≥2 µg/mL were classified as resistant to colistin [27].
2.6. Phylotyping of E. coli Isolates
The primer sequences and their target genes for E. coli phylotyping and the interpretations of the results were conducted according to the Clermont method [28]. E. coli isolates of this study were subjected to phylogroup PCRs targeting 7 housekeeping genes. A quadruplex genotype corresponding to the presence/absence of the four genes (arpA, chuA, yjaA, and TspE4.C2) was determined for each strain. Then, based on the quadruplex genotype obtained, an isolate was either immediately assigned to a phylogroup or subsequently based on the results of the C and E group PCRs. Some strains of E. coli belonging to a group intermediate between the F and B2 phylogroups were classified as phylogroup G. Hence, B2 group and F group isolates were screened for phylogroup G/F PCRs targeting three housekeeping genes trpA, cfaB, and ybgD [29].
3. Results
3.1. Quinolone and Colistin Resistance in ESBL-Producing E. coli and Their Genetic Determinants
Of 269 isolates tested, 198 isolates (73.60%) were resistant to moxifloxacin, 156 isolates (57.99%) were resistant to ciprofloxacin, 132 isolates (49.07%) were resistant to nalidixic acid, 47 isolates (17.47%) were resistant to norfloxacin, 39 isolates (14.49%) were resistant to levofloxacin, and 23 isolates (8.55%) were resistant to ofloxacin (Table 1).

Table 1.
Antibiotic susceptibility profiles of E. coli (n = 269) isolates based on the disk diffusion test.
The occurrence of plasmid-mediated quinolone resistance (PMQR) genes in ESBL E. coli isolates was studied using PCR for qnrA, qnrB, and qnrS. Of 269 isolates tested, 150 (55.76%) isolates were found positive for at least one of these genes conferring resistance to quinolones. The qnrS gene was found in the maximum number of isolates (145 isolates, 53.90%), followed by qnrB (20 isolates, 7.43%). The qnrA gene was not found in any of the isolates tested (Table 2).

Table 2.
Distribution of quinolone and colistin resistance genes in E. coli.
Overall, 235 isolates resistant to at least one of the quinolones tested were screened for the occurrence of genomic mutations in gyrA and parC genes, of which gyrA83 mutation was found in 162 (68.93%) isolates, gyrA87 mutation in 166 (70.63%) isolates, parC80 mutation in 162 (68.93%) isolates, and parC84 mutation in 186 (79.14%) isolates (Table 3). At least one tested mutation was found in 193 (82.12%) isolates of E. coli.

Table 3.
Molecular detection of QRDR mutations in E. coli.
3.2. Incidence of Colistin Resistance Gene mcr-1 and mcr-2 in E. coli
The presumptive colistin-resistant isolates from HiCrome™ colistin-resistant agar were further screened for mcr-1 and mcr-2 using PCR. The n = 38 (14.12%) isolates were found to be harboring the mcr-2 gene, and the mcr-1 gene was not detected in any isolates (Table 2). All (n = 38) mcr-2 positive isolates were tested for colistin minimum inhibitory concentration (MIC) determination by the broth micro dilution method. For 21 isolates (55.26%), the minimum inhibitory concentration (MIC) was 16 µg/mL; it was 8 µg/mL for 10 (26.31%) isolates and 4 µg/mL for 7 (18.42%) isolates.
3.3. Phylogroup Evaluation of E. coli
All (n = 269) E. coli isolates of this study were subjected to phylogroup PCRs targeting seven housekeeping genes. The most prevalent phylogroup was found to be B1 (46.46%), followed by type UN (23.04%), type A (11.52%), type D (7.80%), type C (6.31%), type B2 (2.23%), type F (1.11%), and type E (three isolates; 1.11%). Six B2 group and four F group isolates were screened for phylogroup G/F PCR targeting three housekeeping genes trpA, cfaB, and ybgD. One isolate belonging to phylogroup G was identified (Table 4, Figure 1).

Table 4.
Phylogroup of E. coli isolates tested.
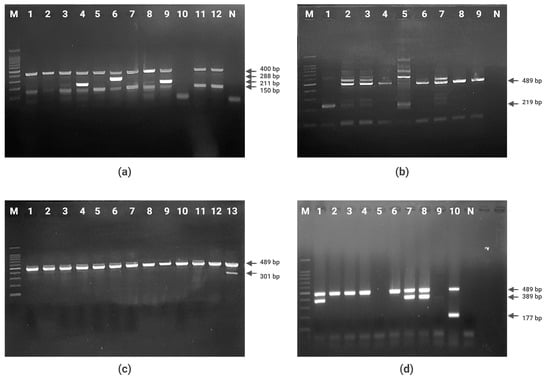
Figure 1.
