Inclusion Bodies in Ionic Liquids
Abstract
1. Introduction
1.1. Inclusion Bodies
1.2. Mild Solubilization Concept
1.3. Refolding and Purification
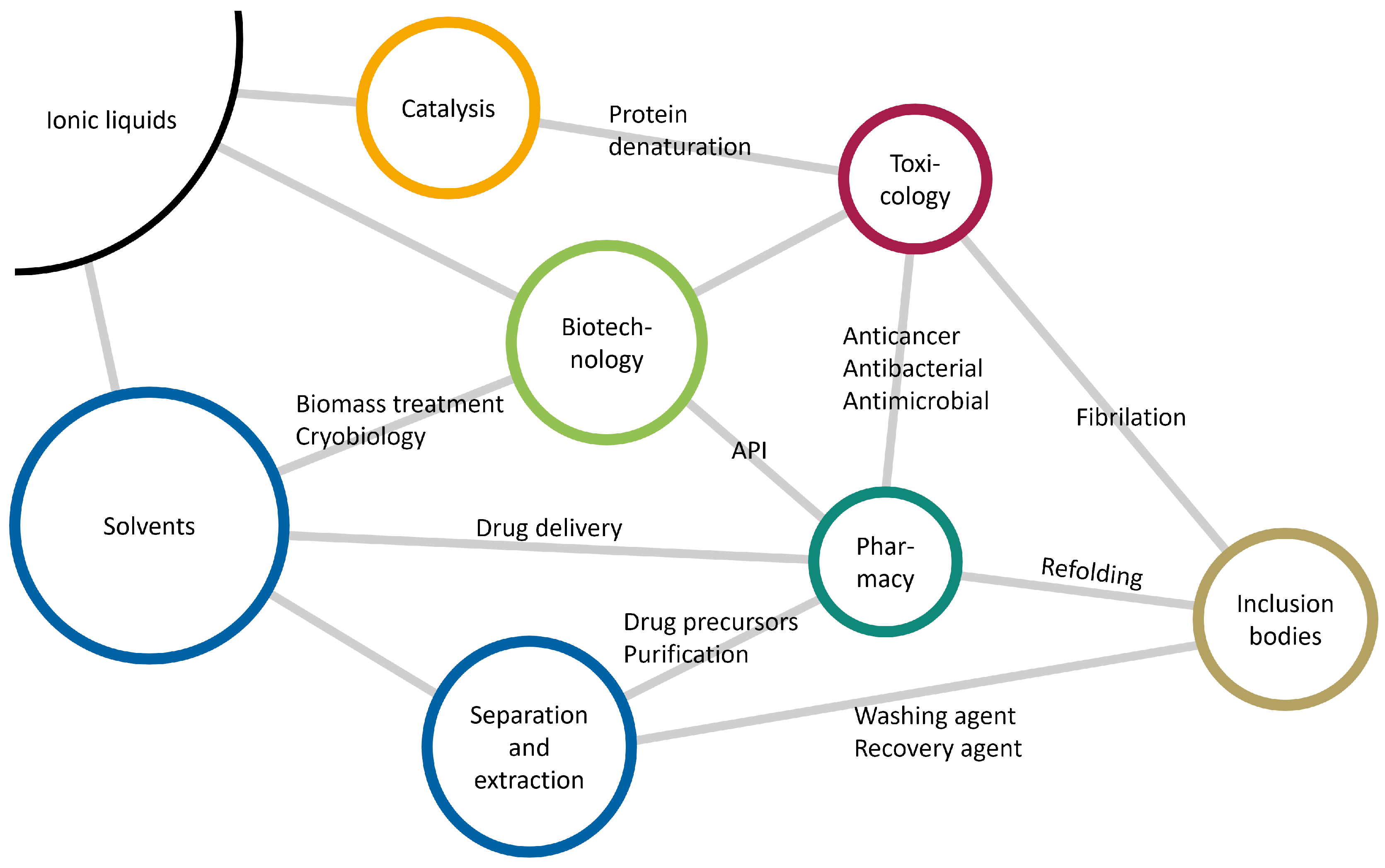
1.4. Ionic Liquids
2. Theoretical Understanding
2.1. From Inclusion Bodies Back to Native State
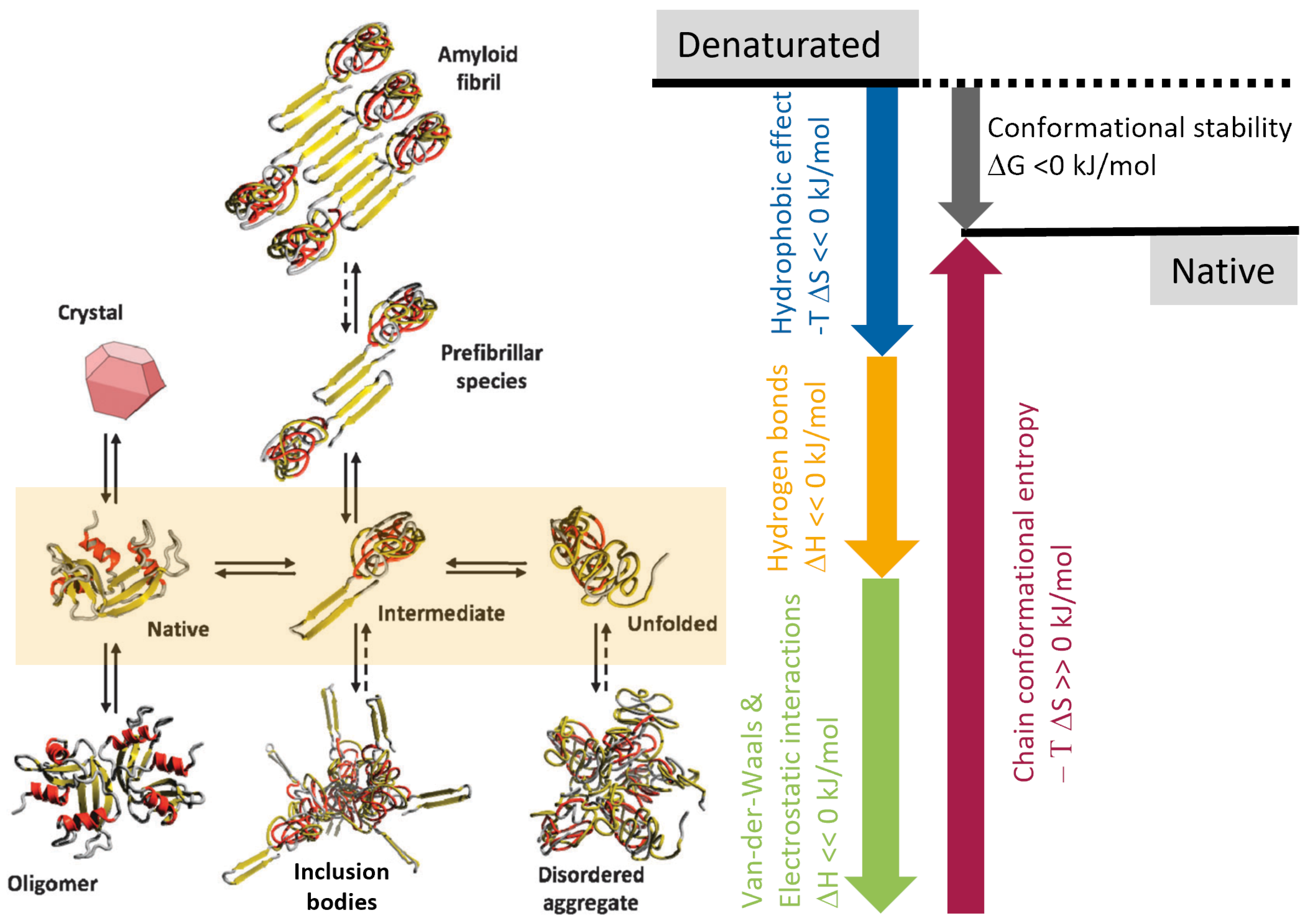
2.1.1. Protein States and Their Aggregation
- Inclusion bodies are dense aggregates of partially misfolded proteins that may still contain a high percentage of -helices and -sheets. They form due to high local concentrations of a specific protein.
- Fibrils, and in particular amyloid fibrils, are highly ordered, stable structures composed of -sheet-rich protein aggregates. These fibrils are characterized by their elongated, fibrous nature. Fibrillation is a specific type of protein aggregation often associated with pathological conditions.
- Disordered aggregates are irregular, amorphous conglomerations of proteins. Unlike proteins in inclusion bodies or fibrils, they lack a defined secondary or tertiary structure and are more heterogeneous. These aggregates form when proteins misfold or (partially) unfold, leading to exposure of hydrophobic residues that facilitate aggregation.
