Microchip Electrophoresis
Definition
:1. Introduction
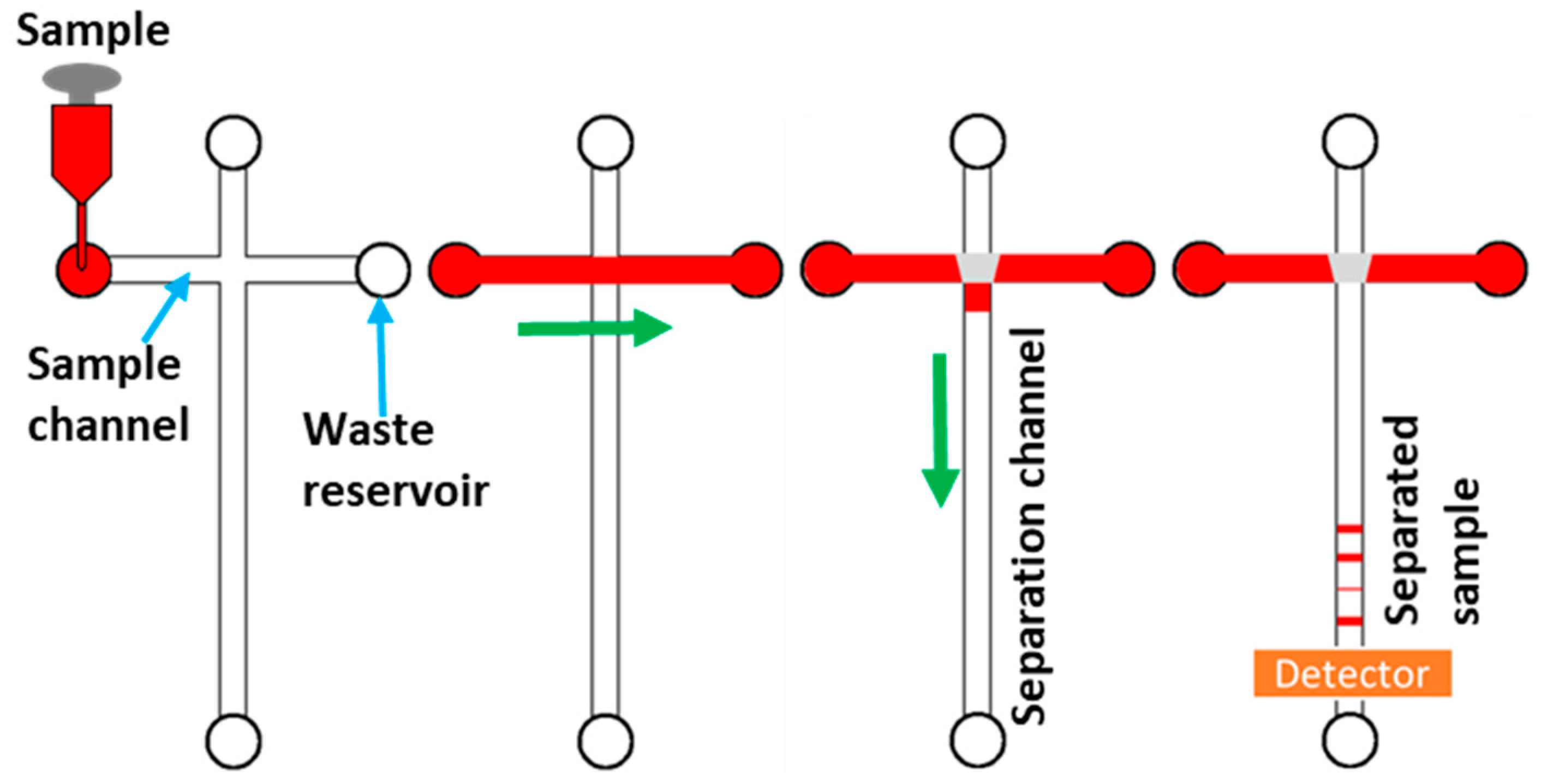
2. Principles of Microchip Electrophoresis
3. Applications of Microchip Electrophoresis
3.1. Microchip Capillary Zone Electrophoresis (Microchip CZE)
3.2. Microchip Gel Electrophoresis
3.3. Microchip Western Blotting (µWB)
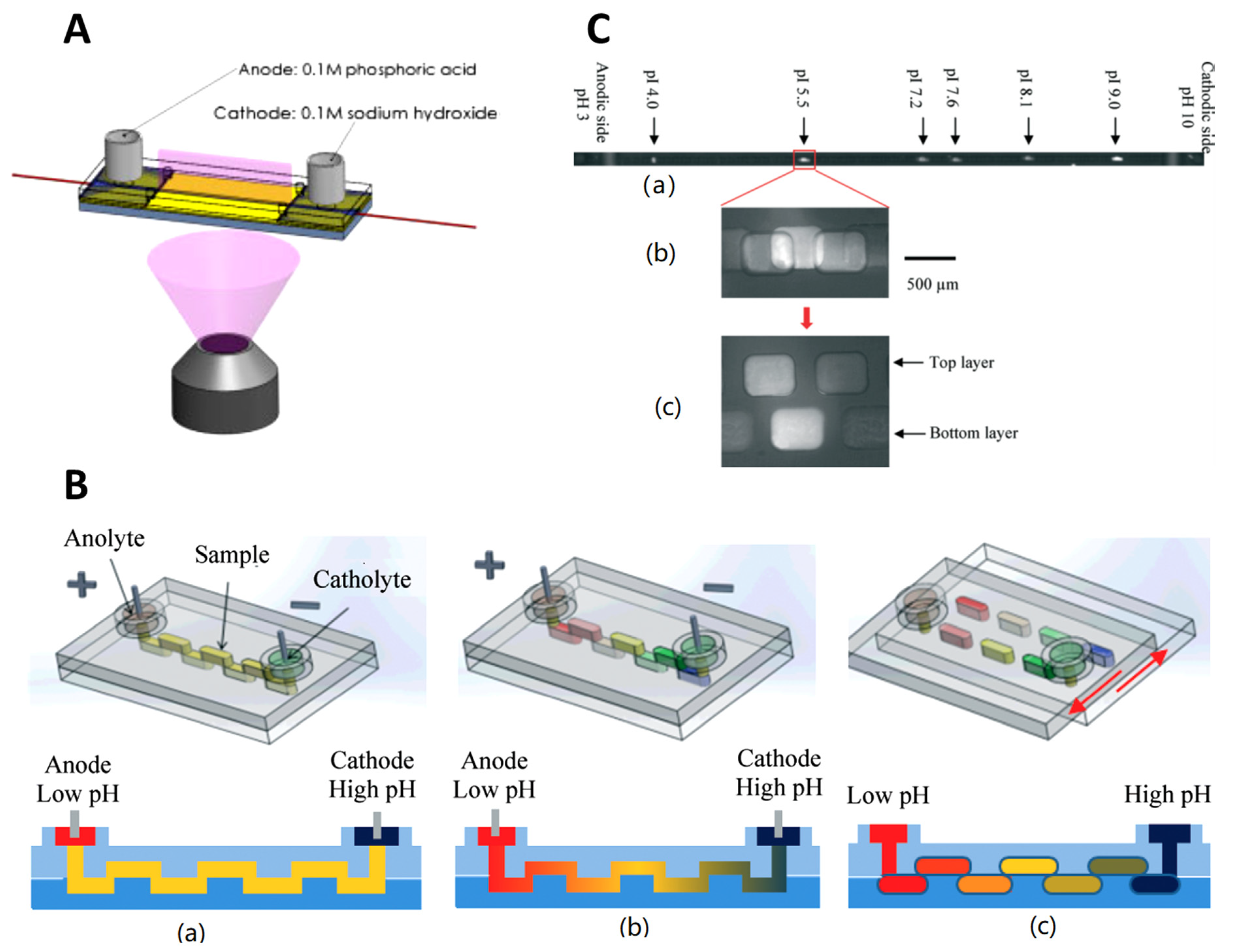
3.4. Microchip Isoelectric Focusing (Microchip IEF)
Funding
Conflicts of Interest
Entry Link on the Encyclopedia Platform
References
- Tiselius, A. A new apparatus for electrophoretic analysis of colloidal mixtures. Trans. Faraday Soc. 1937, 33, 524–531. [Google Scholar] [CrossRef]
- Ostergaard, J.; Jensen, H. Simultaneous evaluation of ligand binding properties and protein size by electrophoresis and Taylor dispersion in capillaries. Anal. Chem. 2009, 81, 8644–8648. [Google Scholar] [CrossRef] [PubMed]
- Migneault, I.; Dartiguenave, C.; Vinh, J.; Bertrand, M.J.; Waldron, K.C. Two Glutaraldehyde-Immobilized Trypsin Preparations for Peptide Mapping by Capillary Zone Electrophoresis, Liquid Chromatography, and Mass Spectrometry. J. Liq. Chromatogr. Relat. Technol. 2008, 31, 789–806. [Google Scholar] [CrossRef]
- Pereira, F.; Hassard, S.; Hassard, J.; deMello, A. CE of dsDNA in low-molecular-weight polyethylene oxide solutions. Electrophoresis 2009, 30, 2100–2109. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.C.; Quesada, M.A.; Mathies, R.A. DNA sequencing using capillary array electrophoresis. Anal. Chem. 2002, 64, 2149–2154. [Google Scholar] [CrossRef]
- Haeberle, S.; Zengerle, R. Microfluidic platforms for lab-on-a-chip applications. Lab Chip 2007, 7, 1094–1110. [Google Scholar] [CrossRef]
- Koutny, L.B.; Schmalzing, D.; Taylor, T.A.; Fuchs, M. Microchip electrophoretic immunoassay for serum cortisol. Anal. Chem. 1996, 68, 18–22. [Google Scholar] [CrossRef]
- Karns, K.; Herr, A.E. Human tear protein analysis enabled by an alkaline microfluidic homogeneous immunoassay. Anal. Chem. 2011, 83, 8115–8122. [Google Scholar] [CrossRef]
- Herr, A.E.; Hatch, A.V.; Throckmorton, D.J.; Tran, H.M.; Brennan, J.S.; Giannobile, W.V.; Singh, A.K. Microfluidic immunoassays as rapid saliva-based clinical diagnostics. Proc. Natl. Acad. Sci. USA 2007, 104, 5268–5273. [Google Scholar] [CrossRef]
- Wang, M.; Roman, G.T.; Perry, M.L.; Kennedy, R.T. Microfluidic chip for high efficiency electrophoretic analysis of segmented flow from a microdialysis probe and in vivo chemical monitoring. Anal. Chem. 2009, 81, 9072–9078. [Google Scholar] [CrossRef]