Agarose gel electrophoresis of PCR products. (a) Quadruplex PCR for arpA, chuA, yjaA, and TspE4.C2 genes (Lane 1–12: strains of E. coli; lane M: 100 bp molecular size marker; lane N: negative control). (b) PCR for phylogroup C identification (Lane 1–9: E. coli strains of A or C phylogroup; lane M: 100 bp molecular size marker; lane N: negative control). (c) PCR for phylogroup E identification (Lane 1–13: E. coli strains of D or E phylogroup; lane M: 100 bp molecular size marker). (d) PCR for phylogroup G identification (Lane 1–10: E. coli strains of F or B2 phylogroup; lane M: 100 bp molecular size marker; lane N: negative control).
In the present study, of 150 quinolone-resistant E. coli isolates harboring one or more qnr genes, 44% belonged to phylogroup B1, followed by 15.33% to phylogroup A, 15.33% to phylogroup UN, 12% to phylogroup D, 8.66% to phylogroup C, 2% to phylogroup B2, 1.33% to phylogroup F, and 0.66% each to phylogroups E and G. Of the 38 E. coli isolates carrying colistin resistance gene mcr-2, 71.05% of the isolates belonged to phylogroup B1, followed by 10.52% to phylogroup A and 5.26% each to phylogroups D and UN, while one isolate each belonged to phylogroups B2, C, and E (Table 4).
4. Discussion
Fluroquinolone resistance is often caused by specific point mutations in the quinolone resistance-determining regions (QRDRs) of either the topoisomerase IV (parC and parE) or DNA gyrase (gyrA and gyrB) genes. The most common mutations identified in E. coli are at Ser83 and/or Asp87 in gyrA and Ser80 and/or Glu84 in parC [25]. In the present study, of the 235 isolates resistant to at least one of the quinolones tested, gyrA and parC mutations were observed in the majority of the isolates. Further, 42 (17.87%) strains were negative for the all four mutations tested. Novel mutations in QRDRs involved in resistance to nalidixic acid, ciprofloxacin, and ofloxacin in E. coli isolated from imported shrimp in USA were at positions 68, 83, and 87 in gyrA and at positions 80 and 84 in parC genes [30]. The qnrA gene was the first plasmid-mediated quinolone resistance gene discovered, followed by qnrB and qnrS, which share 40% and 59% amino acid identity, respectively, with qnrA [23]. Among the plasmid-mediated quinolone-resistant genes tested in this study, 55.76% of isolates were found to be positive for at least one of these genes conferring resistance to quinolones, whereas the qnrS gene was found in the maximum number of isolates (53.90%). In a study from the German food chain, among the qnr-positive E. coli isolates, the most common gene was qnrS (92.2%) followed by qnrB (5.8%), while qnrA and qnrVC were detected in only one isolate each and none had qnrC or qnrD [31]. E. coli isolated from food producing animals predominantly carried the qnrS1 gene (63.2%), followed by qnrB4 (36.7%) [32]. The qnr genes have been detected in several members of the Enterobacteriaceae family, mainly in K. pneumoniae, E. coli, Enterobacter spp., Citrobacter freundii, and Providencia stuartii in different countries [14]. Very few studies have reported the incidence of quinolone resistance in seafood-borne E. coli. Plasmid-mediated qnrA, qnrB, and qnrS have been reported in 73.8% of ESBL-positive E. coli from farmed fish in China, with qnrB being the most dominant gene, followed by qnrS [33]. The relative abundance of different plasmid-mediated quinolone resistance genes can vary in clinical, environmental, and food isolates of E. coli according to different studies from India [34,35]. The present study corroborates the general finding that qnrB and qnrS are the predominant PMQR genes in E. coli, while qnrA is relatively less prevalent. Co-occurrence of β-lactamase genes such as ESBL and MBL with the quinolone resistance genes in bacteria leaves little scope for chemotherapeutic control of such bacteria.
