1. Introduction
Drug repurposing is a process to identify new roles for existing drugs and is generally considered an efficient and economical approach [
1]. Repurposing—also known as re-profiling, re-tasking, repositioning, and rescue of drugs—can help identify new therapies for diseases, at a lower cost and in a shorter time, particularly in those cases where preclinical safety studies have already been conducted. It can play a crucial role in “therapeutic stratification procedures” for patients with rare, complex, or chronic diseases with few to no treatment procedures. The advent of genetic programming and computational approaches has led to the development of novel strategies for drug-repurposing. It is a convenient alternative when an unexpected medical scenario, such as the coronavirus disease 2019 (COVID-19), which was declared a pandemic by WHO (World Health Organization) in March 2020, presents itself and the need for new drugs becomes inevitable [
2,
3].
Outbreaks of COVID-19 have presented unique challenges to healthcare professionals. The urgent need to provide appropriate pharmacological therapeutics leaves medical professionals minimal time for new drug discovery. Drug discovery is a daunting task that takes several years, from conception to availability in the market [
1]. Since its spread from Wuhan, China, COVID-19 has rapidly claimed the lives of hundreds of thousands of people across the world [
1,
4]. As of November 2021, 5.2 million people have died so far due to this disease, and confirmed cases of persons with COVID-19 are 262.8 million [
5]. The need for vaccines and drugs has become more urgent as new virus variants have evolved much faster than anticipated [
6,
7].
Growing evidence suggests that mutations changing the antigenic phenotype of SARS-CoV-2 are in circulation. This requires immediate attention as it affects immune recognition at an alarming degree. SARS-CoV-2 infection uses angiotensin-converting enzyme-2 (ACE2) receptor and transmembrane serine protease (TMPRSS2) to infect human respiratory cells. Mutations that affect the antigenicity of the spike protein of the virus are of significance. The spike protein enables attachment of the virus to the host cell surface receptors like ACE2 and fusion between the virus and cell membranes. It is the principal target of the neutralizing antibodies that are generated after infection [
7]. The various mutations of SARS-CoV-2 evolve with time. Thousands of variants have emerged since the pandemic began. The first major variant, termed Variant of Concern (VOC) 202012/01 N501Y and the second major variant 501Y.V2, were detected in the last quarter of 2020 in the UK and South Africa, respectively. Since then, these variants have been detected in over 40 countries. These variants have increased transmissibility, virulence, and low response rates to available diagnostics and therapeutics [
8].
To repurpose drugs for the treatment of any disease, one must look into the structure of the causative organism. Coronavirus (CoVs) belong to Coronaviridae, which is a single-stranded enveloped positive RNA virus subdivided into alpha (α), beta (β), gamma (γ), and delta (δ). Among them, the β (β-CoV) group is divided into severe acute respiratory syndrome coronavirus (SARS-CoV); severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), which is COVID-19; and Middle East respiratory syndrome coronavirus (MERS-CoV) [
4]. These viruses are fatal and responsible for respiratory, liver, gastrointestinal, and central nervous system damage in humans and animals. SARS-CoV-2 responsible for COVID-19 is more pathogenic than SARS-CoV and MERS-CoV, and it can transmit from human to human, causing fatal illness. It can lead to severe respiratory problems with several symptoms, including fever, dry cough, vomiting, fatigue, diarrhea, and shortness of breath. The virus SARS-CoV-2 likely binds to epithelial cells in the nasal cavity after inhalation and starts propagating. It migrates down the respiratory tract along the conducting airways, triggering an innate immune response. At this stage, the virus can be detected by nasal swabs. Recent studies suggest that initial viral contact occurs in the nasal mucosa through binding of the viral S (spike) protein to the ACE2 (angiotensin-converting enzyme-2) receptor, followed by cleavage of S protein by TMPRSS2 (transmembrane serine protease 2) [
9].
The discovery and licensed use of a drug come with a long gestation period. The development of a drug for any disease, especially for diseases caused by viruses, takes time, even in an accelerated mode; it would take around 3–5 years to market it as a ready-to-use product. The cost of the new drug development process amounts to more than a billion dollars, extending for 10–15 years with a success rate of only 2.01% [
10]. This creates a lag in the productivity of pharmaceutical research to develop a new drug, resulting in a persistent gap between therapeutic needs and available treatments. Despite base evidence being of variable quality, existing drugs have been repurposed in the war against the coronavirus pandemic [
11]. Even though vaccines have been made available for the general populace, there is no specific treatment for the patients suffering from the disease. Thus, investigational molecules that fail to show efficacy for a predetermined medication typically provide alternative therapeutic effects for another disease [
10]. With computational tools and improving knowledge of virology and clinical presentation of COVID-19, researchers are tapping into a broadening pool of potential pharmacological targets.
Currently, several groups of drugs are being investigated against COVID-19. This includes hydroxychloroquine, remdesivir, chloroquine, lopinavir, ritonavir, and so on. These are established drugs used for the treatment of SARS-CoV, MERS-CoV, and other viruses. As there is an emergency requirement to establish potential inhibitors against COVID-19, repurposing drugs is an ideal strategy to gain headway into the same. During repurposing, virtual screening, pharmacophore modelling, other computational methods, and experimental methods are extensively used. As an example of successful stances in drug repurposing, one need not look any further than the treatment modules for the Zika virus, a mosquito-borne flavivirus. Zika virus infection lacks a specific drug or vaccine for its treatment. Over the years, researchers around the globe have screened thousands of previously studied compounds and have identified several compounds with anti-ZIKV activities. Barrows and colleagues identified approximately 24 potential drugs with anti-ZIKV activities from a library of 774 FDA-approved drugs. Based on such studies, niclosamide and azithromycin are the most commonly used drugs in ZIKV treatment, especially for pregnant women. Chloroquine, a commonly used anti-inflammatory and antimalarial drug, is also used to treat the same [
10].