2.1.2. Pathways of (Re-)Folding
2.2. Interaction with the Refolding Buffer
2.2.1. Gibbs Free Energy as a Function of Concentration
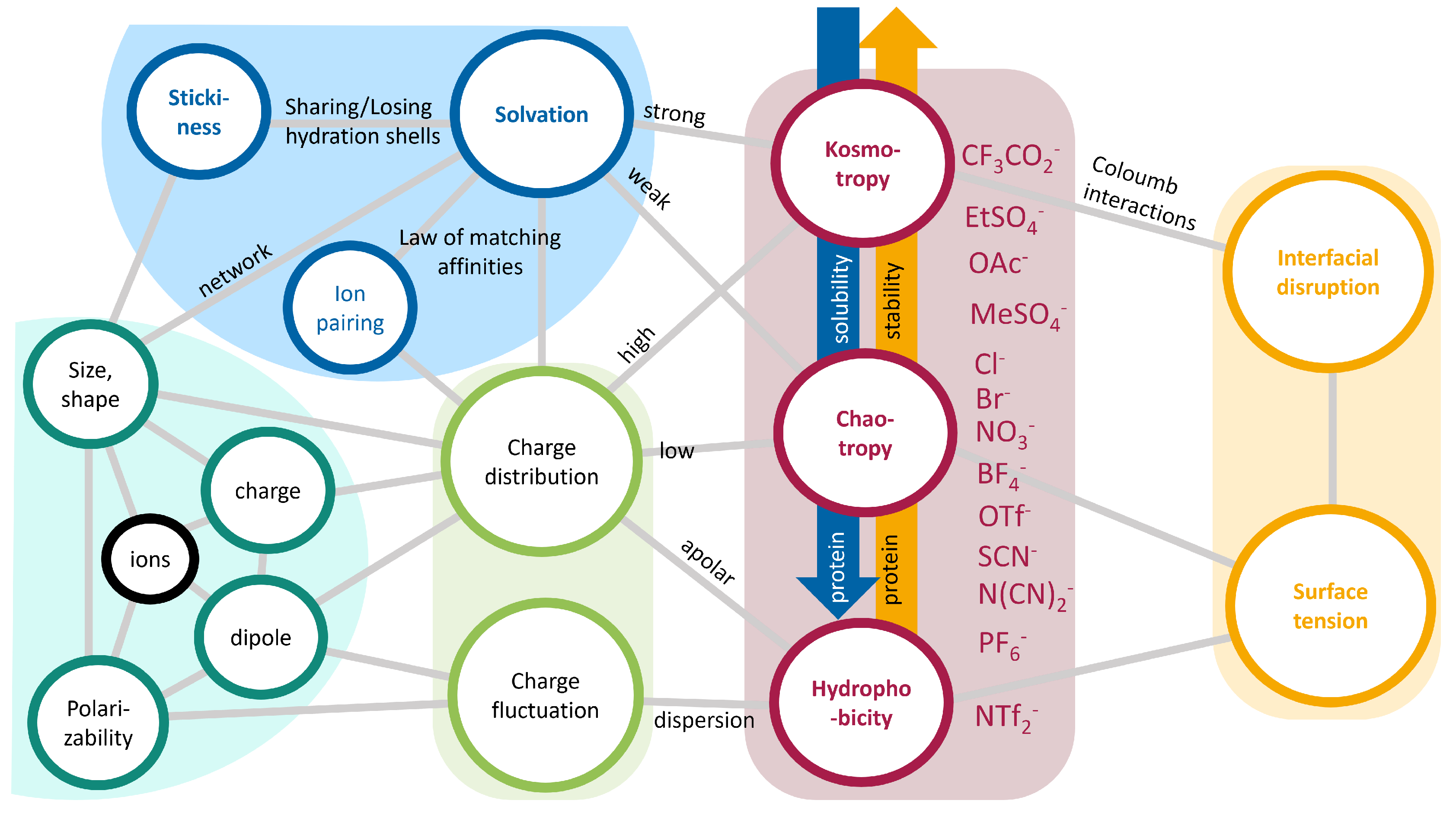
2.2.2. Hofmeister Series
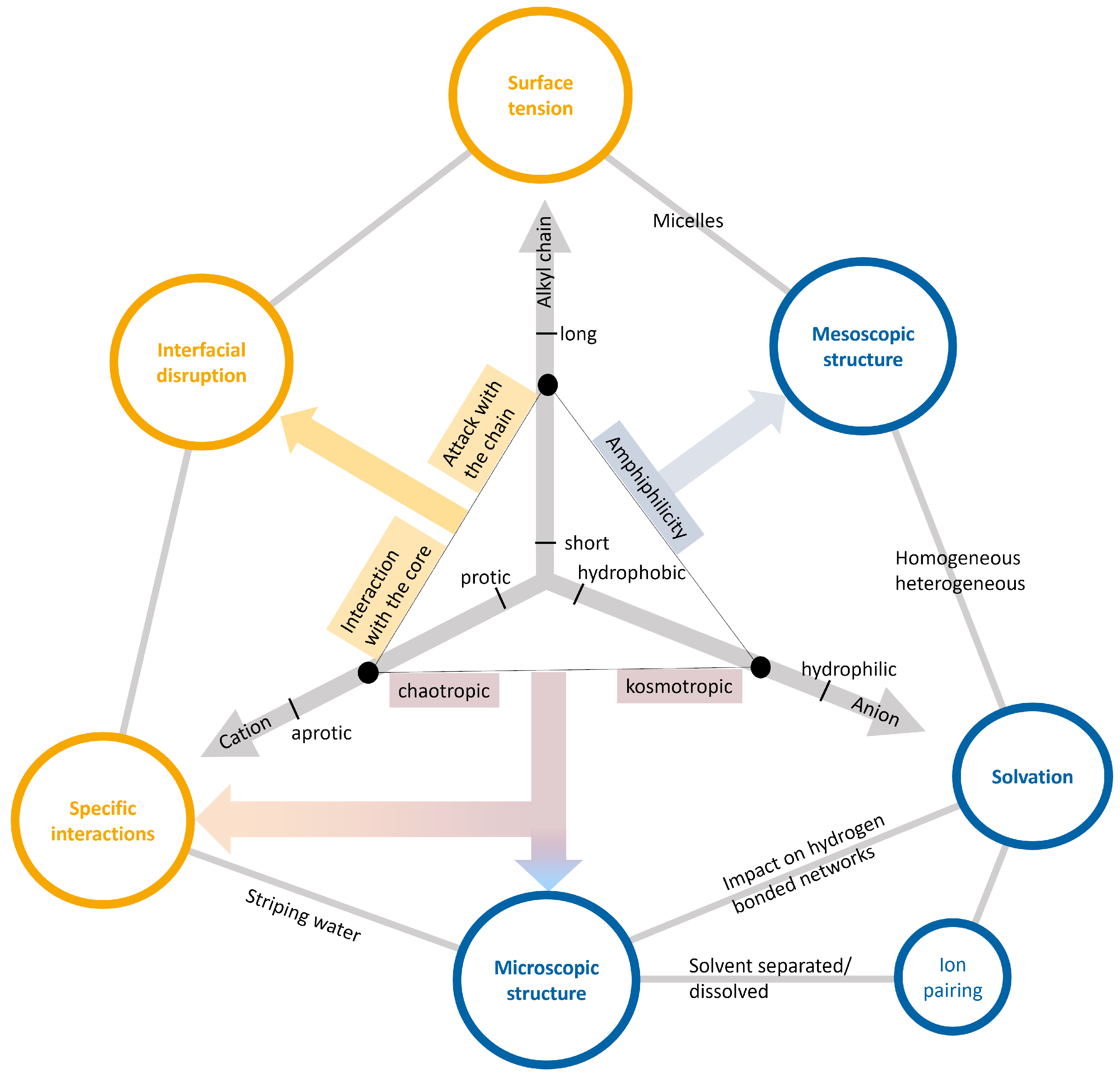
2.3. Ionic Liquids as Co-Solvents
Properties of Ionic Liquids
3. Experimental and Theoretical Studies
3.1. Analytical Techniques to Study Folding States
3.2. Molecular Dynamics Simulations to Study (Re-)Folding
3.3. Screening of Key Parameters
- Studies that use ionic liquids to dissolve aggregated or otherwise difficult-to-solubilize proteins;
- Studies that add ionic liquids to solutions of denatured protein in order to induce the (re)folding of the previously unfolded protein;
- Studies in which ionic liquids act as protein structure stabilizers that protect dissolved native proteins against unfolding and aggregation.
