Effect of SARS-CoV-2 Infection on Renal and Hepatic Function after NSAID and Paracetamol Therapy
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Case–Control Studies
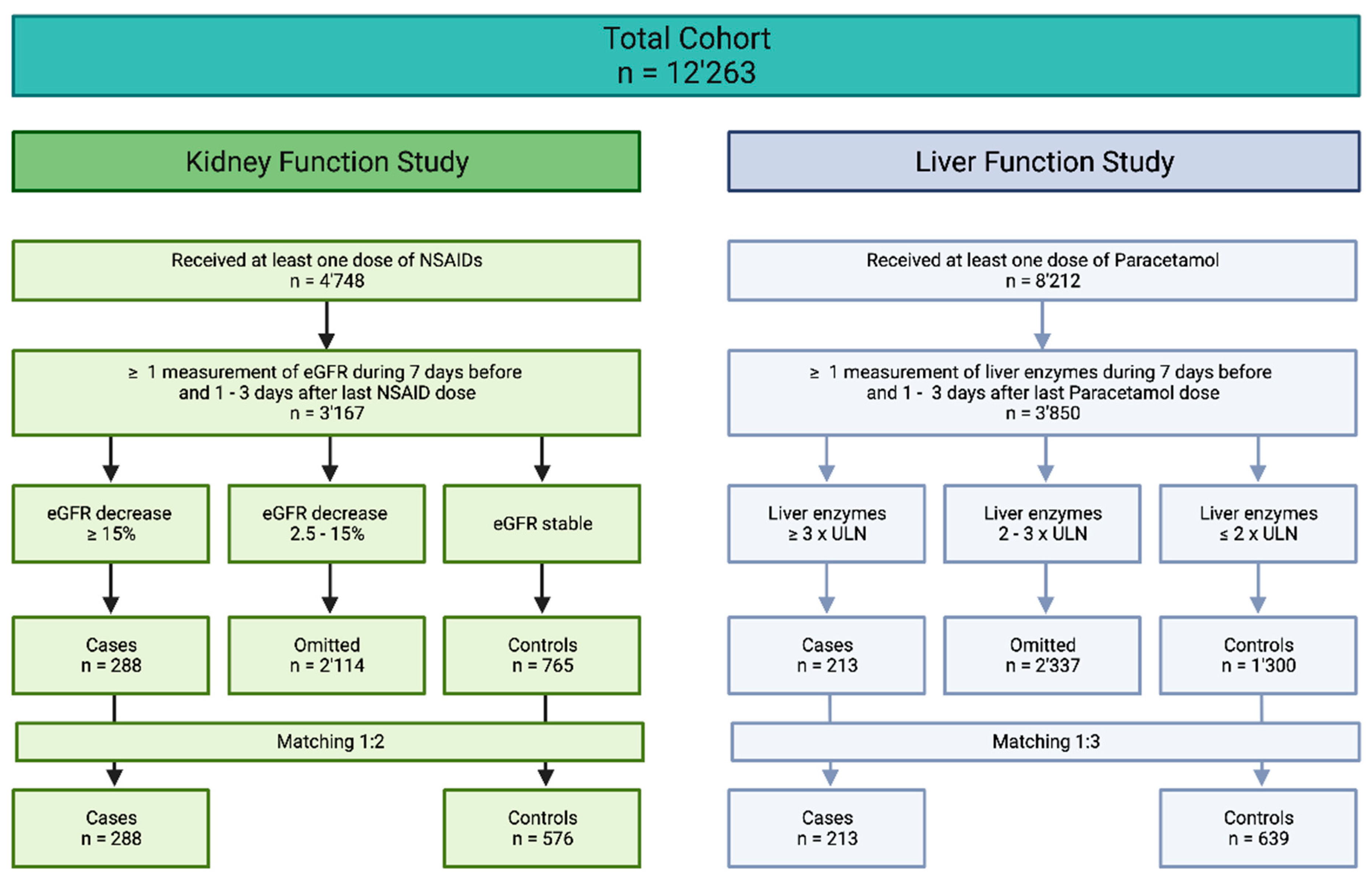
2.2.1. Definitions of Cases and Controls
2.2.2. Data Preparation
2.2.3. Model Building
2.3. Software
3. Results
3.1. Study Population
3.2. Case–Control Study on Renal Function
3.3. Case–Control Study on Hepatic Function
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hörl, W.H. Nonsteroidal Anti-Inflammatory Drugs and the Kidney. Pharmaceuticals 2010, 3, 2291–2321. [Google Scholar] [CrossRef] [PubMed]
- Harris, R.C. COX-2 and the kidney. J. Cardiovasc. Pharmacol. 2006, 47 (Suppl. S1), S37–S42. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Donnan, P.T.; Bell, S.; Guthrie, B. Non-steroidal anti-inflammatory drug induced acute kidney injury in the community dwelling general population and people with chronic kidney disease: Systematic review and meta-analysis. BMC Nephrol. 2017, 18, 256. [Google Scholar] [CrossRef] [PubMed]
- Whelton, A. Nephrotoxicity of nonsteroidal anti-inflammatory drugs: Physiologic foundations and clinical implications. Am. J. Med. 1999, 106, 13s–24s. [Google Scholar] [CrossRef] [PubMed]
- Mazaleuskaya, L.L.; Sangkuhl, K.; Thorn, C.F.; FitzGerald, G.A.; Altman, R.B.; Klein, T.E. PharmGKB summary: Pathways of acetaminophen metabolism at the therapeutic versus toxic doses. Pharmacogenet. Genom. 2015, 25, 416–426. [Google Scholar] [CrossRef] [PubMed]
- Watkins, P.B.; Kaplowitz, N.; Slattery, J.T.; Colonese, C.R.; Colucci, S.V.; Stewart, P.W.; Harris, S.C. Aminotransferase elevations in healthy adults receiving 4 grams of acetaminophen daily: A randomized controlled trial. JAMA 2006, 296, 87–93. [Google Scholar] [CrossRef] [PubMed]
- Pan American Health Organization. Ibuprofen and COVID-19; Pan American Health Organization: Washington, DC, USA, 2020. [Google Scholar]
- Day, M. COVID-19: Ibuprofen should not be used for managing symptoms, say doctors and scientists. BMJ 2020, 368, m1086. [Google Scholar] [CrossRef] [PubMed]
- Moore, N.; Bosco-Levy, P.; Thurin, N.; Blin, P.; Droz-Perroteau, C. NSAIDs and COVID-19: A Systematic Review and Meta-analysis. Drug Saf. 2021, 44, 929–938. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e278. [Google Scholar] [CrossRef]
- Valenzuela, R.; Pedrosa, M.A.; Garrido-Gil, P.; Labandeira, C.M.; Navarro, G.; Franco, R.; Rodriguez-Perez, A.I.; Labandeira-Garcia, J.L. Interactions between ibuprofen, ACE2, renin-angiotensin system, and spike protein in the lung. Implications for COVID-19. Clin. Transl. Med. 2021, 11, e371. [Google Scholar] [CrossRef]