- Dishinger, J.F.; Reid, K.R.; Kennedy, R.T. Quantitative monitoring of insulin secretion from single islets of Langerhans in parallel on a microfluidic chip. Anal. Chem. 2009, 81, 3119–3127. [Google Scholar] [CrossRef] [PubMed]
- Magdeldin, S. Gel Electrophoresis–Principles and Basics; BoD–Books on Demand: Norderstedt, Germany, 2012. [Google Scholar]
- Gel Electrophoresis: Nucleic Acids (Essential Techniques); Wiley-Blackwell: Hoboken, NJ, USA, 1995.
- Stellwagen, E.; Stellwagen, N.C. Determining the electrophoretic mobility and translational diffusion coefficients of DNA molecules in free solution. Electrophoresis 2002, 23, 2794–2803. [Google Scholar] [CrossRef]
- Lin, S.-K. Physical Biochemistry: Principles and Applications. By David Sheehan. Molecules 2000, 5, 1517. [Google Scholar] [CrossRef]
- Landers, J.P. Handbook of Capillary and Microchip Electrophoresis and Associated Microtechniques, 3rd ed.; CRC Press: Boca Raton, FL, USA, 2007. [Google Scholar]
- Dolník, V. Wall coating for capillary electrophoresis on microchips. Electrophoresis 2004, 25, 3589–3601. [Google Scholar] [CrossRef] [PubMed]
- Dolnik, V.; Liu, S.; Jovanovich, S. Capillary electrophoresis on microchip. Electrophoresis 2000, 21, 41–54. [Google Scholar] [CrossRef]
- Llopis, S.L.; Osiri, J.; Soper, S.A. Surface modification of poly(methyl methacrylate) microfluidic devices for high-resolution separations of single-stranded DNA. Electrophoresis 2007, 28, 984–993. [Google Scholar] [CrossRef]
- Qu, H.; Wang, H.; Huang, Y.; Zhong, W.; Lu, H.; Kong, J.; Yang, P.; Liu, B. Stable microstructured network for protein patterning on a plastic microfluidic channel: Strategy and characterization of on-chip enzyme microreactors. Anal. Chem. 2004, 76, 6426–6433. [Google Scholar] [CrossRef]
- Okada, H.; Kaji, N.; Tokeshi, M.; Baba, Y. Rinse and evaporation coating of poly(methyl methacrylate) microchip for separation of sodium dodecyl sulfate-protein complex. J. Chromatogr. A 2008, 1192, 289–293. [Google Scholar] [CrossRef]
- Currie, C.A.; Shim, J.S.; Lee, S.H.; Ahn, C.; Limbach, P.A.; Halsall, H.B.; Heineman, W.R. Comparing polyelectrolyte multilayer-coated PMMA microfluidic devices and glass microchips for electrophoretic separations. Electrophoresis 2009, 30, 4245–4250. [Google Scholar] [CrossRef]
- Ermakov, S.V.; Jacobson, S.C.; Ramsey, J.M. Computer simulations of electrokinetic injection techniques in microfluidic devices. Anal. Chem. 2000, 72, 3512–3517. [Google Scholar] [CrossRef]
