Abstract
Abraham model solute descriptors are reported for the first time for 62 additional C10 through C13 methyl- and ethyl-branched alkanes. The numerical values were determined using published gas chromatographic retention Kováts retention indices for 157 alkane solutes eluted from a squalane stationary phase column. The 95 alkane solutes that have known descriptor values were used to construct the Abraham model KRI versus L-solute descriptor correlation needed in our calculations. The calculated solute descriptors can be used in conjunction with previously published Abraham model correlations to predict a wide range of important physico-chemical and biological properties. The predictive computations are illustrated by estimating the air-to-polydimethylsiloxane partition coefficient for each of the 157 alkane solutes.
1. Introduction
Linear free energy relationships (LFERs) and Quantitative Structure Property Relationships (QSPRs) provide a convenient means to estimate physical and thermodynamic properties in the absence of direct experimental data. Predictive expressions have been developed for a wide range of properties including surface tensions, vapor pressures, boiling point and melting point temperatures, chromatographic retention indices, partition coefficients and enthalpies of solvation. The more successful methods not only provide reasonably accurate estimates of the desired property, but also further our understanding of the molecular interactions and structural features that govern the property of the specific molecule or specific solute–solvent combination under consideration. It is only by thoroughly understanding a process that one obtains the knowledge necessary to hopefully develop a more comprehensive predictive method.
The particular model that we [1,2,3,4] have been promoting during the last 20 years is commonly referred to as the Abraham solvation parameter method [5,6,7,8] that was originally developed to describe solute transfer between two condensed phases:
and solute transfer from the gas phase into a condensed phase:
Solute Property = eeq 1 × E + seq 1 × S + aeq 1 × A + beq 1 × B + veq 1 × V + ceq 1
Solute Property = eeq 2 × E + seq 2 × S + aeq 2 × A + beq 2 × B + leq 2 × L + ceq 2
Logarithms of the water-to-organic solvent, log P, and gas-to-organic solvent partition coefficients, log K, were among the first properties to be correlated. The model was subsequently extended to other solute transfer processes, such as molar solubility ratios [1,2,3,4], blood-to-tissue and gas-to-tissue partition coefficients [9,10], gas–liquid chromatographic and high-performance liquid chromatographic retention factors/indices [11,12,13], and enthalpies of solvation [14,15,16], as well as several important physical and biological response properties [17,18,19]. For each of the aforementioned properties the mathematical form of the predictive expression is retained for transfer processes pertaining to nonionic molecular compounds. Additional terms are needed on the right-hand side of Equations (1) and (2) to describe transfer properties for ionic and zwitterionic species which can interact with surrounding solvent molecules through their ionic moieties. Equations (1) and (2) do not contain provisions for solvent molecules to interact with ionic moieties on the dissolved solute.
As we just alluded the terms on the right-hand side of Equations (1) and (2) represent the different types of solute–solvent molecular interactions that are believed to be present in nonelectrolyte solutions. Each type of interaction is quantified as a product of a solute property multiplied by the complimentary solvent property. Solute properties (commonly referred to as solute descriptors) are identified by the uppercase alphabetical characters and are defined as follows: A and B refer to the respective overall hydrogen-bond donating and accepting capacities of the dissolved solute; E corresponds to the molar refraction of the given solute (in units of (cm3 mol−1)/10) in excess of that of a linear alkane having a comparable molecular size; L is the logarithm of the solute’s gas-to-hexadecane partition coefficient determined at 298.15 K; S represents a combination of the electrostatic polarity and polarizability of the solute; and V denotes the McGowan molecular volume of the solute (in units of (cm3 mol−1)/100) calculated from atomic sizes and chemical bond numbers. The complimentary solvent properties in Equations (1) and (2) are given by the lowercase alphabetical characters (ceq 1, eeq 1, seq 1, aeq 1, beq 1, veq 1, ceq 2, eeq 2, seq 2, aeq 2, beq 2, and leq 2). Numerical values of the solvent properties are determined by regressing measured data for a series of solutes with known descriptor values in accordance with Equations (1) and (2). Once determined, the lowercase alphabetical characters allow one to predict the specified property of additional solutes in the given organic solvent, provided of course that the solute descriptors are known. The Abraham solvation parameter model has been described in greater detail in several published papers, book chapters and review articles [5,20,21,22,23].
Continued development of the Abraham model requires that: (a) additional measurements be performed so that descriptor values and equation coefficients can be calculated for more solutes and transfer processes; and (b) that existing experimental data be more effectively utilized in determining solute descriptors and process coefficients. There is an enormous quantity of published data that could be used in expanding the applicability of the Abraham model. Utilization of existing experimental data is not always easy in that solute descriptors may have to be first calculated in order to obtain a sufficient number of data points to permit the determination of equation coefficients, and vice versa. As noted above, the equation coefficients are determined by regressing measured data for a series of solutes whose descriptor values are known in accordance with Equations (1) and (2).