The methodologies adopted in drug repurposing can be divided into three broad groups depending on the pharmacological, toxicological, and biological activity information available. These are drug-oriented, target-oriented, and disease/therapy-oriented [
10]. This review focuses primarily on the target-oriented methodology of drug repurposing strategies against COVID-19.
3. Strategies in Drug Repurposing for COVID-19
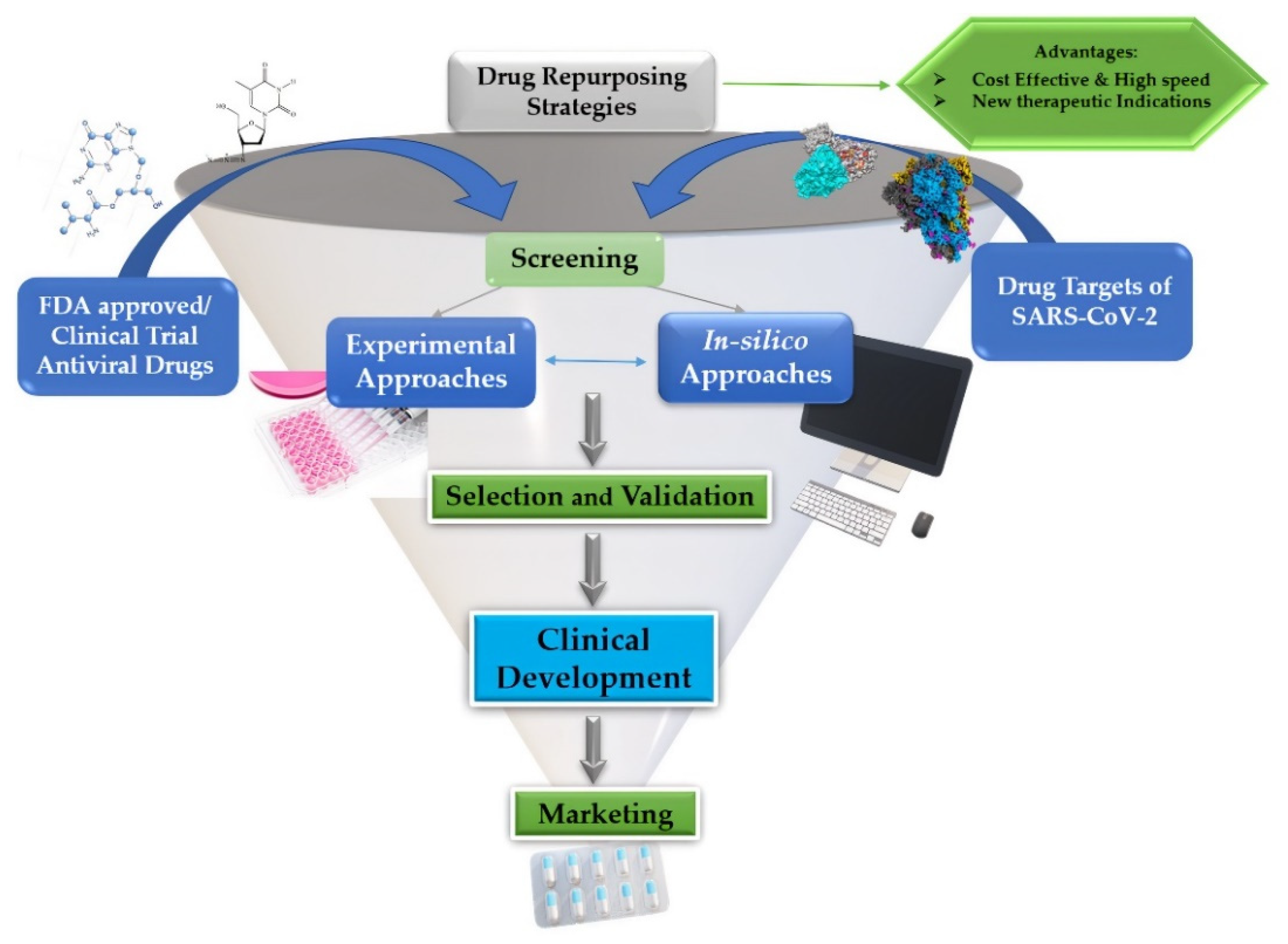
For rapidly establishing drugs for arising and re-emerging viruses, drug repurposing is one of the most substantial preferences nowadays. Given the SARS-CoV-2 pandemic, compared to de novo drug development, drug repositioning appears as a favourable perspective to develop efficient therapeutics. Traditional drug repurposing techniques establish drug effects and their mode of action (MoA). This process employs screening techniques of current pharmacopoeia to reveal novel drug indications of already established drugs [
25]. Drug repurposing for COVID-19 undergoes three steps before getting considered for development and marketing: candidate drug identification; mechanistic evaluation of the drug effect in preclinical models; and evaluation of candidate drugs’ efficacy in phase II clinical trials. Among these three steps, the first step—the screening and identification of drugs with a high potential for repurposing—is the most crucial. As such, drug repositioning has two alternative and complementary approaches: an experiment-based approach and a theoretical or in silico-based approach, as shown in
Figure 1 [
26,
27,
28].
The experiment-based approach, also known as activity-based repositioning, refers to screening original drugs for new pharmacological applications based on experimental assays. It involves protein-target-based and cell-based screens in disease models without requiring any structural information of target proteins. Approaches of experimental repositioning include the target screening approach, cell assay approach, animal model approach, and clinical approach [
29].