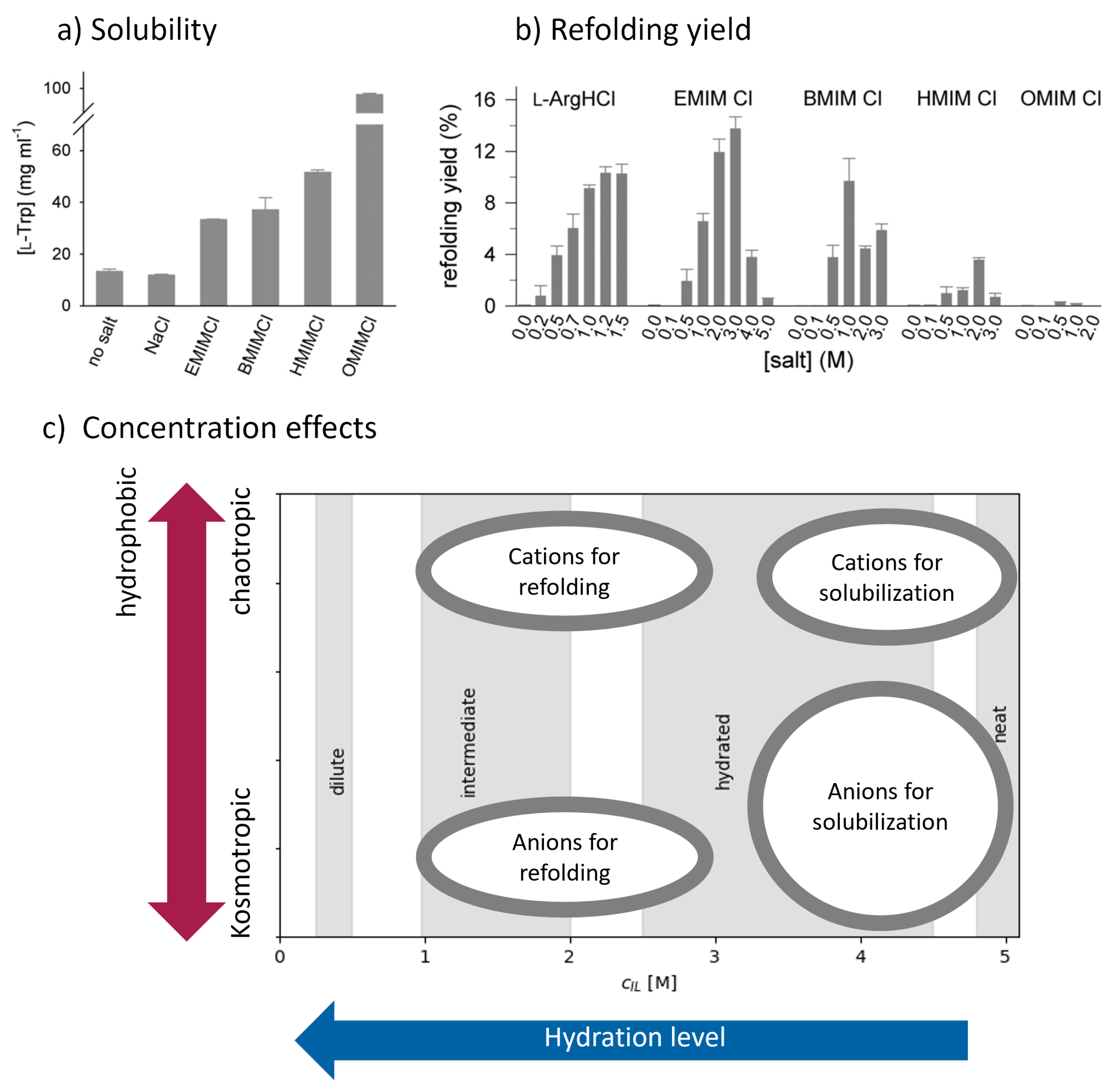
3.3.1. Ionic Liquids as Solubilizing Agents
3.3.2. Ionic Liquids as Refolding Additives
3.3.3. Ionic Liquids as Stabilizers
4. Challenges and Limitations
5. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- Klausser, R.; Kopp, J.; Brichtova, E.P.; Gisperg, F.; Elshazly, M.; Spadiut, O. State-of-the-art and novel approaches to mild solubilization of inclusion bodies. Front. Bioeng. Biotechnol. 2023, 11, 1249196. [Google Scholar] [CrossRef] [PubMed]
- Tsumoto, K.; Umetsu, M.; Kumagai, I.; Ejima, D.; Arakawa, T. Solubilization of active green fluorescent protein from insoluble particles by guanidine and arginine. Biochem. Biophys. Res. Commun. 2003, 312, 1383–1386. [Google Scholar] [CrossRef] [PubMed]
- Umetsu, M.; Tsumoto, K.; Nitta, S.; Adschiri, T.; Ejima, D.; Arakawa, T.; Kumagai, I. Nondenaturing solubilization of β2 microglobulin from inclusion bodies by L-arginine. Biochem. Biophys. Res. Commun. 2005, 328, 189–197. [Google Scholar] [CrossRef] [PubMed]
- Kachhawaha, K.; Singh, S.; Joshi, K.; Nain, P.; Singh, S.K. Bioprocessing of recombinant proteins from Escherichia coli inclusion bodies: Insights from structure-function relationship for novel applications. Prep. Biochem. Biotechnol. 2022, 53, 728–752. [Google Scholar] [CrossRef] [PubMed]
- Buscajoni, L.; Martinetz, M.C.; Berkemeyer, M.; Brocard, C. Refolding in the modern biopharmaceutical industry. Biotechnol. Adv. 2022, 61, 108050. [Google Scholar] [CrossRef]
- Slouka, C.; Kopp, J.; Hutwimmer, S.; Strahammer, M.; Strohmer, D.; Eitenberger, E.; Schwaighofer, A.; Herwig, C. Custom made inclusion bodies: Impact of classical process parameters and physiological parameters on inclusion body quality attributes. Microb. Cell Fact. 2018, 17, 148. [Google Scholar] [CrossRef]
- Humer, D.; Spadiut, O. Wanted: More monitoring and control during inclusion body processing. World J. Microbiol. Biotechnol. 2018, 34, 158. [Google Scholar] [CrossRef]
- Singh, A.; Upadhyay, V.; Upadhyay, A.K.; Singh, S.M.; Panda, A.K. Protein recovery from inclusion bodies of Escherichia coli using mild solubilization process. Microb. Cell Fact 2015, 14, 41. [Google Scholar] [CrossRef]
- Burgess, R.R. Purification of overproduced Escherichia coli RNA polymerase σ factors by solubilizing inclusion bodies and refolding from Sarkosyl. In RNA Polymerase and Associated Factors Part A; Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 1996; Volume 273, pp. 145–149. [Google Scholar] [CrossRef]
- Gani, K.; Bhambure, R.; Deulgaonkar, P.; Mehtaa, D.; Kamble, M. Understanding unfolding and refolding of the antibody fragment (Fab). I. In-vitro study. Biochem. Eng. J. 2020, 164, 107764. [Google Scholar] [CrossRef]
- Khan, S.; Siraj, S.; Shahid, M.; Haque, M.M.; Islam, A. Osmolytes: Wonder molecules to combat protein misfolding against stress conditions. Int. J. Biol. Macromol. 2023, 234, 123662. [Google Scholar] [CrossRef]
- Martinetz, M.; Friedrich, A.; Walther, C.; Tscheließnig, A.L.; Voigtmann, M.; Jung, A.; Brocard, C.; Bluhmki, E.; Smiatek, J. Solubilization of Inclusion Bodies: Insights from Explainable Machine Learning Approaches. Front. Chem. Eng. 2023, 5, 1227620. [Google Scholar] [CrossRef]
- Guncheva, M. Role of ionic liquids on stabilization of therapeutic proteins and model proteins. Protein J. 2022, 41, 369–380. [Google Scholar] [CrossRef] [PubMed]
- Javed, F.; Ullah, F.; Zakaria, M.R.; Akil, H.M. An approach to classification and hi-tech applications of room- temperature ionic liquids (RTILs): A review. J. Mol. Liq. 2018, 271, 403–420. [Google Scholar] [CrossRef]
- Hallett, J.P.; Welton, T. Room-Temperature Ionic Liquids: Solvents for Synthesis and Catalysis. 2. Chem. Rev. 2011, 111, 3508–3576. [Google Scholar] [CrossRef] [PubMed]
- Kaur, G.; Kumar, H.; Singla, M. Diverse applications of ionic liquids: A comprehensive review. J. Mol. Liq. 2022, 351, 118556. [Google Scholar] [CrossRef]
- Watanabe, M.; Thomas, M.L.; Zhang, S.; Ueno, K.; Yasuda, T.; Dokko, K. Application of Ionic Liquids to Energy Storage and Conversion Materials and Devices. Chem. Rev. 2017, 117, 7190–7239. [Google Scholar] [CrossRef] [PubMed]
- Somers, A.E.; Howlett, P.C.; MacFarlane, D.R.; Forsyth, M. A review of ionic liquid lubricants. Lubricants 2013, 1, 3–21. [Google Scholar] [CrossRef]
- Zhou, F.; Liang, Y.; Liu, W. Ionic liquid lubricants: Designed chemistry for engineering applications. Chem. Soc. Rev. 2009, 38, 2590–2599. [Google Scholar] [CrossRef]
- Mohd, N.; Draman, S.F.S.; Salleh, M.S.N.; Yusof, N.B. Dissolution of cellulose in ionic liquid: A review. AIP Conf. Proc. 2017, 1809, 020035. [Google Scholar] [CrossRef]
- Eyckens, D.J.; Henderson, L.C. A Review of Solvate Ionic Liquids: Physical Parameters and Synthetic Applications. Front. Chem. Green Sustain. Chem. 2019, 7, 1. [Google Scholar] [CrossRef]
- Takekiyo, T.; Ishikawa, Y.; Yoshimura, Y. Cryopreservation of Proteins Using Ionic Liquids: A Case Study of Cytochrome c. J. Phys. Chem. B 2017, 121, 7614–7620. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, Y.; Takekiyo, T.; Yoshimura, Y. Recovery and cryopreservation of insulin amyloid using ionic liquids. J. Mol. Liq. 2018, 272, 1019–1024. [Google Scholar] [CrossRef]
- Zaijun, L.; Jie, C.; Haixia, S.; Jiaomai, P. Advance of Room Temperature Ionic Liquid as Solvent for Extraction and Separation. Rev. Analy. Chem. 2007, 26, 109–153. [Google Scholar] [CrossRef]
- Ventura, S.P.; e Silva, F.A.; Quental, M.V.; Mondal, D.; Freire, M.G.; Coutinho, J.A. Ionic-Liquid-Mediated Extraction and Separation Processes for Bioactive Compounds: Past, Present, and Future Trends. Chem. Rev. 2017, 117, 6984–7052. [Google Scholar] [CrossRef] [PubMed]
- Shamshina, J.L.; Kelley, S.P.; Gurau, G.; Rogers, R.D. Chemistry: Develop ionic liquid drugs. Nature 2015, 528, 188–189. [Google Scholar] [CrossRef] [PubMed]
- Egorova, K.S.; Gordeev, E.G.; Ananikov, V.P. Biological Activity of Ionic Liquids and Their Application in pharmaceutics and Medicine. Chem. Rev. 2017, 117, 7132–7189. [Google Scholar] [CrossRef] [PubMed]
- Ventura, S.P.M.; de Barros, R.L.; Sintra, T.; Soares, C.M.F.; Lima, A.S.; Coutinho, J.A.P. Simple screening method to identify toxic/non-toxic ionic liquids: Agar diffusion test adaptation. Ecotoxicol. Environ. Saf. 2012, 83, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Benedetto, A.; Ballone, P. Room Temperature Ionic Liquids Meet Biomolecules: A Microscopic View of Structure and Dynamics. ACS Sustain. Chem. Eng. 2016, 4, 392–412. [Google Scholar] [CrossRef]
- Kumar, A.; Bhakuni, K.; Venkatesu, P. Strategic planning of proteins in ionic liquids: Future solvents for the enhanced stability of proteins against multiple stresses. Phys. Chem. Chem. Phys. 2019, 21, 23269–23282. [Google Scholar] [CrossRef]