- Harrison, D.J.; Manz, A.; Fan, Z.; Luedi, H.; Widmer, H.M. Capillary electrophoresis and sample injection systems integrated on a planar glass chip. Anal. Chem. 1992, 64, 1926–1932. [Google Scholar] [CrossRef]
- Hardt, S.; Schönfeld, F. Microfluidic Technologies for Miniaturized Analysis Systems; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2007. [Google Scholar]
- Giddings, J.C. Unified Separation Science; John Wiley Sons: New York, NY, USA, 1991; pp. 97–101. [Google Scholar]
- Dolnik, V.; Liu, S. Applications of capillary electrophoresis on microchip. J. Sep. Sci. 2005, 28, 1994–2009. [Google Scholar] [CrossRef]
- Novotny, M.; Springston, S.R.; Peaden, P.A.; Fjeldsted, J.C.; Lee, M.L. Capillary supercritical fluid chromatography. Anal. Chem. 1981, 53, 407A–414A. [Google Scholar] [CrossRef]
- Lauer, H.H.; McManigill, D. Capillary zone electrophoresis of proteins in untreated fused silica tubing. Anal. Chem. 1986, 58, 166–170. [Google Scholar] [CrossRef]
- Li, Y.; Champion, M.M.; Sun, L.; Champion, P.A.D.; Wojcik, R.; Dovichi, N.J. Capillary Zone Electrophoresis-Electrospray Ionization-Tandem Mass Spectrometry as an Alternative Proteomics Platform to Ultraperformance Liquid Chromatography-Electrospray Ionization-Tandem Mass Spectrometry for Samples of Intermediate Complexity. Anal. Chem. 2012, 84, 1617–1622. [Google Scholar] [CrossRef] [PubMed]
- Stewart, D.T.; Celiz, M.D.; Vicente, G.; Colón, L.A.; Aga, D.S. Potential use of capillary zone electrophoresis in size characterization of quantum dots for environmental studies. TrAC-Trends Anal. Chem. 2011, 30, 113–122. [Google Scholar] [CrossRef]
- Joneli, J.; Wanzenried, U.; Schiess, J.; Lanz, C.; Caslavska, J.; Thormann, W. Determination of carbohydrate-deficient transferrin in human serum by capillary zone electrophoresis: Evaluation of assay performance and quality assurance over a 10-year period in the routine arena. Electrophoresis 2013, 34, 1563–1571. [Google Scholar] [CrossRef]
- Colyer, C.L.; Mangru, S.D.; Harrison, D.J. Microchip-based capillary electrophoresis of human serum proteins. J. Chromatogr. A 1997, 781, 271–276. [Google Scholar] [CrossRef]
- Hayes, M.A.; Kheterpal, I.; Ewing, A.G. Electroosmotic flow control and surface conductance in capillary zone electrophoresis. Anal. Chem. 1993, 65, 2010–2013. [Google Scholar] [CrossRef]
- Mitra, I.; Marczak, S.P.; Jacobson, S.C. Microchip electrophoresis at elevated temperatures and high separation field strengths. Electrophoresis 2014, 35, 374–378. [Google Scholar] [CrossRef]