In contrast, in silico repositioning involves virtual screening of public databases of huge drug/chemical libraries using computational biology and bioinformatics/cheminformatics tools. In this approach, the identification of potentially bioactive molecules is achieved based upon the molecular interaction between drug molecules and protein targets. Although experimental explorations are functional for drug efficacy determination, they may be time-consuming with small-scale results. Therefore, computational methodologies have enhanced this approach by digging deeper into drug executions, being capable of evaluating the interaction of ligand(s) with respective target proteins, predicting the novel signalling pathways, and making rapid development in lesser time with reduced costs, which is significant for the current pandemic (COVID-19) situation [
30].
3.1. Experimental Approaches
3.1.1. Binding Assays to Identify Target Interactions
Today, analyses of the targets and off-targets of drugs and drug repurposing complement each other. Proteomic techniques such as affinity chromatography and mass spectrometry have been used as approaches to identify binding partners for many drugs. Experimental assays of pre-approved drugs are being adopted in laboratories to bridge the gap between instant diagnosis and long-term research required for the availability of treatment for COVID-19 [
31]. The cellular thermostability assay (CETSA) is one such technique introduced to map target engagement in cells using biophysical principles that predict thermal stabilization of target proteins by drug-like ligands that possess the appropriate cellular affinity [
32]. Examples of execution of this technique include the confirmation of cellular targets for the tyrosine kinase inhibitor (TKI) crizotinib and the detection of quinone reductase 2 (NQO2) as a cellular off-target of acetaminophen (paracetamol) [
33]. In a study by Friman et al., a panel of drugs was screened using the CETSA MS format on HepG2 cells to identify host proteins. The experiment was designed to study off-target effects of remdesivir and chloroquine as repurposed drugs for targeting SARS-CoV-2. This leads to a hopeful venture to further improve or develop fortuitous therapies for SARS-CoV-2 infection [
34].
The promiscuity of protein kinase inhibitors has increased efforts to develop better compounds for preclinical research that can help clinical drug development and repurposing through evidence-based pharmacological assays. It is also noteworthy that fallacies made in various kinase drug discovery approaches have led to seeking out beneficial pathways of cellular effects through early-stage affinity approaches. Numerous kinases have been considered essential mediators of various viral infections, particularly SARS-CoV and MERS-CoV. These same proteins are also predicted to be involved in mediating infection by SARS-CoV-2 as well. Many kinase inhibitors with pharmacologic effects that may be beneficial in mitigating the severe and potentially life-threatening symptoms of COVID-19 are already approved. Kinase inhibitors can be tested in combination with antiviral agents or other targeted therapies that work against COVID-19 to achieve greater efficacy [
35].
Tyrosine-protein kinase (ABL) inhibitors have been demonstrated to inhibit replication of several unrelated viruses like dengue and Ebola in in-vitro cell-based studies. ABL inhibitors, imatinib and dasatinib, were identified as inhibitors of both SARS-CoV and MERS-CoV replication. Nilotinib was identified as an inhibitor of SARS-CoV. Investigation of the mechanism for imatinib against both SARS-CoV and MERS-CoV revealed inhibition of the early stages of the virus life cycle and inhibition of viral replication through blocking the fusion of the coronavirus virion with the endosomal membrane. Importantly, authors show that targeted knockdown of ABL2, but not ABL1, significantly inhibited SARS-CoV and MERS-CoV replication/entry in vitro [
35].
JAK inhibitors belong to a family of DMARDs or disease-modifying antirheumatic drugs, which have been repurposed to treat COVID-19. Chen et al. have summarized the observational studies of the clinical uses of Janus Kinase (JAK) inhibitors, including ruxolitinib, baricitinib, and tofacitinib for COVID-19 patients. They compared the clinical efficacy of JAK inhibitors of different meta-analyses and concluded that they are safe agents and can lead to a better clinical outcome for COVID-19 patients [
36].
Diving into chemical genetics, one can also better understand the relationship between binding and efficacy in the cellular context. Non-kinase targets of small molecules designed initially to inhibit protein kinases are increasingly recognized. Their validation has led to repurposing opportunities in cancer, as Zika virus modulators and potential agents to treat cancer antibiotic-resistant microorganisms [
37]. Small-molecule–kinase binding is analyzed in a kinome-wide fashion using various in vitro and increasingly organism-based assays to generate heat maps of biologically essential interactions. Thus, findings can be rapidly translated into new clinical developments to address drug-resistance outcomes in cancer. Many of these studies stem from industry-driven high-throughput direct binding or catalytic assays [
32]. Karaman and colleagues used an in vitro competition binding assay to evaluate 38 kinase inhibitors against a panel of 317 distinct human protein kinases in one such study. Their analysis identified a total of 3175 binding interactions. Interestingly, some kinase inhibitors such as sorafenib and dasatinib showed higher affinity to secondary kinase targets than their known primary target [
38]. Such studies show that drug repurposing may indeed be a quick and effective combat strategy against the current pandemic.
3.1.2. Phenotypic Screening
Phenotypic screening puts forward drug candidates to proteins in more biologically relevant contexts than screens involving purified proteins. These screens determine cellular function without the requirement of prior knowledge of the relevant targets and signalling pathways, and they offer the possibility of discovering new therapeutic targets [
39]. Phenotypic screening can identify compounds that show disease-relevant effects in model systems without prior knowledge of the target(s). In the context of drug repurposing, if the compounds screened are approved or investigational drugs, this may fast-track repurposing opportunities that can readily be pursued.
Typically, in vitro phenotypic screens use a wide range of cell-based assays in a 96-well format. For example, Chen et al., in 2021, tested a total of 8810 approved and investigational compounds, out of which 319 compounds were found to have anti-SARS-CoV-2 activities [
40]. These included 91 approved drugs and 49 investigational drugs. The anti- SARS-CoV-2 activities of 230 of these confirmed drugs had not been previously reported, including 38 approved drugs from the same. The three most potent FDA-approved drugs with anti-SARS-CoV-2 activities were chlorprothixene, methotrimeprazine, and piperacetazine [
40].