There are many published papers in the chemical literature that report chromatographic retention data for the compounds found in essential oils [24,25,26], beverages [27,28], petroleum and coal-based samples [29,30], insects [31,32] and bee honeys [33]. The published datasets contain a large number of pesticides, flavor and fragrance compounds for which solute descriptors are not available. Unfortunately, many of the published papers do not contain Abraham model correlations for the different stationary phases and column temperatures that were used to achieve the reported chemical separations. Descriptor values are known for too few solutes in several of the datasets for us to obtain a meaningful Abraham correlation. We are in the process of addressing this problem by determining solute descriptors for several of the compounds contained in each dataset that we plan to use in our future studies. The current communication reports Abraham model solute descriptor values for an additional 62 C10 through C13 methyl- and ethyl-branched alkanes based on the published chromatographic data contained in the paper by Heinzen and coworkers [34]. The paper contains retention indices for 157 linear and branched alkanes. Abraham model solute descriptors are currently known for all but 62 of the methyl- and ethyl-branched alkanes given in the paper.
2. Computational Methodology for Calculation of Abraham Model Solute Descriptors
Normally the determination of Abraham model solute descriptors involves constructing a series of mathematical expressions for the measured solute properties of the given solute in a series of solvents and/or for a series of processes for which the lowercase equation coefficients are known. The computational procedure is greatly simplified for the compounds considered in the current study (see Table 1 for the complete list of alkane solutes) because four of the solute descriptors are equal to zero. In other words, E = 0, S = 0, A = 0 and B = 0. Methyl- and ethyl-branched alkane solutes possess no excess molar refraction (E = 0) or polarity/polarizability (S = 0), and are not capable of hydrogen-bond formation (A = 0 and B = 0) with the surrounding solvent molecules. The V solute descriptor is readily calculated from the solute’s molecular structure, the atomic volumes of the constituent atoms contained in the solute molecule and the number of chemical bonds in the solute molecule as described by Abraham and McGowan [35]. The calculated V solute descriptors for the four molecular formulas are: V = 1.5175 for C10H22; V = 1.6585 for C11H24; V = 1.7994 for C12H26; and V = 1.9403 for C13H28. Only the L solute descriptor remains to be calculated from the published chromatographic retention data.

Table 1.
Kováts Gas Chromatographic Retention Indices, KRI and Abraham Model L-Solute Descriptor Values for Linear Alkanes, and Methyl- and Ethyl-branched Alkanes Eluted from a Squalane Stationary Phase Liquid Column.
Isothermal chromatographic retention is often described in terms of either the retention factor [11,36]:
or the Kováts retention index, KRI [37]:
where tr(A) denotes the retention time of analyte A, tm refers to the so-called “hold-up” time measured by an unretained compound on the column and tr(z2) and tr(z1) are the retention times of two linear alkanes having z2 and z1 carbon atoms, respectively. Poole and coworkers [12,13,38,39,40] have reported Abraham model correlations for describing the elution behavior of organic solutes, in terms of log k(A), on a wide range of gas chromatographic stationary phase liquids, and for a wide range of HPLC stationary–mobile combinations, at different temperatures. While the published correlations are extremely useful if one wishes to predict and/or analyze retention factor values, the published gas chromatographic data that we wish to analyze is given in terms of KRI values.
k(A) = (tr(A) − tm)/tm
We provide the basis for using the Abraham model to describe KRI values through the following mathematical manipulations. First, we substitute into Equation (2) the numerical descriptor values of the solutes:
and the numerical values of the two reference linear alkanes:
log ksolute = eeq 2 × Esolute + seq 2 × Ssolute + aeq 2 × Asolute + beq 2 × Bsolute + leq 2 × Lsolute + ceq 2
log kz1 = leq 2 × Lz1 + ceq 2
log kz2 = leq 2 × Lz2 + ceq 2
Remember that the E, S, A and B solute descriptors of the reference linear alkanes are equal to zero, so only the ceq 2 and leq 2 terms contribute. A combination of the Equations (4)–(7) yields the following expression:
which upon suitable algebraic rearrangement will give a mathematical form:
that is consistent with the Abraham model. We have changed the subscripting so as not imply that the numerical values of equation coefficients in Equation (9) are the same as those in Equation (2).