In the present study, the mobile colistin resistance gene mcr-2 was detected in 14.12% of the isolates, whereas the mcr-1 gene was not detected. In Belgium, an E. coli strain carrying the mcr-2 gene was isolated from pigs and cattle [36]. More than 18% of ESBL/AmpC-producing E. coli isolates from poultry in Lebanon carried the mcr-1 gene [37]. The occurrence of mcr-1 and mcr-5 colistin resistance genes in E. coli and Salmonella, respectively, has been reported in the broiler meat supply chain from Indonesia [38]. A study from Barcelona, Spain suggests the occurrence of mcr-1 harboring multidrug-resistant E. coli in sewage from waste water plants [39]. These colistin-resistant E. coli were the commonly circulating pulsotypes in the community, suggesting that environmental contamination with colistin-resistant bacteria was occurring due to the community. Several studies from India have reported a high prevalence of colistin resistance among clinical isolates [40,41]. A study conducted in Chennai, India analyzed raw food materials including poultry meat, mutton meat, fish, fruits, and vegetables collected from food outlets and found that 46.4% of the samples, including seafood, were contaminated with colistin-resistant bacteria [42]. Studies have demonstrated co-transfer of blaNDM and mcr-1 genes through conjugation [40]. These studies emphasize the need to monitor the presence and spread of multidrug-resistant Enterobacteriaceae carrying colistin resistance genotypes in the community and the environment.
An E. coli strain belonging to ST48 and phylogroup A with quinolone resistance harboring the mcr-1 gene on the IncHI2, IncN, and IncX3 plasmids was isolated from Scampi shipped to Norway from Bangladesh [43], indicating that the seafood trade may be a possible means of intercontinental transmission. In the present study, of 150 quinolone-resistant E. coli isolates harboring one or more qnr genes, 44% belonged to phylogroup B1, followed by 15.33% to phylogroup A, 15.33% to phylogroup UN, 12% to phylogroup D, 8.66% to phylogroup C, 2% to phylogroup B2, 1.33% to phylogroup F, and 0.66% each to phylogroups E and G. Among the 38 E. coli isolates carrying the colistin resistance gene mcr-2, 71.05% of the isolates belonged to phylogroup B1, followed by 10.52% to phylogroup A, and 5.26% each to phylogroups D and UN, while one isolate each belonged to phylogroups B2, C, and E. In a study on Escherichia coli in chicken meats, phylogroups A, C, D, and F were found among colistin-resistant E. coli, suggesting that colistin-resistant E. coli strains are genetically more diverse [44]. A study on colistin-resistant E. coli isolated from diseased pigs in France showed that isolates belonged to groups A (63%), D (17.3%), B1 (11.1%), E (5%), B2, C, and F (1.2% each) using the Clermont E. coli phylotyping method [45]. Our results showed that E. coli phylogroups B1 and A harboring plasmid-mediated quinolone and colistin resistance genes are predominant in the seafood supply chain. It has been reported that isolates belonging to the B1 and A clade carry diverse virulence and antibiotic resistance genes with no linkage between phylogeny and antibiotic resistance gene presence [46]. A study by on the mcr-1 resistance gene and plasmidome in E. coli pathogenic strains showed variable phylogroups A, B1, B2, C, D, E, and G with no correlation for particular genotypes with pathotypes [47]. E. coli ST131 from the phylogroup B2 was found in a study of seafood to be blaNDM-positive [19].
5. Conclusions
In conclusion, the present study indicates the predominance of plasmid-mediated quinolone and colistin resistance genes in seafood isolates of E. coli. We did not characterize the plasmids carrying quinolone and colistin resistance genes or study their mobility, which is a limitation of this study. Nevertheless, the occurrence of such ESBL-producing E. coli in seafood and the coastal environment conferring multi-drug resistance is a concern since the genetic traits of antibiotic resistance can easily be disseminated among other members of Enterobacterales. Quinolone and colistin antibiotics are “critically important” in human medicine, and the rapid emergence of resistance to these can severely compromise their clinical efficacy. Phylogenetic typing is a valuable tool for understanding the diversity and sources of ESBL-producing E. coli and the emergence of any new resistant lineage in the environment. The present study emphasizes the need to develop strategies to mitigate the contamination of coastal-marine waters with antibiotic-resistant E. coli from human and animal sources. This study also draws attention to developing reliable and faster detection methods to ascertain the risk of ESBL-producing E. coli in seafood harvest and post-harvest environments. This is necessary to ensure the safety of fish and shellfish for human consumption and control the spread of antibiotic-resistant E. coli in the community.