- Egorova, K.S.; Ananikov, V.P. Fundamental importance of ionic interactions in the liquid phase: A review of recent studies of ionic liquids in biomedical and pharmaceutical applications. J. Mol. Liq. 2018, 272, 271–300. [Google Scholar] [CrossRef]
- Weingärtner, H.; Cabrele, C.; Herrmann, C. How ionic liquids can help to stabilize native proteins. Phys. Chem. Chem. Phys. 2012, 14, 415–426. [Google Scholar] [CrossRef]
- Pillai, V.V.S.; Kumari, P.; Kolagatla, S.; Sakai, V.G.; Rudić, S.; Rodriguez, B.J.; Rubini, M.; Tych, K.M.; Benedetto, A. Controlling Amyloid Fibril Properties Via Ionic Liquids: The Representative Case of Ethylammonium Nitrate and Tetramethylguanidinium Acetate on the Amyloidogenesis of Lysozyme. J. Phys. Chem. Lett. 2022, 13, 7058–7064. [Google Scholar] [CrossRef] [PubMed]
- Kiefhaber, T.; Rudolph, R.; Kohler, H.H.; Buchner, J. Protein Aggregation in vitro and in vivo: A Quantitative Model of the Kinetic Competition between Folding and Aggregation. Nat. Biotechnol. 1991, 9, 825–829. [Google Scholar] [CrossRef]
- Han, Q.; Brown, S.J.; Drummond, C.J.; Greaves, T.L. Protein aggregation and crystallization with ionic liquids: Insights into the influence of solvent properties. J. Colloid Interface Sci. 2022, 608, 1173–1190. [Google Scholar] [CrossRef] [PubMed]
- Schröder, C.; Steinhauser, O.; Sasisanker, P.; Weingärtner, H. Orientational alignment of amyloidogenic proteins in pre-aggregated solutions. Phys. Rev. Lett. 2015, 114, 128101. [Google Scholar] [CrossRef] [PubMed]
- Levinthal, C. Are there pathways for protein folding? J. Chim. Phys. 1968, 65, 44–45. [Google Scholar] [CrossRef]
- Smiatek, J. Aqueous ionic liquids and their effects on protein structures: An overview on recent theoretical and experimental results. J. Phys. Condens. Matter. 2017, 29, 233001. [Google Scholar] [CrossRef] [PubMed]
- Shukla, S.K.; Mikkola, J.P. Use of Ionic Liquids in Protein and DNA Chemistry. Front. Chem. Phys. Chem. Chem. Phys. 2020, 8, 1. [Google Scholar] [CrossRef]
- Wakayama, R.; Uchiyama, S.; Hall, D. Ionic liquids and protein folding—old tricks for new solvents. Biophys. Rev. 2019, 11, 209–225. [Google Scholar] [CrossRef]
- Sindhu, A.; Kumar, S.; Venkatesu, P. Contemporary Advancement of Cholinium-Based Ionic Liquids for Protein Stability and Long-Term Storage: Past, Present, and Future Outlook. ACS Sustain. Chem. Eng. 2022, 10, 4323–4344. [Google Scholar] [CrossRef]
- Jungbauer, M.K.E.A. Technical refolding of proteins: Do we have freedom to operate? Biotechnol. J. 2010, 5, 547–559. [Google Scholar] [CrossRef]
- Morganti, L.; Chura-Chambi, R.M. Using High Pressure and Alkaline pH for Refolding; Springer: New York, NY, USA, 2023; pp. 177–187. [Google Scholar] [CrossRef]
- Coutard, B.; Danchin, E.G.J.; Oubelaid, R.; Canard, B.; Bignon, C. Single pH buffer refolding screen for protein from inclusion bodies. Protein Expr. Purif. 2012, 82, 352–359. [Google Scholar] [CrossRef] [PubMed]
- Ibarra-Molero, B.; Sanchez-Riuz, J.M. A Model-Independent, Nonlinear Extrapolation Procedure for the Characterization of Protein Folding Energetics from Solvent-Denaturation Data. Biochemistry 1996, 35, 14689–14702. [Google Scholar] [CrossRef] [PubMed]
- Canchi, D.R.; García, A.E. Cosolvent effects on protein stability. Annu. Rev. Phys. Chem. 2013, 64, 273–293. [Google Scholar] [CrossRef] [PubMed]
- Pierce, V.; Kang, M.; Aburi, M.; Weerasinghe, S.; Smith, P.E. Recent applications of Kirkwood-Buff theory to biological systems. Cell Biochem. Biophys. 2008, 50, 1–22. [Google Scholar] [CrossRef]
- Tadeo, X.; Blanca López-Méndez, D.C.n.; Trigueros, T.; Millet, O. Protein Stabilization and the Hofmeister Effect: The Role of Hydrophobic Solvation. Biophys. J. 2009, 97, 2595–2603. [Google Scholar] [CrossRef]
- Bolen, D.W.; Rose, G.D. Structure and Energetics of the Hydrogen-Bonded Backbone in Protein Folding. Annu. Rev. Biochem. 2008, 77, 339–362. [Google Scholar] [CrossRef]
- Myers, J.K.; Pace, C.N.; Scholtz, J.M. Denaturant m values and heat capacity changes: Relation to changes in accessible surface areas of protein folding. Protein Sci. 1995, 4, 2138–2148. [Google Scholar] [CrossRef]
- Schellman, J.A. Solvent denaturation. Biopolymers 1978, 17, 1305–1322. [Google Scholar] [CrossRef]
- O’Brien, E.P.; Brooks, B.R.; Thirumalai, D. Molecular origin of constant m-values, denatured state collapse, and residue-dependent transition midpoints in globular proteins. Biochemistry 2009, 48, 3743–3754. [Google Scholar] [CrossRef][Green Version]
- Guinn, E.J.; Kontur, W.S.; Tsodikov, O.V.; Record, M.T., Jr. Probing the protein-folding mechanism using denaturant and temperature effects on rate constants. Proc. Natl. Acad. Sci. USA 2013, 110, 16784–16789. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, E.P.; Ziv, G.; Haran, G.; Thirumalai, D. Effects of denaturants and osmolytes on proteins are accurately predicted by the molecular transfer model. Proc. Natl. Acad. Sci. USA 2008, 105, 13403–13408. [Google Scholar] [CrossRef] [PubMed]
- Senske, M.; Constantinescu-Aruxandei, D.; Havenith, M.; Herrmann, C.; Weingärtner, H.; Ebbinghaus, S. The temperature dependence of the Hofmeister series: Thermodynamic fingerprints of cosolute–protein interactions. Phys. Chem. Chem. Phys. 2016, 18, 29698–29708. [Google Scholar] [CrossRef] [PubMed]
- Pegram, L.M.; Wendorff, T.; Erdmann, R.; Shkel, I.; Bellisssimo, D.; Felitsky, D.J.; Record, M.T., Jr. Why Hofmeister effects of many salts favor protein folding but not DNA helix formation. Proc. Natl. Acad. Sci. USA 2010, 107, 7716–7721. [Google Scholar] [CrossRef] [PubMed]
- Canstantinescu, D.; Weingärtner, H.; Herrmann, C. Protein Denaturation by Ionic Liquids and the Hofmeister Series: A case study of aqueous solutions of ribonuclease A. Angew. Chem. Int. Ed. 2007, 46, 8887–8889. [Google Scholar] [CrossRef] [PubMed]
- Lia, Z.; Han, Q.; Wang, K.; Song, S.; Xue, Y.; Ji, X.; Zhai, J.; Huang, Y.; Zhang, S. Ionic liquids as a tunable solvent and modifier for biocatalysis. Catal. Rev. 2022, 1–47. [Google Scholar] [CrossRef]
- Gregory, K.P.; Elliott, G.R.; Robertson, H.; Kumar, A.; Wanless, E.J.; Webber, G.B.; Craig, V.S.J.; Andersson, G.G.; Page, A.J. Understanding specific ion effects and the Hofmeister series. Phys. Chem. Chem. Phys. 2022, 24, 12682–12718. [Google Scholar] [CrossRef]
- Pitzer, K.S. Thermodynamics of electrolytes. I. Theoretical basis and general equations. J. Phys. Chem. 1973, 77, 268–277. [Google Scholar] [CrossRef]
- Salis, A.; Ninham, B.W. Models and mechanism of Hofmeister effects in electrolyte solutions, and colloid and protein systems revisted. Chem. Soc. Rev. 2014, 43, 7358–7377. [Google Scholar] [CrossRef]
- Leontidis, E. Investigations of the Hofmeister series and other specific ion effects using lipid model systems. Adv. Colloid Interface Sci. 2017, 243, 8–22. [Google Scholar] [CrossRef]