KRI = eeq 9 × E + seq 9 × S + aeq 9 × A + beq 9 × B + leq 9 × L + ceq 9
Equation (9) provides the basis for the mathematical relationship between KRI and the L-solute descriptor. In Table 1 we have assembled the Kováts retention indices for the 157 linear and branched alkanes from the published paper by Heinzen and coworkers [34], along with the known descriptor values from our private database. In total we have 95 experimental values to use in developing our Abraham model KRI versus the L-solute descriptor correlation. The analysis of the numerical values in the second and third columns of Table 1 yielded the following mathematical expression:
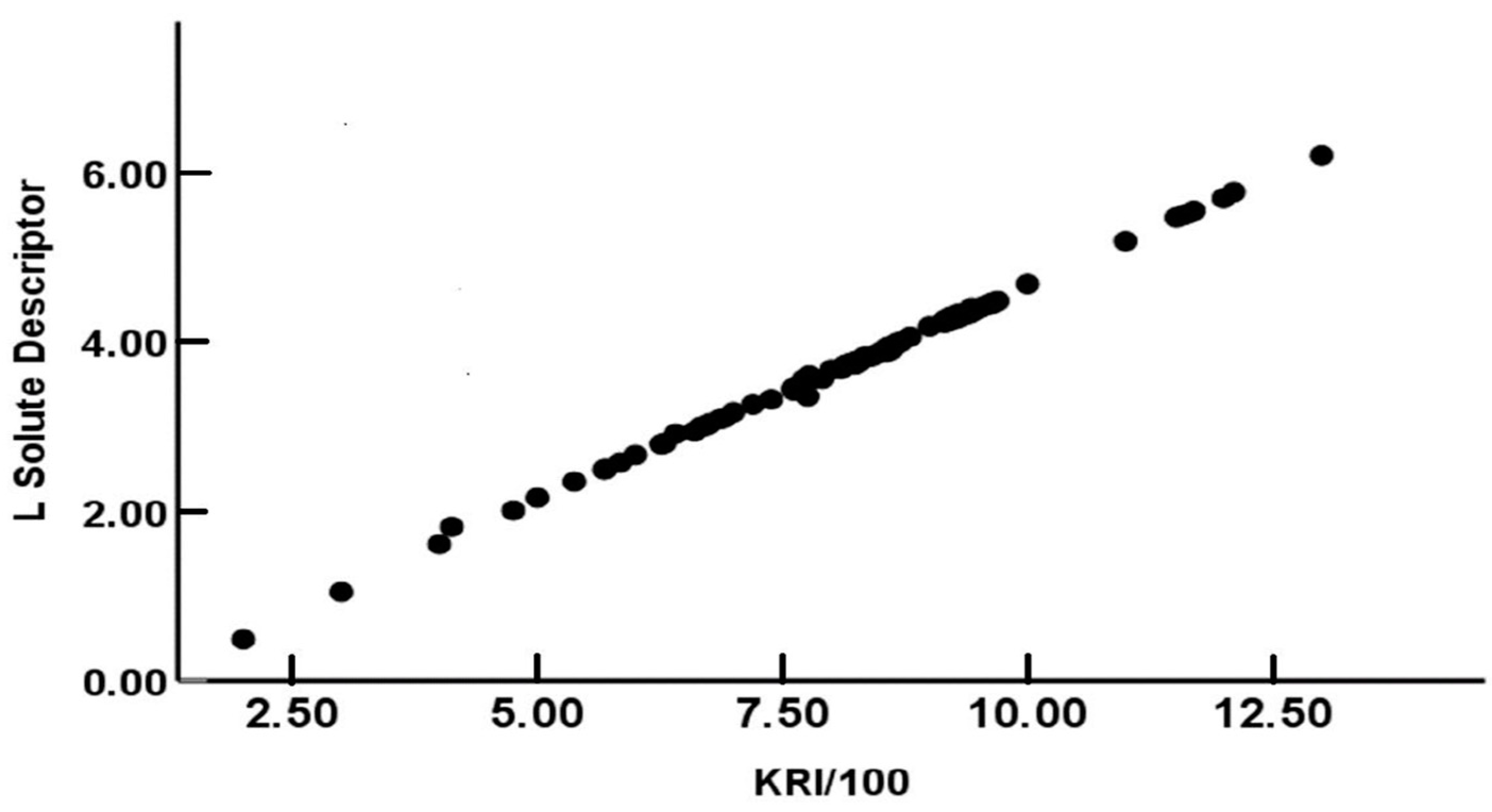
where N represents the number of experimental data points used in obtaining the linear relationship, SD gives the standard deviation of the residuals, R2 and refer to the squared and adjusted squared correlation coefficient, respectively, and F is the Fisher F-statistic. The calculated standard errors in the slope and intercept are given in parentheses immediately following the numerical value of the corresponding equation coefficient. Equation (10) was found to back-calculate the L-descriptor values used in the least-squares analysis to within an average absolute deviation of AAE = 0.025 and an average error of AE = 0.003. Figure 1 depicts the linear plot of the L descriptor values versus KRI/100 values for the 95 data points used in deriving Equation (10). The derived mathematical relationship then allows the calculation of the L solute descriptors of the remaining 62 methyl- and ethyl-branched alkanes for which solute descriptors are not currently available. These calculations are summarized in the last column of Table 1. Solute descriptors for these 62 additional alkanes will now be added to our private database, and will be available to us in future planned studies directed towards determining descriptor values for additional compounds from published chromatographic retention data.