Author Contributions
S.H.K. conceptualized the study. S.H.K., B.B.N., S.K.G. and M.L. designed the experiments. C.K.D. performed the benchwork and collected the data. C.K.D., S.H.K., S.K.G. and M.L. performed data analysis. B.B.N. contributed to data analysis and interpretation of results. C.K.D. wrote the original draft of the manuscript and created the tables and figures. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by ICAR-Central Institution of Fisheries Education, Mumbai through an institutional project CIFE-2012/9.
Data Availability Statement
None to declare.
Acknowledgments
Authors thank the Director, ICAR-CIFE Mumbai for providing necessary facilities for carrying out this work.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Pitout, J.D.D. Multiresistant Enterobacteriaceae: New Threat of an Old Problem. Expert Rev. Anti Infect. Ther. 2008, 6, 657–669. [Google Scholar] [CrossRef] [PubMed]
- Theuretzbacher, U. Global Antibacterial Resistance: The Never-Ending Story. J. Glob. Antimicrob. Resist. 2013, 1, 63–69. [Google Scholar] [CrossRef]
- Tewari, R.; Mitra, S.; Ganaie, F.; Das, S.; Chakraborty, A.; Venugopal, N.; Shome, R.; Rahman, H.; Shome, B.R. Dissemination and Characterisation of Escherichia coli Producing Extended-Spectrum β-Lactamases, AmpC β-Lactamases and Metallo-β-Lactamases from Livestock and Poultry in Northeast India: A Molecular Surveillance Approach. J. Glob. Antimicrob. Resist. 2019, 17, 209–215. [Google Scholar] [CrossRef]
- Varela, M.F.; Stephen, J.; Lekshmi, M.; Ojha, M.; Wenzel, N.; Sanford, L.M.; Hernandez, A.J.; Parvathi, A.; Kumar, S.H. Bacterial Resistance to Antimicrobial Agents. Antibiotics 2021, 10, 593. [Google Scholar] [CrossRef] [PubMed]
- Madec, J.-Y.; Haenni, M.; Nordmann, P.; Poirel, L. Extended-Spectrum β-Lactamase/AmpC- and Carbapenemase-Producing Enterobacteriaceae in Animals: A Threat for Humans? Clin. Microbiol. Infect. 2017, 23, 826–833. [Google Scholar] [CrossRef] [PubMed]
- Vu, T.T.T.; Alter, T.; Roesler, U.; Roschanski, N.; Huehn, S. Investigation of Extended-Spectrum and AmpC β-Lactamase-Producing Enterobacteriaceae from Retail Seafood in Berlin, Germany. J. Food Prot. 2018, 81, 1079–1086. [Google Scholar] [CrossRef] [PubMed]
- Castanheira, M.; Simner, P.J.; Bradford, P.A. Extended-Spectrum β-Lactamases: An Update on Their Characteristics, Epidemiology and Detection. JAC Antimicrob. Resist. 2021, 3, dlab092. [Google Scholar] [CrossRef]
- Price, L.B.; Johnson, J.R.; Aziz, M.; Clabots, C.; Johnston, B.; Tchesnokova, V.; Nordstrom, L.; Billig, M.; Chattopadhyay, S.; Stegger, M.; et al. The Epidemic of Extended-Spectrum-β-Lactamase-Producing Escherichia coli ST131 Is Driven by a Single Highly Pathogenic Subclone, H30-Rx. mBio 2013, 4, e00377-13. [Google Scholar] [CrossRef] [PubMed]
- Belas, A.; Marques, C.; Aboim, C.; Pomba, C. Emergence of Escherichia coli ST131 H30/H30-Rx Subclones in Companion Animals. J. Antimicrob. Chemother. 2019, 74, 266–269. [Google Scholar] [CrossRef]
- Pham, T.D.M.; Ziora, Z.M.; Blaskovich, M.A.T. Quinolone Antibiotics. Medchemcomm 2019, 10, 1719–1739. [Google Scholar] [CrossRef]
- Strahilevitz, J.; Jacoby, G.A.; Hooper, D.C.; Robicsek, A. Plasmid-Mediated Quinolone Resistance: A Multifaceted Threat. Clin. Microbiol. Rev. 2009, 22, 664–689. [Google Scholar] [CrossRef] [PubMed]