- Kuba, K.; Imai, Y.; Rao, S.; Gao, H.; Guo, F.; Guan, B.; Huan, Y.; Yang, P.; Zhang, Y.; Deng, W.; et al. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus–induced lung injury. Nat. Med. 2005, 11, 875–879. [Google Scholar] [CrossRef]
- Webster, F.; Rice, K.; Sud, A. A critical content analysis of media reporting on opioids: The social construction of an epidemic. Soc. Sci. Med. 2020, 244, 112642. [Google Scholar] [CrossRef]
- Webster, F.; Rice, K.; Katz, J.; Bhattacharyya, O.; Dale, C.; Upshur, R. An ethnography of chronic pain management in primary care: The social organization of physicians’ work in the midst of the opioid crisis. PLoS ONE 2019, 14, e0215148. [Google Scholar] [CrossRef]
- Kushner, P.; McCarberg, B.H.; Grange, L.; Kolosov, A.; Haveric, A.L.; Zucal, V.; Petruschke, R.; Bissonnette, S. The use of non-steroidal anti-inflammatory drugs (NSAIDs) in COVID-19. npj Prim. Care Respir. Med. 2022, 32, 35. [Google Scholar] [CrossRef]
- Drake, T.M.; Fairfield, C.J.; Pius, R.; Knight, S.R.; Norman, L.; Girvan, M.; Hardwick, H.E.; Docherty, A.B.; Thwaites, R.S.; Openshaw, P.J.M.; et al. Non-steroidal anti-inflammatory drug use and outcomes of COVID-19 in the ISARIC Clinical Characterisation Protocol UK cohort: A matched, prospective cohort study. Lancet Rheumatol. 2021, 3, e498–e506. [Google Scholar] [CrossRef] [PubMed]
- Lapi, F.; Marconi, E.; Grattagliano, I.; Rossi, A.; Fornasari, D.; Magni, A.; Lora Aprile, P.; Cricelli, C. To clarify the safety profile of paracetamol for home-care patients with COVID-19: A real-world cohort study, with nested case-control analysis, in primary care. Intern. Emerg. Med. 2022, 17, 2237–2244. [Google Scholar] [CrossRef]
- Trypsteen, W.; Van Cleemput, J.; Snippenberg, W.v.; Gerlo, S.; Vandekerckhove, L. On the whereabouts of SARS-CoV-2 in the human body: A systematic review. PLOS Pathog. 2020, 16, e1009037. [Google Scholar] [CrossRef] [PubMed]
- Leo, M.; Galante, A.; Pagnamenta, A.; Ruinelli, L.; Ponziani, F.R.; Gasbarrini, A.; De Gottardi, A. Hepatocellular liver injury in hospitalized patients affected by COVID-19: Presence of different risk factors at different time points. Dig. Liver Dis. 2022, 54, 565–571. [Google Scholar] [CrossRef] [PubMed]
- Perła-Kaján, J.; Jakubowski, H. COVID-19 and One-Carbon Metabolism. Int. J. Mol. Sci. 2022, 23, 4181. [Google Scholar] [CrossRef]
- Zhou, F.; Yu, T.; Du, R.; Fan, G.; Liu, Y.; Liu, Z.; Xiang, J.; Wang, Y.; Song, B.; Gu, X.; et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: A retrospective cohort study. Lancet 2020, 395, 1054–1062. [Google Scholar] [CrossRef]