The compounds that were tested were either mechanism-based bioactive compounds or natural products. They were selected from compound libraries like NCATS Pharmaceutical Collection (NPC), NCATS Mechanism Interrogation Plate (MIPE), NCATS Pharmacologically Active Chemical Toolbox (NPACT), Epigenomic library, Autophagy library, and anti-infective library. A SARS-CoV-2 CPE assay was conducted to check for anti-SARS-CoV-2 activities [
41].
The SARS-CoV-2 CPE is a 72 h assay that measures the phenotypic consequence of viral infection and cell replication [
42]. SARS-CoV-2 induces cell death after 48 to 72 h of infection, and thus cell viability is an indirect measure of viral replication in vitro. Due to its dependence on the host response and its indirect measurement of SARS-CoV-2 infection and replication, the CPE assay may have certain limitations. The phenotypic outcome can also vary depending on culture conditions, the viral multiplicity of infection (MOI), and the number of virions added per cell during infection [
43,
44,
45].
3.2. Computational Approach
For the apprehension of the virus origin and to recognize the evolutionary relationship, other computational procedures are utilizing various computational and AI-based tools. Omics analysis, including proteomics, genomics, metabolomics, epigenomics, and transcriptomics, plays a crucial part, along with classifying the sequence of the virus and its mutational variants, manifesting the sequence and 3D structure of the viral proteins, indispensable for repurposing and discovery of drugs together with the development of vaccines [
46,
47]. Target-based, network or pathway-based, knowledge-based, signature-based, and artificial-intelligence-based techniques are the most common computational methodologies, as shown in
Figure 2.
3.2.1. Drug-Focused Strategies
Polypharmacological techniques focus on how a particular medication affects numerous endpoints in a specific disease pathway or across several disease pathways, and while looking for a novel target for an existing medicine, assessing drug–target binding and interaction might assist in finding additional structurally comparable compounds that have the same target binding capabilities. The polypharmacological strategy also aids in the discovery of undiscovered off-target effects for currently available medications.
3.2.2. Target-Based Approaches
The investigation of substances or stimulants from vast compound libraries as potential drugs for a targeted protein or a biomarker of interest (i.e., protein, receptors) to discern a biochemical reaction is known as target-based screening. Ligand-based inspection or molecular docking [
48,
49] are examples of target-based methods. When contrasted with unbiased research approaches, such strategies have a better chance of uncovering valuable leads. It also takes a minimal time to complete the entire screening procedure. Novel observations are discovered using this method, which involves connecting a drug to a particular disease predicated on its target proteins. As previously stated, a new indication for a medicine can be defined based on both the principal and off-target molecules. Target repositioning is the process of treating a new indication by associating with the same target molecule as discovered initially. Off-target placement occurs when an authorized medicine engages with a second target and can be used to manage a new indication [
50,
51].
Computational analysis for drug repurposing of an azamacrocyclic compound, grazoprevir—an antiviral drug first validated for hepatitis C virus and approved by the FDA—which with viral replication targeting RNA-dependent RNA polymerase (RdRP) and transmembrane serine protease 2 (k) (TMPRSS2), along with angiotensin-converting enzyme 2 (ACE-2) proteins produced an optimal binding affinity. Moving further with the procedure of molecular dynamics simulations as well as molecular mechanics Poisson–Boltzmann surface area evaluation recognized the drug–receptor interactions and the stable association of grazoprevir with all the proteins, which may be a functional therapeutic for COVID-19 treatment [
52].
Another approach using FDA-approved drugs, included 8395 compounds from the DrugBank library. Several natural compounds and various target proteins of SARS-CoV-2—namely, the RNA-binding domain of nucleocapsid phosphoprotein, Nsp12 RNA-dependent RNA polymerase, the Nsp3 ADP ribose phosphatase, the Nsp5 3-chymotrypsin-like protease (3CLpro), Nsp14, the Nsp15 endoribonuclease, and the Nsp16 2′-O-MTase were studied using Docking techniques which observed 124 potential inhibitors after screening. This could be a feasible direction for repurposing of these drugs, together with predicting tipranavir as well as nelfinavir, taking 3Clpro as their target [
53].
An efficient computational drug repurposing study of five neutral drugs—valrubicin, elbasvir, lopinavir, eravacycline, and carfilzomib—identified these drugs as possessing inhibitory activities opposed to the SARS-CoV-2 main protease and suggested them as the best drug repurposing candidates. For the SARS-CoV-2 main protease, streptomycin may also be an inhibitor, and some residues for the receptor–ligand-binding contributing to the protein–ligand binding have also been identified, which may enable the design of novel inhibitors targeting the SARS-CoV-2 main protease [
54].
Approaches Based on Structure
Virtual evaluation can aid in the identification of tiny chemicals capable of binding macromolecular targets with known or expected three-dimensional structures. It enables the screening of millions of substances in a short amount of time, lowering the costs of discovering hits for drug discovery and development, as well as discovering new targets for patented drugs. This method is primarily founded on molecular docking [
55,
56], a computational paradigm that was originally created to comprehend how a pharmacological molecule interacts with a biological equivalent, which is now widely utilized for a variety of activities, including therapeutic repositioning [
57,
58,
59].