- Mirzaii, M.; Jamshidi, S.; Zamanzadeh, M.; Marashifard, M.; Malek Hosseini, S.A.A.; Haeili, M.; Jahanbin, F.; Mansouri, F.; Darban-Sarokhalil, D.; Khoramrooz, S.S. Determination of gyrA and parC Mutations and Prevalence of Plasmid-Mediated Quinolone Resistance Genes in Escherichia coli and Klebsiella pneumoniae Isolated from Patients with Urinary Tract Infection in Iran. J. Glob. Antimicrob. Resist. 2018, 13, 197–200. [Google Scholar] [CrossRef] [PubMed]
- Tang, K.; Zhao, H. Quinolone Antibiotics: Resistance and Therapy. Infect. Drug Resist. 2023, 16, 811–820. [Google Scholar] [CrossRef] [PubMed]
- Nordmann, P.; Poirel, L. Emergence of Plasmid-Mediated Resistance to Quinolones in Enterobacteriaceae. J. Antimicrob. Chemother. 2005, 56, 463–469. [Google Scholar] [CrossRef]
- Mirjalili, M.; Mirzaei, E.; Vazin, A. Pharmacological Agents for the Prevention of Colistin-Induced Nephrotoxicity. Eur. J. Med. Res. 2022, 27, 64. [Google Scholar] [CrossRef]
- Hussein, N.H.; Al-Kadmy, I.M.S.; Taha, B.M.; Hussein, J.D. Mobilized Colistin Resistance (Mcr) Genes from 1 to 10: A Comprehensive Review. Mol. Biol. Rep. 2021, 48, 2897–2907. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Smith, A.L. Emergence of Mobile Colistin Resistance Genes within Los Angeles County Wastewater. Environ. Sci. Technol. Lett. 2023, 10, 316–321. [Google Scholar] [CrossRef]
- Shen, Y.; Zhang, R.; Schwarz, S.; Wu, C.; Shen, J.; Walsh, T.R.; Wang, Y. Farm Animals and Aquaculture: Significant Reservoirs of Mobile Colistin Resistance Genes. Environ. Microbiol. 2020, 22, 2469–2484. [Google Scholar] [CrossRef]
- Singh, A.S.; Nayak, B.B.; Kumar, S.H. High Prevalence of Multiple Antibiotic-Resistant, Extended-Spectrum β-Lactamase (ESBL)-Producing Escherichia coli in Fresh Seafood Sold in Retail Markets of Mumbai, India. Vet. Sci. 2020, 7, 46. [Google Scholar] [CrossRef] [PubMed]
- CLSI Supplement M100; CLSI Performance Standards for Antimicrobial Susceptibility Testing, 33rd ed. CLSI: Wayne, PA, USA, 2023.
- Gay, K.; Robicsek, A.; Strahilevitz, J.; Park, C.H.; Jacoby, G.; Barrett, T.J.; Medalla, F.; Chiller, T.M.; Hooper, D.C. Plasmid-Mediated Quinolone Resistance in Non-Typhi Serotypes of Salmonella enterica. Clin. Infect. Dis. 2006, 43, 297–304. [Google Scholar] [CrossRef] [PubMed]
- Yamane, K.; Wachino, J.; Suzuki, S.; Arakawa, Y. Plasmid-Mediated qepA Gene among Escherichia coli Clinical Isolates from Japan. Antimicrob. Agents Chemother. 2008, 52, 1564–1566. [Google Scholar] [CrossRef] [PubMed]
- Jacoby, G.A.; Walsh, K.E.; Mills, D.M.; Walker, V.J.; Oh, H.; Robicsek, A.; Hooper, D.C. qnrB, Another Plasmid-Mediated Gene for Quinolone Resistance. Antimicrob. Agents Chemother. 2006, 50, 1178–1182. [Google Scholar] [CrossRef] [PubMed]
- Rebelo, A.R.; Bortolaia, V.; Kjeldgaard, J.S.; Pedersen, S.K.; Leekitcharoenphon, P.; Hansen, I.M.; Guerra, B.; Malorny, B.; Borowiak, M.; Hammerl, J.A.; et al. Multiplex PCR for Detection of Plasmid-Mediated Colistin Resistance Determinants, mcr-1, mcr-2, mcr-3, mcr-4 and mcr-5 for Surveillance Purposes. Eurosurveillance 2018, 23, 17-00672. [Google Scholar] [CrossRef] [PubMed]
- Onseedaeng, S.; Ratthawongjirakul, P. Rapid Detection of Genomic Mutations in gyrA and parC Genes of Escherichia coli by Multiplex Allele Specific Polymerase Chain Reaction. J. Clin. Lab. Anal. 2016, 30, 947–955. [Google Scholar] [CrossRef] [PubMed]
- CLSI Document M07-A10; CLSI Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically. Approved Standard—10th ed. CLSI: Wayne, PA, USA, 2015.