L = 0.508(0.017) × (KRI/100) − 0.412(0.002)
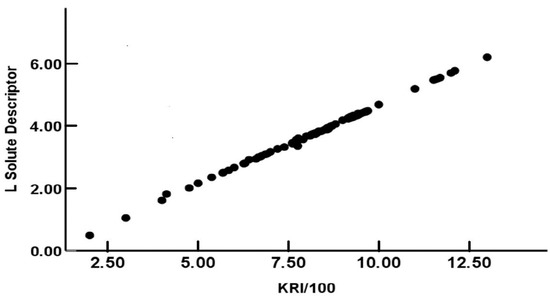
Figure 1.
Comparison of the experimental-based L-solute descriptors in our database and back-calculated values based on Equation (10).
3. Calculation of Air-to-Polydimethylsiloxane Partition Coefficients
In earlier publications [41,42,43], we illustrated the prediction of the standard molar enthalpies of vaporization and the standard molar enthalpies of sublimations at 298 K of more than 100 different large mono-methylated and large poly-methylated alkanes using the newly calculated solute descriptor values. Instead of simply repeating the computational procedure using a different set of alkane molecules, we wish to calculate the logarithm of the air-to-polydimethylsiloxane partition coefficient, log KPDMS-air, using our updated Abraham model correlation:
based on the experimental values for 227 different organic compounds and inert gases. Equation (11) back-calculates the observed 227 data points to within a standard deviation of the residuals of SD = 0.177 log units, which is comparable to the experimental uncertainty associated with many of the data points used in the regression analysis. The equation coefficients differ slightly from an earlier correlation reported by Sprunger and coworkers [44] based on a much smaller number of 142 solute molecules. Polydimethylsiloxane, PDMS, is a coating often found in microextraction devices used to sample and analyze total hydrocarbons present in unknown air samples. Martos and coworkers [45] reported experimental log KPDMS-air for 29 smaller C6–C10 branched alkanes. The authors did not determine the experimental values for the larger C11–C13 alkane molecules considered in the current study.
log KPDMS-air = −0.088 × Esolute + 0.493 × Ssolute + 1.056 × Asolute + 0.487 × Bsolute + 0.829 × Lsolute − 0.027
In the third column of Table 2 we have given the values predicted by Equation (11) using either our existing solute descriptors or the values determined in the current study. The numerical values that are tabulated in the last column were retrieved from the published chemical literature [44,45]. For several of the compounds there were multiple experimental values that were determined by independent research groups. Sometimes the independently determined experimental values differed significantly as was the case for: decane, log KPDMS-air = 3.87 [46] versus log KPDMS-air = 3.50 [44]; undecane, log KPDMS-air = 4.40 [47] versus log KPDMS-air = 3.89 [44]; and 3,3-dimethylpentane, log KPDMS-air = 3.42 [45] versus log KPDMS-air = 3.70 [48]. No attempt was made to select the experimental values that came closest to the calculated values based on Equation (11) as we wished to illustrate that predictive expressions can be used to identify possible outlier values in need of redetermination.

Table 2.
Comparison of the Abraham Model Calculated versus Experimental Logarithms of the Air-to-Polydimethylsiloxane Partition Coefficients, log KPDMS-air, for the Linear Alkanes, and Methyl- and Ethyl-branched Alkanes Considered in the Current Study.
4. Summary
The current study represents a continuation of our ongoing efforts to determine experimental-based solute descriptors from measured solubility, partition coefficient and/or chromatographic retention data. Abraham model solute descriptors are reported for the first time for 62 additional C10 through C13 methyl- and ethyl-branched alkanes. The numerical values were determined using published gas chromatographic retention Kováts retention indices for 157 alkane solutes eluted from a squalane stationary phase column. The 95 alkane solutes that have known descriptor values were used to construct the Abraham model KRI versus L-solute descriptor correlation needed in our calculations. The calculated solute descriptors can be used in conjunction with previously published Abraham model correlations to predict a wide range of important physico-chemical and biological properties, including partition coefficients, vapor pressures, the standard molar enthalpies of vaporization and sublimation, chromatographic retention factors and retention times, nasal pungencies and eye irritation thresholds. Of the aforementioned properties, the chromatographic retention times will likely be the most useful as this can aid in the identification of compounds present in unknown chemical samples. The predictive computations are illustrated by calculating the air-to-polydimethylsiloxane partition coefficients of the 157 alkane solutes. Polydimethylsiloxane, PDMS, is a coating often found in microextraction devices used to sample and analyze total hydrocarbons present in unknown air samples.