Before crystal structures of viral particles were accessible, the first instance of structure-based medication repurposing performed for COVID-19 were reported. The architectures of numerous target viral proteins were predicted using homology modelling approaches. In most cases, the findings were not experimentally validated. The molecules of compounds in a docking-ready manner were traditionally the beginning point for screening infrequent items, with at most a portion allocated to authorized medications, which may supply the biomolecules of the compounds in a docking-ready style. Additionally, an investigation of the structural properties of Mpro discovered significant changes in the structure and size of this protease’s active sites compared to the SARS-CoV protease, indicating that repurposing SARS medicines for COVID-19 may be futile. Ribonucleoside inhibitors, antiretrovirals, nucleotide inhibitors, antimalarials, antineoplastics, immunomodulators, steroid hormones, and protease inhibitors account for the majority of the hits identified in these investigations [
60]. Scientists who established publicly available web servers to forecast targets and multitarget- and multi-site-based virtual screening added an extra input to the structure-based strategy for drug repurposing.
Employing the docking methodology with molecular dynamics simulation, an exploration for interpretation of the inhibitory system for the nucleocapsid protein in SARS-CoV-2 with antiviral molecules together with the stability of protein-ligand complex was established and revealed that saracatinib, nafamostat, rapamycin, trametinib, and camostat were the top hit compounds, suggesting these residues as potential sites of drug targeting for the SARS-CoV-2 N protein that can be utilized for COVID-19 treatment [
61,
62].
For protein inhibition, approximately 50 antiviral drugs were collected in a research approach, examined utilizing the process of molecular docking for drug target proteins, the spike of SARS-CoV-2, RNA-dependent RNA polymerase (RdRp), and main protease. The drugs lopinavir, indinavir, nelfinavir, remdesivir, and saquinavir showed maximum inhibitory potential as drugs for SARS-CoV-2 inhibition, and lopinavir with saquinavir was highest of all after further laboratory evaluation [
63].
Similarly, 15 drugs—quinapril, sofosbuvir, amphotericin B, baloxavir marboxil, fosinopril, danoprevir, clarithromycin, atrovastatin, telmisartan, sulfamethoxazole, virginiamycin, micafungin, tunicamycin, caspofungin, and fidaxomicin—which are FDA-approved, were established as SARS-CoV-2 protein inhibitors in an approach confirmed by docking protocol. They could be explored as drug candidates against COVID-19 in the future and be repurposed against SARS-COV-2 [
64].
Likewise, subsequent research of 6218 clinical trials and FDA-approved drugs were virtually screened for RNA-dependent RNA polymerase and main protease as well, resulting in 15 and 23 possible repurposed drugs for treating COVID-19, selecting Mpro as well as RdRp, respectively. The drugs tipifarnib, omipalisib, and emodin exhibit anti-SARS-CoV-2 activities in human lung cells, while Calu-3 and drug combinations including tipifarnib/remdesivir, tipifarnib/omipalisib, and omipalisib/remdesivir revealed secure synergistic effects for SARS-CoV-2 inhibition [
65].
Another examination for Sar9 Met (O2)11-Substance P along with BV2, which are peptide-based drugs, using the same methodologies of molecular docking then molecular dynamics along with protein–protein interaction simulations, predicted these drugs as inhibitors of the interaction of SARS-CoV-2 spike protein with the human angiotensin-converting enzyme 2 (hACE2) by having a high probability of binding with the SARS-CoV-2 spike protein connecting side on the human angiotensin-converting enzyme 2 (hACE2) receptor, enough for the protein–protein interaction regulation preferred by the virus [
66].
Procedure Based on Pathways or Networks
Pathway- or network-based techniques leverage illness omics data, existing signaling or biochemical functions, and interacting protein connections to reassemble disease-specific circuits that serve as major targets for repositioning medicines by which the medicine displays its effectiveness and drug–target relationships. Networks may be used to describe molecular interactions in biological systems, from developments in high-throughput technologies and bioinformatics tools. As a result, a particular network consisting of a few more individual goals may be identified from a broad network of diverse paths. These approaches benefit from being able to pin down broad signaling networks with a huge range of proteins to a specialized network with only some proteins (or target molecules) [
50,
51,
67].
The goal of network-based pharmaceutical research is to use the capacity of systems to shed light on the processes of the effect of current or new compounds so that innovative therapeutic remedies can be identified. Because of their capacity to incorporate diverse data sources, network-based techniques are critical and frequently employed in medication repositioning. Nodes in the network represent medications, illnesses, or nucleotide sequences in these models, whereas edges reflect node interactions or linkages. The resultant pattern may make structure-guided pharmacological and medical research possible, with the possibility of discovering novel biological targets. Drug–target networks, drug–drug networks, drug–disease networks, and protein interaction networks have all been shown to be effective in identifying new options for drug development or repositioning in previous research. In the human protein–protein interaction network, the network-based approach combines a system-pharmacology-based network medicine framework that analyzes the interaction between the virus–host interactome and therapeutic targets [
68,
69].
Recently, by a network-based algorithm, 282 repurposable drugs, like hydroxychloroquine, chloroquine, heparin tocilizumab, and combination therapies of other drugs, including remdesivir, ritonavir, lopinavir, chloroquine, and hydroxychloroquine, were identified. Moreover, based on their network-based similarity values, 24 potential anti-SARS-CoV repurposable drugs were observed. These drugs include thrombin inhibitors, ACE inhibitors, and monoclonal antibodies such as anti-IL6, anti-TNFα, anti-IL1β, anti-IFNγ, and anti-IL12 [
70].