- Ye, F.; Shi, M.; Huang, Y.; Zhao, S. Noncompetitive immunoassay for carcinoembryonic antigen in human serum by microchip electrophoresis for cancer diagnosis. Clin. Chim. Acta Int. J. Clin. Chem. 2010, 411, 1058–1062. [Google Scholar] [CrossRef]
- Cianciulli, C.; Hahne, T.; Wätzig, H. Capillary gel electrophoresis for precise protein quantitation. Electrophoresis 2012, 33, 3276–3280. [Google Scholar] [CrossRef] [PubMed]
- Bousse, L.; Mouradian, S.; Minalla, A.; Yee, H.; Williams, K.; Dubrow, R. Protein sizing on a microchip. Anal. Chem. 2001, 73, 1207–1212. [Google Scholar] [CrossRef] [PubMed]
- Root, B.E.; Zhang, B.; Barron, A.E. Size-based protein separations by microchip electrophoresis using an acid-labile surfactant as a replacement for SDS. Electrophoresis 2009, 30, 2117–2122. [Google Scholar] [CrossRef] [PubMed]
- Shadpour, H.; Soper, S.A. Two-dimensional electrophoretic separation of proteins using poly(methyl methacrylate) microchips. Anal. Chem. 2006, 78, 3519–3527. [Google Scholar] [CrossRef]
- Shen, Z.; Liu, X.; Long, Z.; Liu, D.; Ye, N.; Qin, J.; Dai, Z.; Lin, B. Parallel analysis of biomolecules on a microfabricated capillary array chip. Electrophoresis 2006, 27, 1084–1092. [Google Scholar] [CrossRef]
- Lee, P.Y.; Costumbrado, J.; Hsu, C.-Y.; Kim, Y.H. Agarose gel electrophoresis for the separation of DNA fragments. J. Vis. Exp. 2012, e3923. [Google Scholar] [CrossRef]
- Kurien, B.T.; Scofield, R.H. Western blotting. Methods 2006, 38, 283–293. [Google Scholar] [CrossRef]
- Herr, A.E.; Singh, A.K. Photopolymerized cross-linked polycrylamide gels for on-chip protein sizing. Anal. Chem. 2004, 76, 4727–4733. [Google Scholar] [CrossRef]
- Kirby, B.J.; Wheeler, A.R.; Zare, R.N.; Fruetel, J.A.; Shepodd, T.J. Programmable modification of cell adhesion and zeta potential in silica microchips. Lab Chip 2003, 3, 5–10. [Google Scholar] [CrossRef]
- Hughes, A.J.; Lin, R.K.C.; Peehl, D.M.; Herr, A.E. Microfluidic integration for automated targeted proteomic assays. Proc. Natl. Acad. Sci. USA 2012, 109, 5972–5977. [Google Scholar] [CrossRef] [PubMed]
- Duncombe, T.A.; Herr, A.E. Photopatterned free-standing polyacrylamide gels for microfluidic protein electrophoresis. Lab Chip 2013, 13, 2115–2123. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Singh, A.K. Rapid protein separations in ultra-short microchannels: Microchip sodium dodecyl sulfate-polyacrylamide gel electrophoresis and isoelectric focusing. J. Chromatogr. A 2004, 1049, 205–209. [Google Scholar] [CrossRef]