- Sharp, K.A.; Nicholls, A.; Fine, R.F.; Honig, B. Reconciling the Magnitude of the Microscopic and Macroscopic Hydrophobic Effects. Science 1991, 252, 106–109. [Google Scholar] [CrossRef] [PubMed]
- Baldwin, R.L. How Hofmeister Ion Interactions affect Protein Stability. Biophys. J. 1996, 71, 2056–2063. [Google Scholar] [CrossRef] [PubMed]
- Collins, K.D.; Washabaugh, M.W. The Hofmeister Effect and the Behaviour of Water at Interfaces. Q. Rev. Biophys. 1985, 18, 323–422. [Google Scholar] [CrossRef] [PubMed]
- Hyde, A.M.; Zultanski, S.L.; Waldman, J.H.; Zhong, Y.L.; Shevlin, M.; Peng, F. General Principles and Strategies for Salting-Out Informed by the Hofmeister Series. Org. Process Res. Dev. 2017, 21, 1355–1370. [Google Scholar] [CrossRef]
- Pearson, R.G. Hard and soft acids and bases. J. Am. Chem. Soc. 1963, 85, 3533–3539. [Google Scholar] [CrossRef]
- Zhao, H. Protein stabilization and enzyme activation in ionic liquids: Specific ion effects. J. Chem. Technol. Biotechnol. 2016, 91, 25–50. [Google Scholar] [CrossRef]
- Voet, A. Quantative Lyotropy. Chem. Rev. 1937, 20, 169–179. [Google Scholar] [CrossRef]
- Schröder, C. Proteins in Ionic Liquids: Current Status of Experiments and Simulations. Top. Curr. Chem. 2017, 375, 25. [Google Scholar] [CrossRef]
- Record, M.T., Jr.; Guinn, E.; Pegram, L.; Cappb, M. Interpreting and Predicting Hofmeister Salt Ion and Solute Effects on Biopolymer and Model Processes Using the Solute Partitioning Model. Faraday Discuss. 2013, 160, 9–120. [Google Scholar] [CrossRef]
- Zhang, Y.; Cremer, P.S. The inverse and direct Hofmeister series for lysozyme. Proc. Natl. Acad. Sci. USA 2009, 106, 15249–15253. [Google Scholar] [CrossRef]
- Boström, M.; Williams, D.R.M.; Ninham, B.W. Why the properties of proteins in salt solutions follow a Hofmeister series. Curr. Opin. Colloid. Interface Sci. 2004, 9, 48–52. [Google Scholar] [CrossRef]
- Ball, P. Water as a biomolecule. ChemPhysChem 2008, 9, 2677–2685. [Google Scholar] [CrossRef] [PubMed]
- Omta, A.W.; Kropman, M.F.; Woutersen, S.; Bakker, H.J. Negligible Effect of Ions on the Hydrogen-Bond Structure in Liquid Water. Science 2003, 301, 347–349. [Google Scholar] [CrossRef] [PubMed]
- Omta, A.W.; Kropman, M.F.; Woutersen, S.; Bakker, H.J. Influence of ions on thehydrogen-bond structure in liquid water. J. Chem. Phys. 2003, 119, 12457–12461. [Google Scholar] [CrossRef]
- Waluyo, I.; Huang, C.; Nordlund, D.; Bergmann, U.; Weiss, T.M.; Pettersson, L.G.M.; Nilsson, A. The structure of water in the hydration shell of cations from x-ray Raman and small angle x-ray scattering measurements. J. Chem. Phys. 2011, 134, 064513. [Google Scholar] [CrossRef] [PubMed]
- Schienbein, P.; Schwaab, G.; Forbert, H.; Havenith, M.; Marx, D. Correlations in the Solute–Solvent Dynamics Reach Beyond the First Hydration Shell of Ions. J. Phys. Chem. Lett. 2017, 8, 2373–2380. [Google Scholar] [CrossRef] [PubMed]
- Sedlák, E.; Stagg, L.; Wittung-Stafshede, P. Effect of Hofmeister ions on protein thermal stability: Roles of ion hydration and peptide groups? Arch. Biochem. Biophys. 2008, 479, 69–73. [Google Scholar] [CrossRef]
- Zhang, Y.; Cremer, P.S. Chemistry of Hofmeister Anions and Osmolytes. Annu. Rev. Phys. Chem. 2010, 61, 63–83. [Google Scholar] [CrossRef]
- Yang, Z. Hofmeister effects: An explanation for the impact of ionic liquids on biocatalysis. J. Biotechnol. 2009, 144, 12–22. [Google Scholar] [CrossRef]
- Sheldon, R.A. Biocatalysis in ionic liquids: State-of-the-union. Green Chem. 2021, 23, 8406–8427. [Google Scholar] [CrossRef]
- Rick, S.W.; Stuart, S.J.; Bader, S.; Berne, B.J. Fluctuating charge force fields for aqueous solutions. J. Mol. Liq. 1995, 65–66, 31–40. [Google Scholar] [CrossRef]
- Mancini, G.; Brancato, G.; Barone, V. Combining the Fluctuating Charge Method, Non-periodic BoundaryConditions and Meta-dynamics: Aqua Ions as Case Studies. J. Chem. Theory Comput. 2014, 10, 1150–1163. [Google Scholar] [CrossRef] [PubMed]
- Kunz, W.; Belloni, L.; Bernard, O.; Ninham, B.W. Osmotic coefficients and surface tensions of aqueous electrolyte solutions: Role of dispersion forces. J. Phys. Chem. B 2004, 108, 2398–2404. [Google Scholar] [CrossRef]
- Wiggins, P.M. Hydrophobic hydration, hydrophobic forces and protein folding. Phys. A 1997, 238, 113–128. [Google Scholar] [CrossRef]
- Némethy, G. Hydrophobic interactions. Angew. Chem. Int. Ed. 1967, 6, 195–206. [Google Scholar] [CrossRef] [PubMed]
- Bica, K.; Deetlefs, M.; Schröder, C.; Seddon, K.R. Polarisabilities of alkylimidazolium ionic liquids. Phys. Chem. Chem. Phys. 2013, 15, 2703–2711. [Google Scholar] [CrossRef] [PubMed]
- Mazzini, V.; Craig, V.S.J. Volcano Plots Emerge from a Sea of Nonaqueous Solvents: The Law of Matching Water Affinities Extends to All Solvents. ACS Cent. Sci. 2018, 4, 1056–1064. [Google Scholar] [CrossRef]
- Collins, K.D. The behavior of ions in water is controlled by their water affinity. Q. Rev. Biophys. 2019, 52, e11. [Google Scholar] [CrossRef]
- Buettner, C.; Cognigni, A.; Schröder, C.; Schröder, K. Surface-active ionic liquids; A review. J. Mol. Liq. 2022, 21, 118160. [Google Scholar] [CrossRef]
- Booth, J.; Abbott, S.; Shimizu, S. Mechanism of hydrophobic drug solubilization by small molecule hydrotropes. Phys. Chem. Chem. Phys. 2012, 116, 14915–14921. [Google Scholar] [CrossRef]
- Zeindlhofer, V.; Khlan, D.; Bica, K.; Schröder, C. Computational analysis of the solvation of coffee ingredients in aqueous ionic liquid mixtures. RSC Adv. 2017, 7, 3495–3504. [Google Scholar] [CrossRef] [PubMed]
- Constatinescu, D.; Herrmann, C.; Weingärtner, H. Patterns of protein unfolding and protein aggregation in ionic liquids. Phys. Chem. Chem. Phys. 2010, 12, 1756–1763. [Google Scholar] [CrossRef] [PubMed]
- Okur, H.; Hladílková, J.; Rembert, K.; Cho, Y.; Heyda, J.; Dzubiella, J.; Cremer, P.; Jungwirth, P. Beyond the Hofmeister Series: Ion-Specific Effects on Proteins and Their Biological Functions. J. Phys. Chem. B 2017, 121, 1997–2014. [Google Scholar] [CrossRef] [PubMed]
- Peruzzi†, N.; Ninham, B.W.; Lo Nostro, P.; Baglioni, P. Hofmeister Phenomena in Nonaqueous Media: The Solubility of Electrolytes in Ethylene Carbonate. J. Phys. Chem. B 2012, 116, 14398–14405. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, L.; Guo, X.; Wu, S. Salting-in counter-current chromatography separation of tanshinones based on room temperature ionic liquids. J. Chromat. A 2018, 1559, 149–155. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, S.; Mohan, R.S.; Scott, J.L. Reactivity of ionic liquids. Tetrahedron 2007, 63, 2363–2389. [Google Scholar] [CrossRef]
- Reslan, M.; Kayser, V. Ionic liquids as biocompatible stabilizers of proteins. Biophys. Rev. 2018, 10, 781–793. [Google Scholar] [CrossRef]