As part of the current study, an expression was derived which shows the mathematical relationship between the equation coefficients given in the Abraham model log k expression versus those used in the KRI correlation. The derived relationship, Equation (8), provides a possible means to conveniently obtain the Abraham model KRI expression from existing log k correlations. All that is needed in the conversion is the L-solute descriptor values of the linear alkanes used in the calculation of Kováts retention indices.
We note at this time that the popularity of the Abraham solvation parameter model has recently prompted several researcher groups [49,50,51,52] to develop either group contribution, quantum chemical or machine learning models to estimate the numerical values of solute descriptors. Our experience in using the different estimation software programs is that the methods do provide fairly good estimates of the descriptor values for simple compounds; however, as the structural complexity of the solute increases, the “quality” of the estimations decreases. For example, we have previously shown that two software programs overestimate the hydrogen-bond acidity (e.g., A solute descriptor) for 4,5-dihydroxyanthraquinone-2-carboxylic acid [4], 1,4-dihydroxyanthraquinone [53] and 1,8-dihydroxyanthraquinone [53] when compared with values based on measured solubility data. The experimental-based A solute descriptor values were much smaller, and suggested the formation of strong intramolecular hydrogen bonds between the hydrogen of the –OH functional groups and the oxygen atom of the neighboring aromatic carbonyl group. The estimation methods do not appear to incorporate this structural feature and possible intramolecular hydrogen-bond formation in their calculation approach. We have further suggested that poor estimations might also result from the inadequate representation of select functional groups in the datasets used in developing/training the various group contribution, machine learning and quantum chemical methods [54,55]. This is not intended as a criticism of the estimation methods, but rather a statement that the methods are only as “good” as the datasets used in their development. Experimental-based solute descriptors need to be determined for molecules that have a greater chemical diversity and structural complexity. To paraphrase the recent comments of Poole and Atapattu [23]—the expansion of the Abraham model capabilities will stall without other researchers’ participation in the determination of solute descriptors. The calculation of the solute descriptors for the 62 additional branched alkanes considered in the current communication is the first step in this endeavor.
Author Contributions
Conceptualization, writing—original draft preparation, W.E.A.J.; formal data analysis, R.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
Data is contained within article.
Conflicts of Interest
The authors have no conflict of interest to declare.
References
- Kim, K.; Shanmugam, N.; Xu, A.; Varadharajan, A.; Cai, S.K.; Huang, E.; Acree, W.E., Jr. Abraham model correlations for describing solute transfer into anisole based on measured activity coefficients and molar solubilities. Phys. Chem. Liq. 2022, 60, 452–462. [Google Scholar] [CrossRef]
- Longacre, L.; Wu, E.; Yang, C.; Zhang, M.; Sinha, S.; Varadharajan, A.; Acree, W.E., Jr. Development of Abraham model correlations for solute transfer into the tert-butyl acetate mono-solvent and updated equations for both ethyl acetate and butyl acetate. Liquids 2022, 2, 258–288. [Google Scholar] [CrossRef]
- Varadharajan, A.; Sinha, S.; Xu, A.; Daniel, A.; Kim, K.; Shanmugam, N.; Wu, E.; Yang, C.; Zhang, M.; Acree, W.E., Jr. Development of Abraham model correlations for describing solute transfer into transcutol based on molar solubility ratios for pharmaceutical and other organic compounds. J. Solut. Chem. 2022, 52, 70–90. [Google Scholar] [CrossRef]
- Sinha, S.; Yang, C.; Wu, E.; Acree, W.E., Jr. Abraham solvation parameter model: Examination of possible intramolecular hydrogen-bonding using calculated solute descriptors. Liquids 2022, 2, 131–146. [Google Scholar] [CrossRef]