- Adams, D. The Hitch Hiker’s Guide to the Galaxy; Pan Books: London, UK, 1979. [Google Scholar]
- Thöni, S.; Keller, F.; Denicolò, S.; Buchwinkler, L.; Mayer, G. Biological variation and reference change value of the estimated glomerular filtration rate in humans: A systematic review and meta-analysis. Front. Med. 2022, 9, 1009358. [Google Scholar] [CrossRef]
- Carobene, A.; Aarsand, A.K.; Bartlett, W.A.; Coskun, A.; Diaz-Garzon, J.; Fernandez-Calle, P.; Guerra, E.; Jonker, N.; Locatelli, M.; Plebani, M.; et al. The European Biological Variation Study (EuBIVAS): A summary report. Clin. Chem. Lab. Med. 2022, 60, 505–517. [Google Scholar] [CrossRef]
- Pugliese, G.; Solini, A.; Bonora, E.; Orsi, E.; Zerbini, G.; Giorgino, F.; Cavalot, F.; Pontiroli, A.E.; Baroni, M.G.; Morano, S.; et al. The Chronic Kidney Disease Epidemiology Collaboration (CKD-EPI) equation provides a better definition of cardiovascular burden associated with CKD than the Modification of Diet in Renal Disease (MDRD) Study formula in subjects with type 2 diabetes. Atherosclerosis 2011, 218, 194–199. [Google Scholar] [CrossRef] [PubMed]
- Aithal, G.P.; Watkins, P.B.; Andrade, R.J.; Larrey, D.; Molokhia, M.; Takikawa, H.; Hunt, C.M.; Wilke, R.A.; Avigan, M.; Kaplowitz, N.; et al. Case definition and phenotype standardization in drug-induced liver injury. Clin. Pharmacol. Ther. 2011, 89, 806–815. [Google Scholar] [CrossRef] [PubMed]
- Ho, D.; Imai, K.; King, G.; Stuart, E.A. MatchIt: Nonparametric Preprocessing for Parametric Causal Inference. J. Stat. Softw. 2011, 42, 1–28. [Google Scholar] [CrossRef]
- Stekhoven, D.J.; Bühlmann, P. MissForest—Non-parametric missing value imputation for mixed-type data. Bioinformatics 2011, 28, 112–118. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Casas, R.; Jones, E.A.; Moreno-Otero, R. Spectrum of anemia associated with chronic liver disease. World J. Gastroenterol. 2009, 15, 4653–4658. [Google Scholar] [CrossRef] [PubMed]
- Peck-Radosavljevic, M. Thrombocytopenia in liver disease. Can. J. Gastroenterol. 2000, 14 (Suppl. D), 60d–66d. [Google Scholar] [CrossRef]
- Giannini, E.G. Review article: Thrombocytopenia in chronic liver disease and pharmacologic treatment options. Aliment. Pharmacol. Ther. 2006, 23, 1055–1065. [Google Scholar] [CrossRef]
- Loomis, A.K.; Kabadi, S.; Preiss, D.; Hyde, C.; Bonato, V.; St Louis, M.; Desai, J.; Gill, J.M.; Welsh, P.; Waterworth, D.; et al. Body Mass Index and Risk of Nonalcoholic Fatty Liver Disease: Two Electronic Health Record Prospective Studies. J. Clin. Endocrinol. Metab. 2016, 101, 945–952. [Google Scholar] [CrossRef]