Another computational antiviral drug repurposing procedure identified 16 potential anti-HCoV repurposable drugs, including melatonin, sirolimus, and mercaptopurine, together with three possible combinations of drugs, namely sirolimus plus dactinomycin, toremifene plus emodin, and mercaptopurine plus melatonin, using a system-pharmacology-based network medicine system, in between the HCoV–host interactome along with target proteins in the human protein–protein interlinkage network. The significant druggability of this HCoV–host interactome facilitates the creation of a drug repurposing approach towards 2019-nCoV/SARS-CoV-2 therapy by precisely addressing protein molecules linked with HCoVs. These network accessibility evaluations identify possible repurposable options for HCoV treatment and prevention. Furthermore, phylogenetic recognition of 15 HCoV whole genomes to examine the evolutionary connection of 2019-nCoV/SARS-CoV-2 with several other HCoVs revealed that the complete genome of 2019-nCoV/SARS-CoV-2 shows 99.99 percent nucleotide sequence similarity. From the six previous common pathogenic HCoVs [
71,
72,
73], the 2019-nCoV/SARS-CoV-2 has the greatest nucleotide sequence similarity (79.7 percent) with SARS-CoV, indicating a maintained evolutionary connection with 2019-nCoV/SARS-CoV-2 and SARS-CoV. When compared to SARS-CoV, the envelope as well as nucleocapsid proteins of 2019-nCoV/SARS-CoV-2 are the evolutionarily conserved areas, with 96 percent and 89.6 percent sequence identity, respectively [
74].
Knowledge-Based Methods
Blinded (unintentional, serendipitous findings) and target-based techniques, while successful, cannot be used to investigate novel drug–target relationships. Knowledge-based strategies integrate existing data about a medicine to predict heretofore unrecognized procedures, namely the availability of unrecognized drug targets for older pharmaceuticals, unknown drug–drug correlations, and novel biomarkers. Bioinformatics, cheminformatics, or text analytics are three kinds of knowledge-based techniques. Knowledge-based approaches improve prediction confidence by incorporating a large quantity of data for drug repurposing. The most promising repurposing conclusion comes from a mix of biological, chemical, and diagnostic features, which can lay the groundwork for introducing a strong target for an already-approved medicine, as well as a detailed understanding of its mechanism of action.
A pharmaceutical development system uses bioinformatics mining (computational biology) to identify novel inter-relationships and perhaps new property rights in a specific study phenomenon by using data obtained privately or from publicly released sources. When a group uses its own biological data for repurposing research, it is common for them to look at different disease conditions. On the other hand, generalist groups do not focus their efforts on a single illness; instead, they create a panel of disease-relevant models and then screen chemicals that can be reused globally. Specialists rely on their ability and specialty within a particular disease area to test vast numbers of chemicals against a selected set of disorders.
Two key aspects must be thoroughly examined in order to know how to appropriately repurpose these safe pharmacological molecules to acceptable applications. To begin with, each given small-molecule drug candidate can potentially interact with a wide range of proteins. Second, complex disorders are typically the outcome of complicated intra- and inter-cellular molecular interactions that are standardized and organized regularly. Often, information concerning drug–protein relationships is insufficient, and most of the computerized data analysis for repurposing originates through the combination of many data sources. Very notably, this occurs at the intersection of conventional clinical care, chemistry, biology, and toxicological rules.
In a drug repurposing strategy, bioinformatics-based techniques have been effective in discovering novel links among biological variables such as genes, biological processes, and illnesses. Another approach uses transcriptional data for therapeutic repurposing, which involves finding drugs with opposing transcriptional profiles that target a disease. Considering the current technique’s unimpressive effectiveness, emerging transcription-based approaches such as CuGuCtD are improving our knowledge and ability to determine if a chemical has the capability to regulate gene expression genes in the very same manner as the chemicals that are currently managing that illness does [
50,
75].
For the need of medication, repositioning may even arise following positive phase I clinical study consequences, including the quality and system of an acceptable clinical situation for phase II or III studies in cheminformatics. The identification of relevant biomarkers confirms requests for activation assumption. A disorderly behavior might be owing to a lack of knowledge of the target protein’s involvement in illness occurrence and development if the therapy is a proven hit to the targets.
Techniques Based on Signatures
Gene signatures produced from illness omics data even without therapy are used in signature-based medication repurposing techniques to find undiscovered off-targets or disease causes. Signature-based techniques use deterministic data at the molecular levels, such as a gene mutation and successive protein production, to unveil unidentified modes of action of bioactive compounds, using strategies such as normalized gene co-expression network analysis (WGCNA) to CMap, as well as the Library of Integrated Network-Based Cellular Signatures (LINCS) [
74].
The benefit of these approaches is that they may be used to investigate undiscovered pharmacological modes of action. In contrast to knowledge-based mechanisms, signature-based strategies use computational methodologies to examine pharmacological processes at a microscopic level, such as changes in gene expression. As microarray and next-generation sequencing techniques improve, massive amounts of genomics data relevant to medication repurposing studies gather, which might be utilized to uncover gene signatures for investigating unknown disease-altering pathways. Because individual gene signatures determine a medicine’s efficacy, a gene signature database can aid drug repurposing using computational approaches.
AI-Based Approaches
AI is a portion of computer-aided drug design (CADD), which, alongside the emergence of AI, is inexorable, limiting the restrictions of a computational methodology in drug repositioning and drug discovery. The machine learning cognitive approach has progressed to deep learning approaches that encourage better data processing, dependable outcomes in the shortest feasible time, and significant expense.
In critical situations, such as the present COVID-19 worldwide outbreak, AI has proven to be a valuable addition to medication repositioning. For example, BenevolentAI, London, UK, an AI predictive approach based on an extensive collection of structured medical data that incorporates multiple linkages retrieved from research journals using deep learning, has benefited in discovering possible COVID-19 medication repositioning options [
57,
74]. The AI sector contributes to the battle against COVID-19 by creating publicly available web servers and tools, such as the Confederation of Laboratories for Artificial Intelligence Research in Europe (CLAIRE AISBL), which comprises 381 AI-related laboratories and institutes around Europe [
76].