- Abraham, M.H. Scales of solute hydrogen-bonding: Their construction and application to physicochemical and biochemical processes. Chem. Soc. Rev. 1993, 22, 73–83. [Google Scholar] [CrossRef]
- Abraham, M.H.; Ibrahim, A.; Zissimos, A.M. Determination of sets of solute descriptors from chromatographic measurements. J. Chromatogr. A 2004, 1037, 29–47. [Google Scholar] [CrossRef]
- Abraham, M.H.; Smith, R.E.; Luchtefeld, R.; Boorem, A.J.; Luo, R.; Acree, W.E., Jr. Prediction of solubility of drugs and other compounds in organic solvents. J. Pharm. Sci. 2010, 99, 1500–1515. [Google Scholar] [CrossRef]
- Abraham, M.H.; Acree, W.E., Jr. Descriptors for the prediction of partition coefficients of 8-hydroxyquinoline and its derivatives. Sep. Sci. Technol. 2014, 49, 2135–2141. [Google Scholar] [CrossRef]
- Abraham, M.H.; Ibrahim, A.; Zhao, Y.; Acree, W.E., Jr. A data base for partition of volatile organic compounds and drugs from blood/plasma/serum to brain, and an LFER analysis of the data. J. Pharm. Sci. 2006, 95, 2091–2100. [Google Scholar] [CrossRef]
- Abraham, M.H.; Ibrahim, A. Air to fat and blood to fat distribution of volatile organic compounds and drugs: Linear free energy analyses. Eur. J. Med. Chem. 2006, 41, 1430–1438. [Google Scholar] [CrossRef]
- Twu, P.; Zhao, Q.; Pitner, W.R.; Acree, W.E., Jr.; Baker, G.A.; Anderson, J.L. Evaluating the solvation properties of functionalized ionic liquids with varied cation/anion composition using the solvation parameter model. J. Chromatogr. A 2011, 1218, 5311–5318. [Google Scholar] [CrossRef] [PubMed]
- Poole, C.F.; Lenca, N. Applications of solvation parameter model in reversed-phase liquid chromatography. J. Chromatogr. A 2017, 1486, 2–19. [Google Scholar] [CrossRef]
- Poole, C.F. Gas chromatography system constant database for 52 wall-coated, open-tubular columns covering the temperature range 60–140 °C. J. Chromatogr. A 2019, 1604, 460482. [Google Scholar] [CrossRef] [PubMed]
- Magsumov, T.I.; Sedov, I.A.; Acree, W.E., Jr. Development of Abraham model correlations for enthalpies of solvation of solutes dissolved in N-methylformamide, 2-pyrrolidone and N-methylpyrrolidone. J. Mol. Liq. 2021, 323, 1–17. [Google Scholar]
- Stolov, M.A.; Zaitseva, K.V.; Varfolomeev, M.A.; Acree, W.E., Jr. Enthalpies of solution and enthalpies of solvation of organic solutes in ethylene glycol at 298.15 K: Prediction and analysis of intermolecular interaction contributions. Thermochim. Acta 2017, 648, 91–99. [Google Scholar] [CrossRef]
- Varfolomeev, M.A.; Stolov, M.A.; Nagrimanov, R.N.; Rakipov, I.T.; Acree, W.E., Jr.; Abraham, M.H. Analysis of solute-pyridine intermolecular interactions based on experimental enthalpies of solution and enthalpies of solvation of solutes dissolved in pyridine. Thermochim. Acta 2018, 660, 11–17. [Google Scholar] [CrossRef]
- Abraham, M.H.; Sanchez-Moreno, R.; Cometto-Muniz, J.E.; Cain, W.S. A quantitative structure-activity analysis on the relative sensitivity of the olfactory and the nasal trigeminal chemosensory systems. Chem. Senses 2007, 32, 711–719. [Google Scholar] [CrossRef]
- Abraham, M.H.; Gola, J.M.R.; Cometto-Muniz, J.E. An assessment of air quality reflecting the chemosensory irritation impact of mixtures of volatile organic compounds. Environ. Int. 2016, 86, 84–91. [Google Scholar]
- Abraham, M.H.; Acree, W.E., Jr.; Mintz, C.; Payne, S. Effect of anesthetic structure on inhalation anesthesia: Implications for the mechanism. J. Pharm. Sci. 2008, 97, 2373–2384. [Google Scholar]
- Endo, S.; Goss, K.-U. Applications of polyparameter linear free energy relationships in environmental chemistry. Environ. Sci. Technol. 2014, 48, 12477–12491. [Google Scholar]
- Poole, C.F.; Ariyasena, T.C.; Lenca, N. Estimation of the environmental properties of compounds from chromatographic measurements and the solvation parameter model. J Chromatogr. A 2013, 1317, 85–104. [Google Scholar] [CrossRef] [PubMed]
- Jalan, A.; Ashcraft, R.W.; West, R.H.; Green, W.H. Predicting solvation energies for kinetic modeling. Ann. Rep. Prog. Chem. Sect. C Phys. Chem. 2010, 106, 211–258. [Google Scholar] [CrossRef]