- EUCAST European Committee on Antimicrobial Susceptibility: Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version; 2019. 2019.
- Clermont, O.; Christenson, J.K.; Denamur, E.; Gordon, D.M. The Clermont Escherichia coli Phylo-Typing Method Revisited: Improvement of Specificity and Detection of New Phylo-Groups. Environ. Microbiol. Rep. 2013, 5, 58–65. [Google Scholar] [CrossRef]
- Clermont, O.; Dixit, O.V.A.; Vangchhia, B.; Condamine, B.; Dion, S.; Bridier-Nahmias, A.; Denamur, E.; Gordon, D. Characterization and Rapid Identification of Phylogroup G in Escherichia coli, a Lineage with High Virulence and Antibiotic Resistance Potential. Environ. Microbiol. 2019, 21, 3107–3117. [Google Scholar] [CrossRef] [PubMed]
- Nawaz, M.; Sung, K.; Kweon, O.; Khan, S.; Nawaz, S.; Steele, R. Characterisation of Novel Mutations Involved in Quinolone Resistance in Escherichia coli Isolated from Imported Shrimp. Int. J. Antimicrob. Agents 2015, 45, 471–476. [Google Scholar] [CrossRef] [PubMed]
- Juraschek, K.; Deneke, C.; Schmoger, S.; Grobbel, M.; Malorny, B.; Käsbohrer, A.; Schwarz, S.; Meemken, D.; Hammerl, J.A. Phenotypic and Genotypic Properties of Fluoroquinolone-Resistant, Qnr-Carrying Escherichia coli Isolated from the German Food Chain in 2017. Microorganisms 2021, 9, 1308. [Google Scholar] [CrossRef] [PubMed]
- Belotindos, L.P.; Tsunoda, R.; Villanueva, M.A.; Nakajima, C.; Mingala, C.N.; Suzuki, Y. Characterisation of Plasmids Harbouring qnrA1, qnrS1, and qnrB4 in E. coli Isolated in the Philippines from Food-Producing Animals and Their Products. J. Glob. Antimicrob. Resist. 2022, 30, 38–46. [Google Scholar] [CrossRef]
- Jiang, H.-X.; Tang, D.; Liu, Y.-H.; Zhang, X.-H.; Zeng, Z.-L.; Xu, L.; Hawkey, P.M. Prevalence and Characteristics of β-Lactamase and Plasmid-Mediated Quinolone Resistance Genes in Escherichia coli Isolated from Farmed Fish in China. J. Antimicrob. Chemother. 2012, 67, 2350–2353. [Google Scholar] [CrossRef]
- Chandran, S.P.; Diwan, V.; Tamhankar, A.J.; Joseph, B.V.; Rosales-Klintz, S.; Mundayoor, S.; Lundborg, C.S.; Macaden, R. Detection of Carbapenem Resistance Genes and Cephalosporin, and Quinolone Resistance Genes along with oqxAB Gene in Escherichia coli in Hospital Wastewater: A Matter of Concern. J. Appl. Microbiol. 2014, 117, 984–995. [Google Scholar] [CrossRef]
- Shetty, S.S.; Deekshit, V.K.; Jazeela, K.; Vittal, R.; Rohit, A.; Chakraborty, A.; Karunasagar, I. Plasmid-Mediated Fluoroquinolone Resistance Associated with Extra-Intestinal Escherichia coli Isolates from Hospital Samples. Indian. J. Med. Res. 2019, 149, 192–198. [Google Scholar] [CrossRef]
- Ewers, C.; Göpel, L.; Prenger-Berninghoff, E.; Semmler, T.; Kerner, K.; Bauerfeind, R. Occurrence of Mcr-1 and Mcr-2 Colistin Resistance Genes in Porcine Escherichia coli Isolates (2010-2020) and Genomic Characterization of mcr-2-Positive E. coli. Front. Microbiol. 2022, 13, 1076315. [Google Scholar] [CrossRef]
- Mikhayel, M.; Leclercq, S.O.; Sarkis, D.K.; Doublet, B. Occurrence of the Colistin Resistance Gene mcr-1 and Additional Antibiotic Resistance Genes in ESBL/AmpC-Producing Escherichia coli from Poultry in Lebanon: A Nationwide Survey. Microbiol. Spectr. 2021, 9, e0002521. [Google Scholar] [CrossRef] [PubMed]