- Hagen, M.L.; Harper, J.B.; Croft, A.K. Recent advances in the use of ionic liquids as solvents for protein-based materials and chemistry. Curr. Opin. Green Sustain. Chem. 2022, 36, 100637. [Google Scholar] [CrossRef]
- Walden, P. Molecular weights and electrical conductivity of several fused salts. Bull. Acad. Imper. Sci. 1914, 1800. [Google Scholar]
- Summers, C.A.; Flowers, R.A. Protein renaturation by the liquid organic salt ethylammonium nitrate. Prot. Sci. 2000, 9, 2001–2008. [Google Scholar] [CrossRef]
- Byrne, N.; Angell, C.A. The solubility of hen lysozyme in ethylammonium nitrate/H2O mixtures and a novel approach to protein crystallization. Molecules 2010, 15, 793–803. [Google Scholar] [CrossRef] [PubMed]
- Rogers, R.D.; Seddon, K.R. Ionic liquids–solvents of the future? Science 2003, 302, 792–793. [Google Scholar] [CrossRef] [PubMed]
- Rogers, R.D. Reflections on ionic liquids. Nature 2007, 447, 917–918. [Google Scholar] [CrossRef] [PubMed]
- Bica, K.; Gaertner, P. Applications of chiral ionic liquids. Eur. J. Org. Chem. 2008, 2008, 3235–3250. [Google Scholar] [CrossRef]
- Yanes, E.G.; Gratz, S.R.; Baldwin, M.J.; Robison, S.E.; Stalcup, A.M. Capillary electrophoretic application of 1-alkyl-3-methylimidazolium-based ionic liquids. Anal. Chem. 2001, 73, 3838–3844. [Google Scholar] [CrossRef] [PubMed]
- Hatti-Kaul, R. Aqueous Two-Phase Systems: Methods and Protocols; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2008; Volume 11. [Google Scholar]
- Earle, M.J.; McCormac, P.B.; Seddon, K.R. Diels–Alder reactions in ionic liquids. A safe recyclable alternative to lithium perchlorate–diethyl ether mixtures. Green Chem. 1999, 1, 23–25. [Google Scholar] [CrossRef]
- Canongia Lopes, J.N.; Pádua, A.A. Nanostructural organization in ionic liquids. J. Phys. Chem. B 2006, 110, 3330–3335. [Google Scholar] [CrossRef]
- Veríssimo, N.V.; Vicente, F.A.; de Oliveira, R.C.; Likozar, B.; de Souza Oliveira, R.P.; Pereira, J.F.B. Ionic liquids as protein stabilizers for biological and biomedical applications: A review. Biotechnol. Adv. 2022, 61, 108055. [Google Scholar] [CrossRef]
- Han, D.; Tang, B.; Ri Lee, Y.; Ho Row, K. Application of ionic liquid in liquid phase microextraction technology. J. Sep. Sci. 2012, 35, 2949–2961. [Google Scholar] [CrossRef]
- Almeida, H.F.D.; Freire, M.G.; Fernandes, A.M.; da Silva, J.A.L.; Morgado, P.; Shimizu, K.; Filipe, E.J.M.; Lopes, J.N.C.; Santos, L.M.N.B.F.; Coutinho, J.A.P. Cation Alkyl Side Chain Length and Symmetry Effects on the Surface Tension of Ionic Liquids. Langmuir 2014, 30, 6408–6418. [Google Scholar] [CrossRef]
- Marcus, Y.; Hefter, G. Ion Pairing. Chem. Rev. 2006, 106, 4585–4621. [Google Scholar] [CrossRef] [PubMed]
- Stange, P.; Fumino, K.; Ludwig, R. Ion Speciation of Protic Ionic Liquids in Water: Transition from Contact to Solvent-Separated Ion Pairs. Angew. Chem. Int. Ed. 2013, 52, 2990–2994. [Google Scholar] [CrossRef] [PubMed]
- Kirchner, B.; Malberg, F.; Firaha, D.S.; Hollóczki, O. Ion pairing in ionic liquids. J. Phys. Condens. Matter 2015, 27, 463002. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, J.N.; Zhou, Y.; Guo, Z.; Pérez, B. Genetic and chemical approaches for surface charge engineering of enzymes and their applicability in biocatalysis: A review. Biotechnol. Bioeng. 2019, 116, 1795–1812. [Google Scholar] [CrossRef] [PubMed]
- Diddens, D.; Lesch, V.; Heuer, A.; Smiatek, J. Aqueous ionic liquids and their influence on peptide conformations: Denaturation and dehydration mechanisms. Phys. Chem. Chem. Phys. 2017, 19, 20430–20440. [Google Scholar] [CrossRef]
- Okur, H.I.; Kherb, J.; Cremer, P.S. Cations Bind Only Weakly to Amides in Aqueous Solutions. J. Am. Chem. Soc. 2013, 135, 5062–5067. [Google Scholar] [CrossRef]
- Oehlke, A.; Hofmann, K.; Spange, S. New aspects on polarity of 1-alkyl-3-methylimidazolium salts as measured by solvatochromic probes. New J. Chem. 2006, 30, 533–536. [Google Scholar] [CrossRef]
- Han, Q.; Wang, X.; Byrne, N. Understanding the influence of key ionic liquid properties on the hydrolytic activity of thermomyces lanuginosus lipase. ChemCatChem 2016, 8, 1551–1556. [Google Scholar] [CrossRef]
- Lim, G.S.; Klähn, M. On the stability of proteins solvated in imidazolium-based ionic liquids studied with replica exchange molecular dynamics. J. Phys. Chem. B 2018, 122, 9274–9288. [Google Scholar] [CrossRef]
- Tomé, L.I.; Jorge, M.; Gomes, J.R.; Coutinho, J.A. Molecular dynamics simulation studies of the interactions between ionic liquids and amino acids in aqueous solution. J. Phys. Chem. B 2012, 116, 1831–1842. [Google Scholar] [CrossRef]
- Figueiredo, A.M.; Sardinha, J.; Moore, G.R.; Cabrita, E.J. Protein destabilisation in ionic liquids: The role of preferential interactions in denaturation. Phys. Chem. Chem. Phys. 2013, 15, 19632–19643. [Google Scholar] [CrossRef] [PubMed]
- Bui-Le, L.; Clarke, C.J.; Bröhl, A.; Brogan, A.P.S.; Arpino, J.A.J.; Polizzi, K.M.; Hallett, J.P. Revealing the complexity of ionic liquid–protein interactions through a multi-technique investigation. Commun. Chem. 2020, 3, 55. [Google Scholar] [CrossRef] [PubMed]
- Tokunaga, Y.; Takeuchi, K. Role of NMR in High Ordered Structure Characterization of Monoclonal Antibodies. Int. J. Mol. Sci. 2021, 22, 46. [Google Scholar] [CrossRef] [PubMed]
- Kamatari, Y.O.; Kitahara, R.; Yamada, H.; Yokoyama, S.; Akasaka, K. High-pressure NMR spectroscopy for characterizing folding intermediates and denatured states of proteins. Methods 2004, 34, 133–143. [Google Scholar] [CrossRef] [PubMed]
- Schwaighofer, A.; Lendl, B. Infrared Spectroscopy for Structure Analysis of Protein Inclusion Bodies. In Inclusion Bodies. Methods in Molecular Biology; Kopp, J., Spadiut, P., Eds.; Humana: New York, NY, USA, 2023; Volume 2617. [Google Scholar] [CrossRef]
- Tatkiewicz, W.; Elizondo, E.; Moreno, E.; Díez-Gil, C.; Ventosa, N.; Veciana, J.; Ratera, I. Methods for Characterization of Protein Aggregates. In Insoluble Proteins. Methods in Molecular Biology; García-Fruitós, E., Ed.; Humana Press: New York, NY, USA, 2014; Volume 1258. [Google Scholar] [CrossRef]
- Sukumaran, S.; Hauser, K.; Maier, E.; Benz, R.; Mäntele, W. Tracking the Unfolding and Refolding Pathways of Outer Membrane Protein PorinfromParacoccus denitrificans. Biochemistry 2006, 45, 3972–3980. [Google Scholar] [CrossRef]
- Jackson, M.; Mantsch, H.H. The Use and Misuse of FTIR Spectroscopy in the Determination of Protein Structure. Crit. Rev. Biochem. Mol. Biol. 1995, 30, 95–120. [Google Scholar] [CrossRef]
- Ami, D.; Natalello, A.; Taylor, G.; Tonon, G.; Doglia, S.M. Structural analysis of protein inclusion bodies by Fourier transform infrared microspectroscopy. Biochim. Biophys. Acta: Proteins Proteom. 2006, 1764, 793–799. [Google Scholar] [CrossRef]