- Poole, C.F.; Atapattu, S.N. Recent advances for estimating environmental properties for small molecules from chromatographic measurements and the solvation parameter model. J. Chromatogr. A 2023, 1687, 463682. [Google Scholar] [CrossRef]
- Jalali-Heravi, M.; Ebrahimi-Najafabadi, H. Modeling of retention behaviors of most frequent components of essential oils in polar and non-polar stationary phases. J. Sep. Sci. 2011, 34, 1538–1546. [Google Scholar] [CrossRef] [PubMed]
- Babushok, V.I.; Zenkevich, I.G. Retention indices for most frequently reported essential oil compounds in GC. Chromatographia 2008, 69, 257–269. [Google Scholar] [CrossRef]
- Qin, L.-T.; Liu, S.-S.; Chen, F.; Wu, Q.-S. Development of validated quantitative structure-retention relationship models for retention indices of plant essential oils. J. Sep. Sci. 2013, 36, 1553–1560. [Google Scholar] [CrossRef]
- Cunha, S.C.; Faria, M.A.; Fernandes, J.O. Solid phase extraction in combination with comprehensive two-dimensional gas chromatography coupled to time-of-flight mass spectrometry for the detailed investigation of volatiles in South African red wines. Anal. Chim. Acta 2011, 701, 98–111. [Google Scholar]
- Kotseridis, Y.; Baumes, R. Identification of impact odorants in Bordeaux red grape juice, in the commercial yeast used for its fermentation, and in the produced wine. J. Agric. Food Chem. 2000, 48, 400–406. [Google Scholar] [CrossRef]
- Antle, P.; Zeigler, C.; Robbat, A. Retention behavior of alkylated polycyclic aromatic sulfur heterocycles on immobilized ionic liquid stationary phases. J. Chromatogr. A 2014, 1361, 255–264. [Google Scholar] [CrossRef]
- Woloszyn, T.F.; Jurs, P.C. Prediction of gas chromatographic retention data for hydrocarbons from naphthas. Anal. Chem. 1993, 65, 582–587. [Google Scholar] [CrossRef]
- Pomonis, J.G.; Hakk, H.; Fatland, C.L. Synthetic methyl- and dimethylalkanes. Kovats indexes, carbon-13 NMR and mass spectra of some methylpentacosanes and 2,X-dimethylheptacosanes. J. Chem. Ecol. 1989, 15, 2319–2333. [Google Scholar]
- Junkes, B.S.; Amboni, R.D.M.C.; Heinzen, V.E.F.; Yunes, R.A. Quantitative structure-retention relationships (QSRR), using the optimum semi-empirical topological index, for methyl-branched alkanes produced by insects. Chromatographia 2002, 55, 707–713. [Google Scholar] [CrossRef]
- de Lima Morais da Silva, P.; de Lima, L.S.; Caetano, I.K.; Torres, Y.R. Comparative analysis of the volatile composition of honeys from Brazilian stingless bees by static headspace GC-MS. Food Res. Int. 2017, 102, 536–543. [Google Scholar] [CrossRef] [PubMed]
- Heinzen, V.E.F.; Soares, M.F.; Yunes, R.A. Semi-empirical topological method for the prediction of the chromatographic retention of cis- and trans-alkene isomers and alkanes. J. Chromatogr. A 1999, 849, 495–506. [Google Scholar] [CrossRef] [PubMed]
- Abraham, M.H.; McGowan, J.C. The use of characteristic volumes to measure cavity terms in reversed phase liquid chromatography. Chromatographia 1987, 23, 243–246. [Google Scholar] [CrossRef]
- Baltazar, Q.Q.; Leininger, S.K.; Anderson, J.L. Binary ionic liquid mixtures as gas chromatography stationary phases for improving the separation selectivity of alcohols and aromatic compounds. J. Chromatogr. A 2008, 1182, 119–127. [Google Scholar] [CrossRef]
- Garcia-Dominguez, J.A.; Lebron-Aguilar, R.; Quintanilla-Lopez, J.E. An accurate and easy procedure to obtain isothermal Kovats retention indices in gas chromatography. J. Sep. Sci. 2006, 29, 2785–2792. [Google Scholar] [CrossRef]
- Poole, C.F.; Atapattu, S.N. Analysis of the solvent strength parameter (linear solvent strength model) for isocratic separations in reversed-phase liquid chromatography. J. Chromatogr. A 2022, 1675, 463153. [Google Scholar] [CrossRef]
- Poole, C.F. Gas chromatography system constant database over an extended temperature range for nine open-tubular columns. J. Chromatogr. A 2019, 1590, 130–145. [Google Scholar] [CrossRef]