- Lo, C.T.; Throckmorton, D.J.; Singh, A.K.; Herr, A.E. Photopolymerized diffusion-defined polyacrylamide gradient gels for on-chip protein sizing. Lab Chip 2008, 8, 1273–1279. [Google Scholar] [CrossRef] [PubMed]
- Hou, C.; Herr, A.E. Ultrashort separation length homogeneous electrophoretic immunoassays using on-chip discontinuous polyacrylamide gels. Anal. Chem. 2010, 82, 3343–3351. [Google Scholar] [CrossRef] [PubMed]
- Hassan, S.U.; Morgan, H.; Zhang, X.; Niu, X. Droplet Interfaced Parallel and Quantitative Microfluidic-Based Separations. Anal. Chem. 2015, 87, 3895–3901. [Google Scholar] [CrossRef]
- Nan, H.; Yoo, D.J.; Kang, S.H. Fast parallel detection of feline panleukopenia virus DNA by multi-channel microchip electrophoresis with programmed step electric field strength. J. Sep. Sci. 2013, 36, 350–355. [Google Scholar] [CrossRef]
- Aborn, J.H.; El-Difrawy, S.A.; Novotny, M.; Gismondi, E.A.; Lam, R.; Matsudaira, P.; Mckenna, B.K.; O’Neil, T.; Streechon, P.; Ehrlich, D.J. A 768-lane microfabricated system for high-throughput DNA sequencing. Lab Chip 2005, 5, 669–674. [Google Scholar] [CrossRef]
- Hames, B.D. An Introduction to Polyacrylamide Gel Electrophoresis. Gel Electrophoresis of Proteins: A Practical Approach, 3rd ed.; Oxford University Press: New York, NY, USA, 1988. [Google Scholar]
- Pan, W.; Chen, W.; Jiang, X. Microfluidic western blot. Anal. Chem. 2010, 82, 3974–3976. [Google Scholar] [CrossRef]
- Jin, S.; Anderson, G.J.; Kennedy, R.T. Western blotting using microchip electrophoresis interfaced to a protein capture membrane. Anal. Chem. 2013, 85, 6073–6079. [Google Scholar] [CrossRef]
- Tia, S.Q.; He, M.; Kim, D.; Herr, A.E. Multianalyte on-chip native western blotting. Anal. Chem. 2011, 83, 3581–3588. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Diaz, R.; Wehr, T.; Zhu, M. Capillary isoelectric focusing. Electrophoresis 1997, 18, 2134–2144. [Google Scholar] [CrossRef]
- Castro, E.R.; Manz, A. Present state of microchip electrophoresis: State of the art and routine applications. J. Chromatogr. A 2015, 1382, 66–85. [Google Scholar] [CrossRef] [PubMed]
- Mao, Q.; Pawliszyn, J. Demonstration of isoelectric focusing on an etched quartz chip with UV absorption imaging detection. Analyst 1999, 124, 637–641. [Google Scholar] [CrossRef]