- Sheli, K.B.; Ghorbani, M.; Hekmat, A.; Soltanian, B.; Mohammadian, A.; Jalalirad, R. Structural characterization of recombinant streptokinase following recovery from inclusion bodies using different chemical solubilization treatments. Biotechnol. Rep. 2018, 19, e00259. [Google Scholar] [CrossRef]
- Miles, A.J.; Janes, R.W.; Wallace, B.A. Tools and Methods for Circular Dichroism Spectroscopy of Proteins: A Tutorial Review. Chem. Soc. Rev. 2021, 50, 8400–8413. [Google Scholar] [CrossRef]
- Gatti-Lafranconi, P.; Natalello, A.; Ami, D.; Doglia, S.M.; Lotti, M. Concepts and Tools to Exploit the Potential of Bacterial Inclusion Bodies in Protein Science and Biotechnology. FEBS J. 2011, 278, 2408–2418. [Google Scholar] [CrossRef]
- Pignataro, M.F.; Herrera, M.G.; Dodero, V.I. Evaluation of Peptide/Protein Self-Assembly and Aggregation by Spectroscopic Methods. Molecules 2020, 25, 4854. [Google Scholar] [CrossRef] [PubMed]
- Stetefeld, J.; McKenna, S.A.; Patel, T.R. Dynamic Light Scattering: A Practical Guide and Applications in Biomedical Sciences. Biophys. Rev. 2016, 8, 409–427. [Google Scholar] [CrossRef] [PubMed]
- Bansal, R.; Gupta, S.; Rathore, A.S. Analytical Platform for Monitoring Aggregation of Monoclonal Antibody Therapeutics. Pharma. Res. 2019, 36, 152. [Google Scholar] [CrossRef] [PubMed]
- Ruggeri, F.S.; Šneideris, T.; Vendruscolo, M.; Knowlesa, T.P. Atomic force microscopy for single molecule characterization of protein aggregation. Arch. Biochem. Biophys. 2019, 664, 134–148. [Google Scholar] [CrossRef] [PubMed]
- Williams-Noonan, B.J.; Kamboukos, A.; Todorova, N.; Yarovsky, I. Self-assembling peptide biomaterials: Insights from spontaneous and enhanced sampling molecular dynamics simulations. Chem. Phys. Rev. 2023, 4, 021304. [Google Scholar] [CrossRef]
- Lin, Y.; Penna, M.; Spicer, C.D.; Higgins, S.G.; Gelmi, A.; Kim, N.; Wang, S.T.; Wojciechowski, J.P.; Pashuck, E.T.; Yarovsky, I.; et al. High-Throughput Peptide Derivatization toward Supramolecular Diversification in Microtiter Plates. ACS Nano 2021, 15, 4034–4044. [Google Scholar] [CrossRef]
- Thota, N.; Luo, Z.; Hu, Z.; Jiang, J. Self-Assembly of Amphiphilic Peptide (AF)6H5K15: Coarse-Grained Molecular Dynamics Simulation. J. Phys. Chem. B 2013, 117, 9690–9698. [Google Scholar] [CrossRef]
- Fu, I.W.; Markegard, C.B.; Chu, B.K.; Nguyen, H.D. Role of Hydrophobicity on Self-Assembly by Peptide Amphiphiles via Molecular Dynamics Simulations. Langmuir 2014, 30, 7745–7754. [Google Scholar] [CrossRef]
- Joshi, S.Y.; Deshmukh, S.A. A review of advancements in coarse-grained molecular dynamics simulations. Mol. Sim. 2021, 47, 786–803. [Google Scholar] [CrossRef]
- Bussi, G.; Laio, A.; Parrinello, M. Equilibrium Free Energies from Nonequilibrium Metadynamics. Phys. Rev. Lett. 2006, 96, 090601. [Google Scholar] [CrossRef]
- Laio, A.; Gervasio, F.L. Metadynamics: A method to simulate rare events and reconstruct the free energy in biophysics, chemistry and material science. Rep. Prog. Phys. 2008, 71, 126601. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–592. [Google Scholar] [CrossRef] [PubMed]
- Sivapragasam, M.; Moniruzzaman, M.; Goto, M. Recent advances in exploiting ionic liquids for biomolecules: Solubility, stability and applications. Biotechnol. J. 2016, 11, 1000–1013. [Google Scholar] [CrossRef] [PubMed]
- Taoka, M.; Horita, K.; Takekiyo, T.; Uekita, T.; Yoshimura, Y.; Ichimura, T. An ionic liquid-based sample preparation method for next-stage aggregate proteomic analysis. Anal. Chem. 2019, 91, 13494–13500. [Google Scholar] [CrossRef] [PubMed]
- Fujita, K.; Nakano, R.; Nakaba, R.; Nakamura, N.; Ohno, H. Hydrated ionic liquids enable both solubilisation and refolding of aggregated concanavalin A. Chem. Commun. 2019, 55, 3578–3581. [Google Scholar] [CrossRef] [PubMed]
- Biswas, A.; Shogren, R.; Stevenson, D.; Willett, J.; Bhowmik, P.K. Ionic liquids as solvents for biopolymers: Acylation of starch and zein protein. Carbohydr. Polym. 2006, 66, 546–550. [Google Scholar] [CrossRef]
- Fujita, K.; Ohno, H. Enzymatic activity and thermal stability of metallo proteins in hydrated ionic liquids. Biopolymers 2010, 93, 1093–1099. [Google Scholar] [CrossRef]
- Fujita, K.; MacFarlane, D.R.; Forsyth, M. Protein solubilising and stabilising ionic liquids. Chem. Commun. 2005, 38, 4804–4806. [Google Scholar] [CrossRef]
- Fujita, K.; Kajiyama, M.; Liu, Y.; Nakamura, N.; Ohno, H. Hydrated ionic liquids as a liquid chaperon for refolding of aggregated recombinant protein expressed in Escherichia coli. Chem. Commun. 2016, 52, 13491–13494. [Google Scholar] [CrossRef]
- Fujita, K.; Kobayashi, K.; Ito, A.; Yanagisawa, S.; Ichida, K.; Takeda, K.; Nakamura, N.; Ohno, H. Improved renaturation process of aggregated recombinant proteins through the design of hydrated ionic liquids. J. Mol. Liq. 2023, 377, 121440. [Google Scholar] [CrossRef]
- Chen, J.; Vongsanga, K.; Wang, X.; Byrne, N. What happens during natural protein fibre dissolution in ionic liquids. Materials 2014, 7, 6158–6168. [Google Scholar] [CrossRef] [PubMed]
- Byrne, N.; Angell, C.A. Formation and dissolution of hen egg white lysozyme amyloid fibrils in protic ionic liquids. Chem. Commun. 2009, 9, 1046–1048. [Google Scholar] [CrossRef] [PubMed]
- Mangialardo, S.; Gontrani, L.; Leonelli, F.; Caminiti, R.; Postorino, P. Role of ionic liquids in protein refolding: Native/fibrillar versus treated lysozyme. RSC Adv. 2012, 2, 12329–12336. [Google Scholar] [CrossRef]
- Takekiyo, T.; Kato, M.; Yoshimura, Y. Pressure-induced structural changes of alanine oligopeptides in aqueous solutions. High Press. Res. 2019, 39, 202–209. [Google Scholar] [CrossRef]
- Yamamoto, E.; Yamaguchi, S.; Nagamune, T. Protein refolding by N-alkylpyridinium and N-alkyl-N-methylpyrrolidinium ionic liquids. Appl. Biochem. Biotechnol. 2011, 164, 957–967. [Google Scholar] [CrossRef] [PubMed]
- Bae, S.W.; Chang, W.J.; Koo, Y.M.; Ha, S.H. Enhanced refolding of lysozyme with imidazolium-based room temperature ionic liquids: Effect of hydrophobicity and sulfur residue. Sci. China Chem. 2012, 55, 1657–1662. [Google Scholar] [CrossRef]
- Bae, S.W.; Eom, D.; Mai, N.L.; Koo, Y.M. Refolding of horseradish peroxidase is enhanced in the presence of metal cofactors and ionic liquids. Biotechnol. J. 2016, 11, 464–472. [Google Scholar] [CrossRef]
- Buchfink, R.; Tischer, A.; Patil, G.; Rudolph, R.; Lange, C. Ionic liquids as refolding additives: Variation of the anion. J. Biotechnol. 2010, 150, 64–72. [Google Scholar] [CrossRef]
- Singh, U.K.; Patel, R. Dynamics of ionic liquid-assisted refolding of denatured cytochrome c: A study of preferential interactions toward renaturation. Mol. Pharm. 2018, 15, 2684–2697. [Google Scholar] [CrossRef]