- Poole, C.F.; Lenca, N. Gas chromatography on wall-coated open-tubular columns with ionic liquid stationary phases. J. Chromatogr. A 2014, 1357, 87–109. [Google Scholar] [CrossRef]
- Liu, G.; Eddula, S.; Jiang, C.; Huang, J.; Tirumala, P.; Xu, A.; Acree, W.E., Jr.; Abraham, M.H. Abraham solvation parameter model: Prediction of enthalpies of vaporization and sublimation of mono-methyl branched alkanes using measured gas chromatographic data. Eur. Chem. Bull. 2020, 9, 273–284. [Google Scholar] [CrossRef]
- Tirumala, P.; Huang, J.; Eddula, S.; Jiang, C.; Xu, A.; Liu, G.; Acree, W.E., Jr.; Abraham, M.H. Calculation of Abraham model L-descriptor and standard molar enthalpies of vaporization and sublimation for C9–C26 mono-alkyl alkanes and polymethyl alkanes. Eur. Chem. Bull. 2020, 9, 317–328. [Google Scholar] [CrossRef]
- Wu, E.; Sinha, S.; Yang, C.; Zhang, M.; Acree, W.E., Jr. Abraham solvation parameter model: Calculation of L solute descriptors for large C11 to C42 methylated alkanes from measured gas-liquid chromatographic retention data. Liquids 2022, 2, 85–105. [Google Scholar] [CrossRef]
- Sprunger, L.M.; Proctor, A.; Acree, W.E., Jr.; Abraham, M.H. Characterization of the sorption of gaseous and organic solutes onto polydimethylsiloxane solid-phase microextraction surfaces using the Abraham model. J. Chromatogr. A 2007, 1175, 162–173. [Google Scholar] [CrossRef]
- Martos, P.A.; Saraullo, A.; Pawliszyn, J. Estimation of air/coating distribution coefficients for solid phase microextraction using retention indexes from linear temperature-programmed capillary gas chromatography. application to the sampling and analysis of total petroleum hydrocarbons in air. Anal. Chem. 1997, 69, 402–408. [Google Scholar] [CrossRef]
- Legind, C.N.; Karlson, U.; Burken, J.G.; Reichenberg, F.; Mayer, P. Determining chemical activity of (semi)volatile compounds by headspace solid-phase microextraction. Anal. Chem. 2007, 79, 2869–2876. [Google Scholar] [CrossRef] [PubMed]
- Martos, P.A.; Pawliszyn, J. Calibration of solid phase microextraction for air analyses based on physical chemical properties of the coating. Anal. Chem. 1997, 69, 206–215. [Google Scholar] [CrossRef]
- Pawliszyn, J. Solid Phase Microextraction: Theory and Practice; John Wiley & Sons: Hoboken, NJ, USA, 1997. [Google Scholar]
- Chung, Y.; Vermeire, F.H.; Wu, H.; Walker, P.J.; Abraham, M.H.; Green, W.H. Group contribution and machine learning approaches to predict Abraham solute parameters, solvation free energy, and solvation enthalpy. J. Chem. Inf. Model 2022, 62, 433–446. [Google Scholar] [CrossRef]
- Ulrich, N.; Ebert, A. Can deep learning algorithms enhance the prediction of solute descriptors for linear solvation energy relationship approaches? Fluid Phase Equilib. 2022, 555, 113349. [Google Scholar] [CrossRef]
- Ulrich, N.; Endo, S.; Brown, T.N.; Watanabe, N.; Bronner, G.; Abraham, M.H.; Goss, K.-U. UFZ-LSER Database v 3.2.1, Leipzig, Germany, Helmholtz Centre for Environmental Research-UFZ. 2017. Available online: http://www.ufz.de/lserd (accessed on 1 December 2022).
- Xiao, Z.J.; Chen, J.W.; Wang, Y.; Wang, Z.Y. In silico package models for deriving values of solute parameters in linear solvation energy relationships. SAR QSAR Environ. Res. 2023; in press. [Google Scholar] [CrossRef]
- Acree, W.E., Jr.; Smart, K.; Abraham, M.H. Abraham model solute descriptors reveal strong intramolecular hydrogen bonding in 1,4-dihydroxyanthraquinone and 1,8-dihydroxyanthraquinone. Phys. Chem. Liq. 2018, 56, 416–420. [Google Scholar] [CrossRef]
- Sinha, S.; Varadharajan, A.; Xu, A.; Wu, E.; Acree, W.E., Jr. Determination of Abraham model solute descriptors for hippuric acid from measured molar solubilities in several organic mono-solvents of varying polarity and hydrogen-bonding ability. Phys. Chem. Liq. 2022, 60, 563–571. [Google Scholar] [CrossRef]
- Yao, E.; Zhou, A.; Wu, S.; Shanmugam, N.; Varadharajan, A.; Sinha, S.; Wu, E.; Acree, W.E., Jr. Determination of Abraham model solute descriptors for N-hydroxyphthalimide: An organic compound having a N-hydroxy (N-OH) functional group. J. Solut. Chem. 2023; submitted for publication. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).