Drug repositioning is one of the study subjects in this setting, with data and computational tools available to scientists and researchers. Recently, researchers have developed a deep learning strategy for finding commercialized medications that may have antiviral properties towards coronaviruses [
76,
77]. The approach was designed to swiftly evaluate many chemicals using given learning datasets to identify those with potential anti-SARS-CoV-2 activity. However, publications based on AI research are insufficient for drug repurposing and the expansion of a big pool of biological and chemical data. Robust computation and systems for data mining are required to ensure a quick, high-efficiency, and low-cost drug discovery procedure. Computational methodologies combined with AI offer great potential for drug repositioning, especially in light of the COVID-19 epidemic, which desperately needs a cure. Furthermore, this method is as good as or better in certain circumstances than in vitro experiments.
4. Drugs Repurposed against SARS CoV-2 Drug Targets
Drug repositioning is not only a current scientific trend but spans across several decades. Drugs with a specific clinical indication have been further tested to discover alternative clinical indications for different diseases. The most common drug repurposing examples are NSAIDs (anti-inflammatory drugs) being used as anticancer agents. Chloroquine, an antimalarial drug, and azithromycin, an antibacterial antibiotic are under development as antiviral drugs against COVID-19 [
50]. Drugs currently being tested for repositioning in COVID-19 can be distinguished as drugs potentially able to inhibit one or more steps of the coronavirus lifecycle and those that can counteract the effects of SARS-CoV-2 infection, such as the amplified immune response and the massive cytokine release, both of which lead to severe complications such as coagulopathy and acute respiratory distress syndrome (ARDS). The first is remdesivir, first developed in 2009 to treat hepatitis C, then repurposed to treat Ebola. Although ineffective in treating both diseases, later animal studies found that it effectively managed other coronaviruses such as SARS and MERS. It has proven effective in shortening recovery time from COVID-19 in some patients if administered early. However, it is to be used with only the most severely affected patients in critical care units. Another group of drugs that have previously been widely used among critically ill patients with SARS and MERS are glucocorticoids, powerful anti-inflammatory drugs that inhibit the production and survival of T-cells and macrophages. Although controversial, glucocorticoids have been used to treat patients critically ill with COVID-19 [
23,
78]. A comprehensive list of repurposed drugs used against drug targets of SARS-CoV2 is provided in
Table 1.
4.1. Nafcillin
Nafcillin is a semi-synthetic, narrow-spectrum antibiotic, a beta lactamase-resistant penicillin. The bactericidal action of penicillin inhibits cell wall synthesis due to the presence of the beta-lactam ring. However, certain bacteria develop resistance against the beta-lactam ring by synthesizing beta-lactam inhibitors (i.e., beta-lactamase or penicillinase). Penicillinase resistance drugs were introduced to combat this resistance [
25]. Currently, nafcillin is being used to treat penicillinase-producing staphylococcal species, particularly methicillin-sensitive Staphylococcus aureus (MSSA). Nafcillin is also being used to treat non-specific lower respiratory tract infections and community-acquired pneumonia (CAP) [
26]. Nafcillin is not known to cause life-threatening adverse side effects. An analysis by Das et al. shows the highest binding affinity with the TMD domain of monomeric E-protein [
78]. Thus, nafcillin can be considered for redirecting its purpose for the treatment of SARS-CoV-2 infection as it could also combat bacterial co-infection in a COVID patient, which produces the same symptoms as seen in SARS-CoV-2 infection.
4.2. Nabumetone
Nabumetone is an FDA-approved non-selective anti-inflammatory drug (NSAID) that is currently being used for its anti-inflammatory and antipyretic effects. It is a prodrug that goes through biotransformation within the liver to produce the active component, 6-methoxy-2-naphthyl acetic acid (6MNA), that inhibits the synthesis of prostaglandins by acting on cyclooxygenase (COX) I and II. Prostaglandins are responsible for initiating fever by signaling the hypothalamus to increase body temperature. Prostaglandin acts as an inflammatory mediator acting on blood vessels to promote an inflammatory response. NSAIDs mediate anti-inflammatory effects by preventing vasodilation, reducing capillary permeability and cytokine release from endothelial cells. Altogether, these effects impede the migration of immunocompetent cells to the injury site, thereby preventing uncontrolled immune system activation and inflammation [
26].
4.3. Octacosanol
Octacosanol is the main component of plant-extracted natural wax and is a low-molecular-weight primary aliphatic alcohol. Its role is mainly investigated for the treatment of Parkinson’s disease. It is approved as a nutraceutical by the FDA and is marketed as the main component of policosanol (PC), a generic term for a natural mixture of primary alcohols isolated originally from sugarcane wax.
4.4. Cinametic Acid
Cinametic acid is an FDA-approved food additive, mainly obtained from oil of cinnamon and other plant sources. Among the many therapeutic functions of cinnamic acid, one of its roles has also been linked to inhibiting angiotensin-converting enzyme (ACE). ACE converts angiotensin (Ang) I to Ang II. Ang II is responsible for constricting blood vessels and increasing blood pressure or hypertension, one of the risk factors for COVID-19, via binding to angiotensin1 receptor (AT1R) and activating a cascade of signaling pathways. The role of cinametic acid in inhibiting ACE will hamper conversion of Ang I to Ang II, which can reduce hypertension. Further, Ang II gets converted to Ang-(I-VII) by ACE2 in the absence of ACE.
4.5. Ascorbyl Palmitate
Ascorbyl palmitate is an FDA-approved small molecule. Mainly, it is a fat-soluble form of vitamin C formed by the ester of ascorbic acid and palmitic acid. Being an amphipathic molecule, it has the advantage of being more stable and easily enters into cell membranes.