- Reese, J.T.; Coleman, B.; Chan, L.; Blau, H.; Callahan, T.J.; Cappelletti, L.; Fontana, T.; Bradwell, K.R.; Harris, N.L.; Casiraghi, E.; et al. NSAID use and clinical outcomes in COVID-19 patients: A 38-center retrospective cohort study. Virol. J. 2022, 19, 84. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Dai, Z.; Chen, F.; Gao, S.; Pei, J.; Lai, L. Deep Learning for Drug-Induced Liver Injury. J. Chem. Inf. Model. 2015, 55, 2085–2093. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Ye, T.; Wang, R.; Zhang, C.; Zhang, X.; Sun, G.; Sun, X. An In Silico Model for Predicting Drug-Induced Hepatotoxicity. Int. J. Mol. Sci. 2019, 20, 1897. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Ding, L.; Zou, Y.; Hu, S.Q.; Huang, H.G.; Kong, W.B.; Zhang, J. Predicting drug-induced liver injury in human with Naïve Bayes classifier approach. J. Comput. Aided Mol. Des. 2016, 30, 889–898. [Google Scholar] [CrossRef]
- Wang, M.W.H.; Goodman, J.M.; Allen, T.E.H. Machine Learning in Predictive Toxicology: Recent Applications and Future Directions for Classification Models. Chem. Res. Toxicol. 2021, 34, 217–239. [Google Scholar] [CrossRef]
| Drug | Low Dose | Medium Dose | High Dose | Supratherapeutic |
|---|---|---|---|---|
| Acetylsalicylic acid | ≤300 mg | 300–1000 mg | 1000–4000 mg | >4000 mg |
| Ibuprofen | ≤400 mg | 400–1200 mg | 1200–2400 mg | >2400 mg |
| Diclofenac | ≤50 mg | 50–150 mg | 150–200 mg | >200 mg |
| Naproxen | ≤500 mg | 500–1250 mg | 1250–1500 mg | >1500 mg |
| Coxibe | ≤60 mg | 60–90 mg | 90–400 mg | >400 mg |
| Paracetamol | ≤500 mg | 500–2000 mg | 2000–4000 mg | >4000 mg |
| Feature | Overall (n = 864) | Control (n = 576) | Case (n = 288) | p-Value | |
|---|---|---|---|---|---|
| Min. eGFR [mL/min] mean (SD) [range] | before NSAIDS | 91.53 (11.61) [75.0–148.76] | 90.46 (12.19) [75.0–148.76] | 91.17 (11.81) [75.0–138.93] | 0.211 |
| after NSAIDS | 90.05 (24.47) [6.57–156.43] | 101.32 (15.26) [73.84–156.43] | 59.28 (17.46) [6.57–110.60] | <0.001 * | |
| Sex male (%) | 317 (36.7) | 213 (37.0) | 104 (36.1) | 0.861 | |
| Age mean (SD) | 65.51 (14.30) | 65.0 (14.08) | 66.54 (14.72) | 0.136 | |
| Positive COVID-19 status (%) | 27 (3.1) | 24 (4.2) | 3 (1.0) | 0.023 * | |
| Max. weight [kg] mean (SD) | 77.93 (18.98) | 76.83 (19.39) | 80.02 (18.04) | 0.023 | |
| Mean glucose [mmol/L] mean (SD) | 7.24 (2.18) | 7.50 (2.21) | 7.32 (2.19) | 0.121 | |
| Max. hemoglobin [g/L] mean (SD) | 131.10 (19.36) | 128.88 (21.68) | 130.36 (20.17) | 0.132 | |
| Mean leukocytes [109/L] mean (SD) | 10.47 (9.83) | 9.56 (3.95) | 10.17 (8.35) | 0.135 | |
| Min. INR mean (SD) | 1.09 (0.20) | 1.11 (0.25) | 1.09 (0.22) | 0.298 | |