- Karim, M.R.; Zakaria, Z.; Hassan, L.; Faiz, N.M.; Ahmad, N.I. The Occurrence and Molecular Detection of mcr-1 and mcr-5 Genes in Enterobacteriaceae Isolated from Poultry and Poultry Meats in Malaysia. Front. Microbiol. 2023, 14, 1208314. [Google Scholar] [CrossRef]
- Ovejero, C.M.; Delgado-Blas, J.F.; Calero-Caceres, W.; Muniesa, M.; Gonzalez-Zorn, B. Spread of mcr-1-Carrying Enterobacteriaceae in Sewage Water from Spain. J. Antimicrob. Chemother. 2017, 72, 1050–1053. [Google Scholar] [CrossRef] [PubMed]
- Manohar, P.; Shanthini, T.; Ayyanar, R.; Bozdogan, B.; Wilson, A.; Tamhankar, A.J.; Nachimuthu, R.; Lopes, B.S. The Distribution of Carbapenem- and Colistin-Resistance in Gram-Negative Bacteria from the Tamil Nadu Region in India. J. Med. Microbiol. 2017, 66, 874–883. [Google Scholar] [CrossRef] [PubMed]
- Gandra, S.; Tseng, K.K.; Arora, A.; Bhowmik, B.; Robinson, M.L.; Panigrahi, B.; Laxminarayan, R.; Klein, E.Y. The Mortality Burden of Multidrug-Resistant Pathogens in India: A Retrospective, Observational Study. Clin. Infect. Dis. 2019, 69, 563–570. [Google Scholar] [CrossRef]
- Ghafur, A.; Shankar, C.; GnanaSoundari, P.; Venkatesan, M.; Mani, D.; Thirunarayanan, M.A.; Veeraraghavan, B. Detection of Chromosomal and Plasmid-Mediated Mechanisms of Colistin Resistance in Escherichia coli and Klebsiella pneumoniae from Indian Food Samples. J. Glob. Antimicrob. Resist. 2019, 16, 48–52. [Google Scholar] [CrossRef]
- Anyanwu, M.U.; Jaja, I.F.; Nwobi, O.C. Occurrence and Characteristics of Mobile Colistin Resistance (mcr) Gene-Containing Isolates from the Environment: A Review. Int. J. Environ. Res. Public Health 2020, 17, 1028. [Google Scholar] [CrossRef] [PubMed]
- Lemlem, M.; Aklilu, E.; Mohamed, M.; Kamaruzzaman, N.F.; Zakaria, Z.; Harun, A.; Devan, S.S.; Kamaruzaman, I.N.A.; Reduan, M.F.H.; Saravanan, M. Phenotypic and Genotypic Characterization of Colistin-Resistant Escherichia coli with mcr-4, mcr-5, mcr-6, and mcr-9 Genes from Broiler Chicken and Farm Environment. BMC Microbiol. 2023, 23, 392. [Google Scholar] [CrossRef] [PubMed]
- Delannoy, S.; Le Devendec, L.; Jouy, E.; Fach, P.; Drider, D.; Kempf, I. Characterization of Colistin-Resistant Escherichia coli Isolated from Diseased Pigs in France. Front. Microbiol. 2017, 8, 2278. [Google Scholar] [CrossRef] [PubMed]
- Rafique, M.; Potter, R.F.; Ferreiro, A.; Wallace, M.A.; Rahim, A.; Ali Malik, A.; Siddique, N.; Abbas, M.A.; D’Souza, A.W.; Burnham, C.-A.D.; et al. Genomic Characterization of Antibiotic Resistant Escherichia coli Isolated From Domestic Chickens in Pakistan. Front. Microbiol. 2019, 10, 3052. [Google Scholar] [CrossRef] [PubMed]
- Macori, G.; Nguyen, S.V.; Naithani, A.; Hurley, D.; Bai, L.; El Garch, F.; Woehrlé, F.; Miossec, C.; Roques, B.; O’Gaora, P.; et al. Characterisation of Early Positive Mcr-1 Resistance Gene and Plasmidome in Escherichia coli Pathogenic Strains Associated with Variable Phylogroups under Colistin Selection. Antibiotics 2021, 10, 1041. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).