- Shameli, S.M.; Elbuken, C.; Ou, J.; Ren, C.L.; Pawliszyn, J. Fully integrated PDMS/SU-8/quartz microfluidic chip with a novel macroporous poly dimethylsiloxane (PDMS) membrane for isoelectric focusing of proteins using whole-channel imaging detection. Electrophoresis 2011, 32, 333–339. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Pereira, F.; deMello, A.J.; Morgan, H.; Niu, X. Droplet-based in situ compartmentalization of chemically separated components after isoelectric focusing in a Slipchip. Lab Chip 2014, 14, 555–561. [Google Scholar] [CrossRef] [PubMed]
- Walowski, B.; Hüttner, W.; Wackerbarth, H. Generation of a miniaturized free-flow electrophoresis chip based on a multi-lamination technique-isoelectric focusing of proteins and a single-stranded DNA fragment. Anal. Bioanal. Chem. 2011, 401, 2465–2471. [Google Scholar] [CrossRef]
- Ishibashi, R.; Kitamori, T.; Shimura, K. Two-step perpendicular free-solution isoelectric focusing in a microchamber array chip. Lab Chip 2010, 10, 2628–2631. [Google Scholar] [CrossRef]
- O’Farrell, P.H. High resolution two-dimensional electrophoresis of proteins. J. Biol. Chem. 1975, 250, 4007–4021. [Google Scholar]
- Mackintosh, J.A.; Choi, H.Y.; Bae, S.H.; Veal, D.A.; Bell, P.J.; Ferrari, B.C.; Van Dyk, D.D.; Verrills, N.M.; Paik, Y.K.; Karuso, P. A fluorescent natural product for ultra sensitive detection of proteins in one-dimensional and two-dimensional gel electrophoresis. Proteomics 2003, 3, 2273–2288. [Google Scholar] [CrossRef]
- Cid, C.; Garcia-Descalzo, L.; Casado-Lafuente, V.; Amils, R.; Aguilera, A. Proteomic analysis of the response of an acidophilic strain of Chlamydomonas sp. (Chlorophyta) to natural metal-rich water. Proteomics 2010, 10, 2026–2036. [Google Scholar] [CrossRef] [PubMed]
- Gu, P.; Liu, K.; Chen, H.; Nishida, T.; Fan, Z.H. Chemical-Assisted Bonding of Thermoplastics/Elastomer for Fabricating Microfluidic Valves. Anal. Chem. 2011, 83, 446–452. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Liu, J.; Lee, C.S.; DeVoe, D.L. Microfluidic 2-D PAGE using multifunctional in situ polyacrylamide gels and discontinuous buffers. Lab Chip 2009, 9, 592–599. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hassan, S.-u. Microchip Electrophoresis. Encyclopedia 2021, 1, 30-41. https://doi.org/10.3390/encyclopedia1010006
Hassan S-u. Microchip Electrophoresis. Encyclopedia. 2021; 1(1):30-41. https://doi.org/10.3390/encyclopedia1010006
Chicago/Turabian StyleHassan, Sammer-ul. 2021. "Microchip Electrophoresis" Encyclopedia 1, no. 1: 30-41. https://doi.org/10.3390/encyclopedia1010006
APA StyleHassan, S. -u. (2021). Microchip Electrophoresis. Encyclopedia, 1(1), 30-41. https://doi.org/10.3390/encyclopedia1010006