| Mean CRP [mg/L] mean (SD) | 58.57 (73.41) | 40.20 (61.07) | 52.69 (70.19) | 0.001 * | |
| Mean sodium [mg/L] mean (SD) | 137.11 (7.04) | 136.70 (7.87) | 136.98 (7.33) | 0.441 | |
| Mean potassium [mg/L] mean (SD) | 3.96 (0.35) | 4.00 (0.40) | 3.98 (0.37) | 0.125 | |
| Hypertension (%) | 38 (4.4) | 28 (4.4) | 7 (3.3) | 0.618 | |
| Ischemic heart disease (%) | 18 (2.1) | 14 (2.4) | 4 (1.4) | 0.448 | |
| Pulmonary heart disease (%) | 6 (0.7) | 5 (0.9) | 1 (0.3) | 0.664 | |
| Diseases circulatory system (%) | 11 (1.3) | 6 (1.0) | 5 (1.7) | 0.592 | |
| Diabetes (%) | 13 (1.5) | 8 (1.4) | 5 (1.7) | 0.921 | |
| Obesity (%) | 4 (0.5) | 2 (0.3) | 2 (0.7) | 0.859 | |
| Other virus infections | 0 | 0 | 0 | NA | |
| Feature | Overall (n = 852) | Control (n = 639) | Case (n = 213) | p-Value | |
|---|---|---|---|---|---|
| Max. ALT [IU/L] mean (SD) [range] | Before paracetamol | 26.2 (16.33) [4.0–90.0] | 25.08 (15.55) [4.0–89.0] | 33.09 (19.12) [6.0–90.0] | <0.001 * |
| After paracetamol | 73.29 (270.20) [3.0–8223.0] | 31.7 (19.64) [3.0–90.0] | 327.1 (665.58) [13.0–8223.0] | <0.001 * | |
| Max. ALP [IU/L] mean (SD) [range] | Before paracetamol | 89.39 (33.36) [22.0–200.0] | 88.09 (32.21) [22.0–200] | 97.33 (38.81) [28.0–194.0] | 0.001 * |
| After paracetamol | 131.8 (132.96) [24.0–1784.0] | 97.87 (38.06) [24.0–200.0] | 343.1 (258.95) [41.0–1784.0] | <0.001 * | |
| Sex male (%) | 321 (37.7) | 242 (37.9) | 79 (37.1) | 0.903 | |
| Age mean (SD) | 60.97 (17.26) | 60.94 (17.12) | 60.65 (17.47) | 0.832 | |
| Positive COVID-19 status (%) | 31 (3.6) | 26 (4.1) | 5 (2.3) | 0.342 | |
| Min. BMI [kg/m2] mean (SD) | 25.54 (5.59) | 25.85 (5.60) | 24.61 (5.47) | 0.009 * | |
| Mean glucose [mmol/L] mean (SD) | 7.39 (2.72) | 7.47 (2.85) | 7.16 (2.27) | 0.173 | |
| Max. erythrocytes [g/L] mean (SD) | 4.12 (0.87) | 4.19 (0.85) | 3.91 (0.91) | <0.001 * | |
| Max. thrombocytes [109/L] mean (SD) | 238.37 (160.03) | 247.26 (169.65) | 211.61 (123.31) | 0.005 * | |
| Min. INR mean (SD) | 1.15 (0.29) | 1.15 (0.32) | 1.14 (0.17) | 0.446 | |
| Mean ALT [IU/L] mean (SD) | 27.50 (15.83) | 26.05 (14.53) | 31.88 (18.57) | <0.001 * | |
| Min. ALP [IU/L] mean (SD) | 85.55 (31.60) | 82.92 (28.96) | 93.46 (37.64) | <0.001 * | |
| Mean sodium [mg/L] mean (SD) | 135.58 (10.17) | 135.43 (10.35) | 136.03 (9.62) | 0.455 | |
| Mean potassium [mg/L] mean (SD) | 4.00 (0.52) | 3.98 (0.46) | 4.07 (0.67) | 0.033 * | |
| Max. eGFR [mL/min] mean (SD) | 81.56 (36.18) | 82.4 (32.58) | 78.93 (45.30) | 0.222 | |
| Hypertension (%) | 35 (4.1) | 28 (4.4) | 7 (3.3) | 0.618 | |
| Ischemic heart disease (%) | 15 (1.8) | 12 (1.9) | 3 (1.4) | 0.880 | |
| Pulmonary heart disease (%) | 9 (1.1) | 7 (1.1) | 2 (0.9) | 1.000 | |