4.6. Guaifenesin
Guaifenesin is an FDA-approved over-the-counter (OTC) or non-prescription expectorant for treatment of cough and the common cold. It aids in the clearance of mucous and other respiratory tract secretion by increasing the volume of trachea and bronchi and reducing mucus viscosity, otherwise leading to congestion, chronic bronchitis, and COPD, commonly seen in ARDS. As a result of this, the action of guaifenesin results in a more productive cough, thus combating the condition of ARDS. This is also expected to happen if administered to COVID-19 patients as it can potentially disrupt the formation of the pentameric structure of E-protein, which causes ARD [
78].
4.7. Remdesivir
Remdesivir, a nucleoside analogue prodrug, was developed for use against the Ebola virus, is currently under trial at many medical institutions, and is known to be effective against MERS-CoV. It has demonstrated a better safety profile than other drugs in treating acute Ebola viral infections. It gets activated into triphosphate, inhibits viral RNA polymerase, and has manifested in vitro and in vivo activity against MERS-CoV and SARS-CoV-2. It effectively treated a severe patient with severe pneumonia who needed mechanical ventilation but not inotropic agents for support of circulation. Findings have been mixed in studies being conducted regarding the efficacy of remdesivir for COVID-19 treatment. A multinational cohort study supported by Gilead Sciences showed clinical improvement for 68% of severe COVID-19 patients treated with compassionate use of remdesivir [
79].
4.8. Molnupiravir
Molnupiravir is an isopropyl ester prodrug, which initially emerged as a possible treatment of influenza viruses, and encephalitic alphaviruses such as Venezuelan, Eastern, and Western equine encephalitic viruses. It is derived from the ribonucleoside analog β-D-N4-hydroxycytidine (NHC) triphosphate that converts to its active form molnupiravir (MTP) in the cell. This drug appears to work by the mechanism of “error catastrophe”; this is essentially the concept that by increasing the rate of mutation in the viral genome beyond a biologically tolerable threshold, the virus will no longer be able to exist. It is a broad-spectrum antiviral drug with a two-step mutagenesis mechanism. It targets the virally encoded RdRp of the SARS-CoV-2 and competitively inhibits the cytidine and uridine triphosphates and incorporates MTP. The RdRp utilizes the NHC triphosphate to incorporate either A or G in the active centers. This, in turn, helps in escaping proofreading of a mutated RNA. The resulting mutagenesis is lethal to the virus [
80].
4.9. Nirmatrelvir
Nirmatrelvir is an irreversible inhibitor of SARS-CoV-2 drug target Mpro. It is co-formulated with ritonavir, allowing an oral route of administration (known as Paxlovid). When treatment is initiated during the first days after symptom onset, it results in roughly 90% protection against severe COVID-19 patients [
81].
4.10. Ganciclovir
Ganciclovir, originally used to treat cytomegalovirus (CMV) infection, has shown effectiveness in a study for COVID-19 treatment at 0.25 g intravenously every 12 h. Several other studies have also found such antiviral drugs to reduce viral load and avert possible respiratory impediments [
82].
Table 1.
A list of drugs that are being repurposed as viable treatment options against drug targets of SARS-CoV2.
Table 1.
A list of drugs that are being repurposed as viable treatment options against drug targets of SARS-CoV2.
| Targets | PDB ID | Repurposed Drugs | References |
|---|
| Envelope (E) protein | 7K3G | Ascorbyl palmitate, cinametic acid, lauric acid, guaifenesin, nabumetone, nafcillin, octacosanol, palmidrol, and salmeterol | [78] |
| Main protease (Mpro). | 7C2Q | Dipyridamole, candesartan cilexetil, candesartan, oxytetracycline, valganciclovir hydrochloride, roxatidine acetate, omeprazole, sulfacetamide, cimetidine, disulfiram, atazanavir, hydroxychloroquine, chloroquine, indinavir montelukast sodium, and maribavir | [83] |
| Membrane (M) protein | 3I6G | Colchicine, remdesivir, bafilomycin A1, temozolomide, and colchicine derivatives | [84] |
| Nucleocapsid (N) protein | 6M3M | Apamycin, camostat, nafamostat, saracatinib, trametinib, cefuroxime, ceftriaxone, cefotaxime | [85] |
| RNA-dependent RNA polymerase (RdRp) | 7D4F | molnupiravir, grazoprevir, ganciclovir, atazanavir, daclatasvir, acyclovir, etravirine, entecavir, efavirenz, asunaprevir, abacavir dolutegravir, lomibuvir, penciclovir, trifluridine, danoprevir, ritonavir, saquinavir, raltegravir, and lamivudine | [86] |
| S-protein | 6VXX | Pemirolast, isoniazid pyruvate, nitrofurantoin, and eriodictyol | [87] |
6. Conclusions
The effectiveness of drug repurposing strategies is determined by whether such agents will compare with virus-specific vaccines or small molecules. A broad-spectrum strategy may have fundamental flaws, much like antibiotics, as more virulent, drug-resistant strains emerge. Drugs like remdesivir and chloroquine have essentially suffered from fallacies against COVID-19 as preclinical assumptions overestimate their clinical efficacy. This shows that even repurposed drugs may require extensive clinical trials. However, a global health emergency of the magnitude of the COVID-19 pandemic calls for a bold medical intervention, where speed is of utmost importance. Drug repurposing cuts down a substantial amount of research time and the effective cost. Such strategies may also uncover the effectiveness of drugs that have otherwise failed to demonstrate their efficacy against the original target.
While efforts are underway to seek new indications for existing compounds, regulatory committees must take rapid action to minimize any hurdles, such as updating drug licensure guidelines through repurposing. Furthermore, it is not just a battle against COVID-19, but it is a revolution for the very concept of antiviral drugs and their clinical implications.Drug repurposing endeavours shall proffer multiple uses of a single drug for multiple diseases, thus preparing humanity for a progressive future.