- Castro, L.S.; Pereira, P.; Passarinha, L.A.; Freire, M.G.; Pedro, A.Q. Enhanced performance of polymer-polymer aqueous two-phase systems using ionic liquids as adjuvants towards the purification of recombinant proteins. Sep. Purif. Technol. 2020, 248, 117051. [Google Scholar] [CrossRef]
- Takekiyo, T.; Yamada, N.; Nakazawa, C.T.; Amo, T.; Asano, A.; Yoshimura, Y. Formation of α-synuclein aggregates in aqueous ethylammonium nitrate solutions. Biopolymers 2020, 111, e23352. [Google Scholar] [CrossRef] [PubMed]
- Byrne, N.; Wang, L.M.; Belieres, J.P.; Angell, C.A. Reversible folding–unfolding, aggregation protection, and multi-year stabilization, in high concentration protein solutions, using ionic liquids. Chem. Commun. 2007, 26, 2714–2716. [Google Scholar] [CrossRef] [PubMed]
- Sankaranarayanan, K.; Sathyaraj, G.; Nair, B.; Dhathathreyan, A. Reversible and irreversible conformational transitions in myoglobin: Role of hydrated amino acid ionic liquid. J. Phys. Chem. B. 2012, 116, 4175–4180. [Google Scholar] [CrossRef] [PubMed]
- Kutsch, M.; Hortmann, P.; Herrmann, C.; Weibels, S.; Weingärtner, H. Dissecting ion-specific from electrostatic salt effects on amyloid fibrillation: A case study of insulin. Biointerphases 2016, 11, 019008. [Google Scholar] [CrossRef] [PubMed]
- Belchior, D.C.; Quental, M.V.; Pereira, M.M.; Mendonça, C.M.; Duarte, I.F.; Freire, M.G. Performance of tetraalkylammonium-based ionic liquids as constituents of aqueous biphasic systems in the extraction of ovalbumin and lysozyme. Sep. Purif. Technol. 2020, 233, 116019. [Google Scholar] [CrossRef]
- Fujita, K.; MacFarlane, D.R.; Forsyth, M.; Yoshizawa-Fujita, M.; Murata, K.; Nakamura, N.; Ohno, H. Solubility and stability of cytochrome c in hydrated ionic liquids: Effect of oxo acid residues and kosmotropicity. Biomacromolecules 2007, 8, 2080–2086. [Google Scholar] [CrossRef] [PubMed]
- Fujita, K.; Ohno, H. Hydrated Ionic Liquids: Perspective for Bioscience. Chem. Rec. 2023, e202200282. [Google Scholar] [CrossRef]
- Ohno, H.; Fujita, K.; Kohno, Y. Is seven the minimum number of water molecules per ion pair for assured biological activity in ionic liquid-water mixtures? Phys. Chem. Chem. Phys. 2015, 17, 14454–14460. [Google Scholar] [CrossRef]
- Spange, S.; Lungwitz, R.; Schade, A. Correlation of molecular structure and polarity of ionic liquids. J. Mol. Liq. 2014, 192, 137–143. [Google Scholar] [CrossRef]
- Mester, P.; Wagner, M.; Rossmanith, P. Ionic liquids designed as chaotrope and surfactant for use in protein chemistry. Sep. Purif. Technol. 2012, 97, 211–215. [Google Scholar] [CrossRef]
- Lange, C.; Patil, G.; Rudolph, R. Ionic liquids as refolding additives: N’-alkyl and N’-(ω-hydroxyalkyl) N-methylimidazolium chlorides. Protein Sci. 2005, 14, 2693–2701. [Google Scholar] [CrossRef] [PubMed]
- Patel, R.; Mir, M.U.H.; Singh, U.K.; Beg, I.; Islam, A.; Khan, A.B. Refolding of urea denatured cytochrome c: Role of hydrophobic tail of the cationic gemini surfactants. J. Colloid Interface Sci. 2016, 484, 205–212. [Google Scholar] [CrossRef] [PubMed]



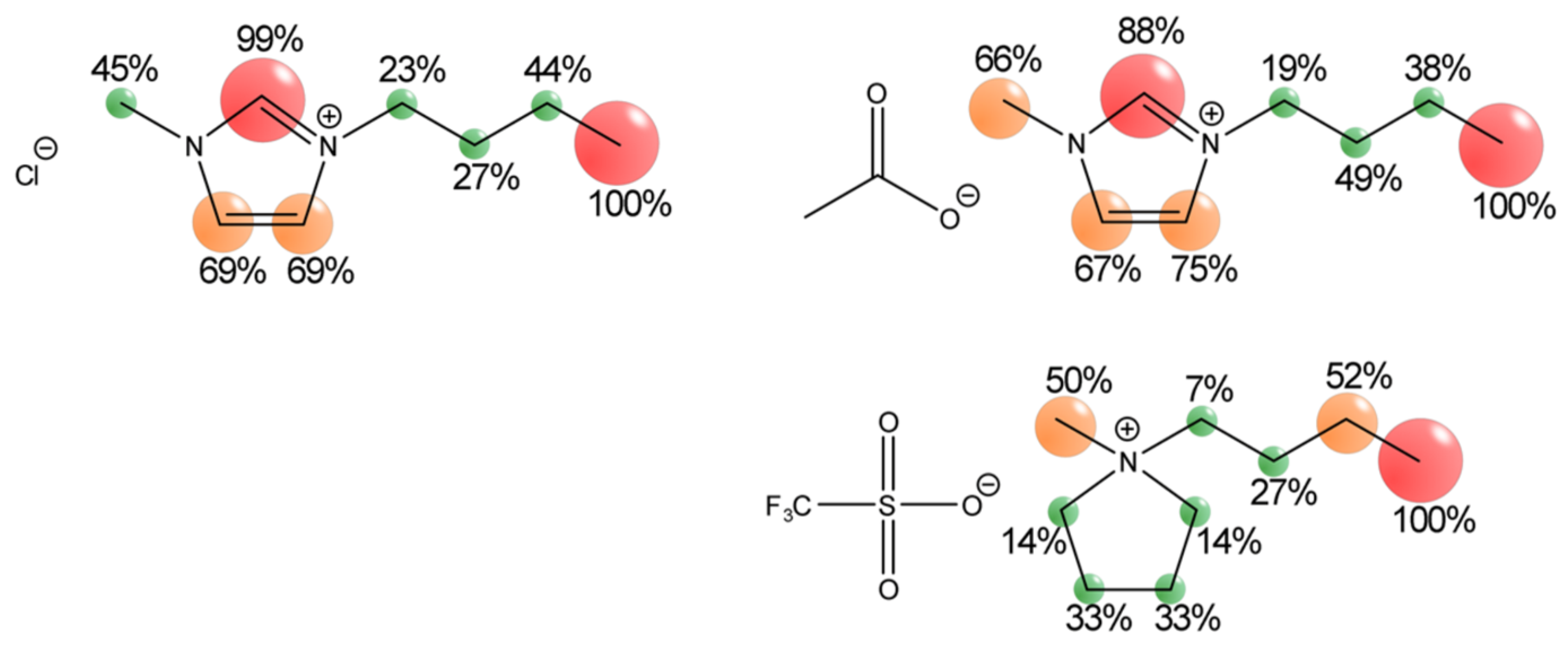

| Cations | Anions | ||
|---|---|---|---|
| 1-ethyl-3-methylimidazolium | EMIM | triflate | OTf |
| 1-butyl-3-methylimidazolium | BMIM | bis(trifluoromethylsulfonyl)imide | NTf |
| 1-hexyl-3-methylimidazolium | HMIM | dihydrogen phosphate | dhp |
| 1-methyl-3-octylimidazolium | OMIM | acetate | OAc |
| 1-decyl-3-methylimidazolium | DMIM | methylsulfate | MeSO |
| cholinium | chol | ethyl sulfate | EtSO |
| hexylsulfate | HexSO | ||
| Ethyl-ammonium nitrate | EAN | ||
| Propyl-ammonium nitrate | PAN | ||
| Butyl-ammonium nitrate | BAN | ||
| Sodium dodecylsulfate | SDS | ||
| guanidinium chloride | GndHCl | ||
| L-Arginine monohydrochloride | [L-Arg][HCl] | ||
| Protein State | Cholinium | Ammonium | Imidazolium | Pyrrolidinium | Phosphonium | Sulfates | Phosphates | Carboxylates | Halides | Nitrate | Protein | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Solubilization | heat aggregated | egg white protein [149] | ||||||||||||
| heat aggregated | concanavalin A [150] | |||||||||||||
| native | Zein [151] | |||||||||||||
| native | metalloproteins [152] | |||||||||||||
| native | cytochrome C [153] | |||||||||||||
| inclusion body | Coprinopsis cinerea cellulase 6A [154] | |||||||||||||
| inclusion body | Coprinopsis cinerea cellulase 6A [155] | |||||||||||||
| natural fibres | wool, silk [156] | |||||||||||||
| amyloid fibrils | lysozyme [157,158] | |||||||||||||
| amyloid fibrils | bovine insulin amyloid [23,159] | |||||||||||||
| Refolding | chemically denatured | lysozyme [160] | ||||||||||||
| chemically denatured | lysozyme [161] | |||||||||||||
| chemically denatured | horseradish peroxidase [162] | |||||||||||||
| chemically denatured | ScFvOx, lysozyme, recomb. plasminogen activator [163] | |||||||||||||
| chemically denatured | cytochrome C [164] | |||||||||||||
| chemically denatured | interferon alpha 2b [165] | |||||||||||||
| Stabilization | native>fibrillation | alpha-synuclein [166] | ||||||||||||
| native>fibrillation | lysozyme [33] | |||||||||||||
| native | ribonuclease A [55,94] | |||||||||||||
| native | lysozyme [167] | |||||||||||||
| native | horse heart myoglobin [168] | |||||||||||||
| native | insulin [169] | |||||||||||||
| native | metalloproteins [152] | |||||||||||||
| native | ovalbumin, lysozyme [170] | |||||||||||||
| native | cytochrome C [153] | |||||||||||||
| native | cytochrome C [171] |
| Strengths | Weaknesses |
|---|---|
| effective solubilizer | effectiveness depends on protein |
| customizability | complex synthesis and high costs |
| protein activity enhancer | toxicity |
| hydrophilic↔hydrophobic | |
| multipurpose co-solvent | |
| Opportunities | Threats |
| alternative solvents | other biocompatible solvents |
| enhanced protein yields | low reproducibility |
| advanced purification | environmental impact |
| protein-based products | regulatory challenges |
| concentration-dependent effects |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Szabadi, A.; Klausser, R.; Spadiut, O.; Schröder, C. Inclusion Bodies in Ionic Liquids. Liquids 2024, 4, 1-31. https://doi.org/10.3390/liquids4010001
Szabadi A, Klausser R, Spadiut O, Schröder C. Inclusion Bodies in Ionic Liquids. Liquids. 2024; 4(1):1-31. https://doi.org/10.3390/liquids4010001
Chicago/Turabian StyleSzabadi, András, Robert Klausser, Oliver Spadiut, and Christian Schröder. 2024. "Inclusion Bodies in Ionic Liquids" Liquids 4, no. 1: 1-31. https://doi.org/10.3390/liquids4010001
APA StyleSzabadi, A., Klausser, R., Spadiut, O., & Schröder, C. (2024). Inclusion Bodies in Ionic Liquids. Liquids, 4(1), 1-31. https://doi.org/10.3390/liquids4010001