| Diseases circulatory system (%) | 18 (2.1) | 14 (2.2) | 4 (1.9) | 1.000 | |
| Diabetes (%) | 17 (2.0) | 9 (1.4) | 8 (3.8) | 0.066 | |
| Obesity (%) | 12 (1.4) | 9 (1.4) | 3 (1.4) | 1.000 | |
| Other virus infections | 1 (0.1) | 0 (0.0) | 1 (0.5) | 0.563 | |
| LogReg | DT | RF | Knn | AdaBoost | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Training | Testing | Training | Testing | Training | Testing | Training | Testing | Training | Testing | |
| Renal function | ||||||||||
| Accuracy (95% CI) | 0.57 (0.53–0.61) | 0.56 (0.48–0.63) | 0.77 (0.74–0.81) | 0.6 (0.52–0.67) | 0.69 (0.66–0.73) | 0.62 (0.55–0.69) | 0.68 (0.64–0.72) | 0.65 (0.57–0.72) | 0.98 (0.96–0.99) | 0.73 (0.65–0.79) |
| Balanced Accuracy | 0.58 | 0.55 | 0.71 | 0.52 | 0.65 | 0.58 | 0.54 | 0.51 | 0.97 | 0.64 |
| Sensitivity | 0.61 | 0.54 | 0.52 | 0.3 | 0.54 | 0.44 | 0.12 | 0.11 | 0.94 | 0.39 |
| Specificity | 0.55 | 0.57 | 0.9 | 0.75 | 0.77 | 0.71 | 0.96 | 0.91 | 1 | 0.9 |
| F1 Score | 0.49 | 0.45 | 0.6 | 0.33 | 0.54 | 0.43 | 0.2 | 0.16 | 0.96 | 0.48 |
| Hepatic function | ||||||||||
| Accuracy (95% CI) | 0.68 (0.65–0.72) | 0.59 (0.51–0.67) | 0.8 (0.77–0.83) | 0.69 (0.62–0.76) | 0.78 (0.74–0.81) | 0.62 (0.54–0.69) | 0.76 (0.73–0.79) | 0.75 (0.68–0.81) | 0.83 (0.8–0.86) | 0.74 (0.67–0.8) |
| Balanced Accuracy | 0.68 | 0.55 | 0.65 | 0.5 | 0.73 | 0.53 | 0.55 | 0.52 | 0.69 | 0.55 |
| Sensitivity | 0.66 | 0.48 | 0.35 | 0.12 | 0.63 | 0.36 | 0.11 | 0.07 | 0.4 | 0.17 |
| Specificity | 0.69 | 0.63 | 0.96 | 0.88 | 0.83 | 0.7 | 0.98 | 0.98 | 0.98 | 0.93 |
| F1 Score | 0.51 | 0.37 | 0.47 | 0.16 | 0.59 | 0.32 | 0.19 | 0.12 | 0.54 | 0.24 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pahud de Mortanges, A.; Liakoni, E.; Schöning, V.; Hammann, F. Effect of SARS-CoV-2 Infection on Renal and Hepatic Function after NSAID and Paracetamol Therapy. COVID 2024, 4, 910-920. https://doi.org/10.3390/covid4070063
Pahud de Mortanges A, Liakoni E, Schöning V, Hammann F. Effect of SARS-CoV-2 Infection on Renal and Hepatic Function after NSAID and Paracetamol Therapy. COVID. 2024; 4(7):910-920. https://doi.org/10.3390/covid4070063
Chicago/Turabian StylePahud de Mortanges, Aurélie, Evangelia Liakoni, Verena Schöning, and Felix Hammann. 2024. "Effect of SARS-CoV-2 Infection on Renal and Hepatic Function after NSAID and Paracetamol Therapy" COVID 4, no. 7: 910-920. https://doi.org/10.3390/covid4070063
APA StylePahud de Mortanges, A., Liakoni, E., Schöning, V., & Hammann, F. (2024). Effect of SARS-CoV-2 Infection on Renal and Hepatic Function after NSAID and Paracetamol Therapy. COVID, 4(7), 910-920. https://doi.org/10.3390/covid4070063






