Improving and Externally Validating Mortality Prediction Models for COVID-19 Using Publicly Available Data
Abstract
:1. Introduction
2. Materials and Methods
2.1. Systematic Search for Publicly Available Data
2.2. Statistical Analysis
2.3. Missing Data
2.4. Univariate Analysis
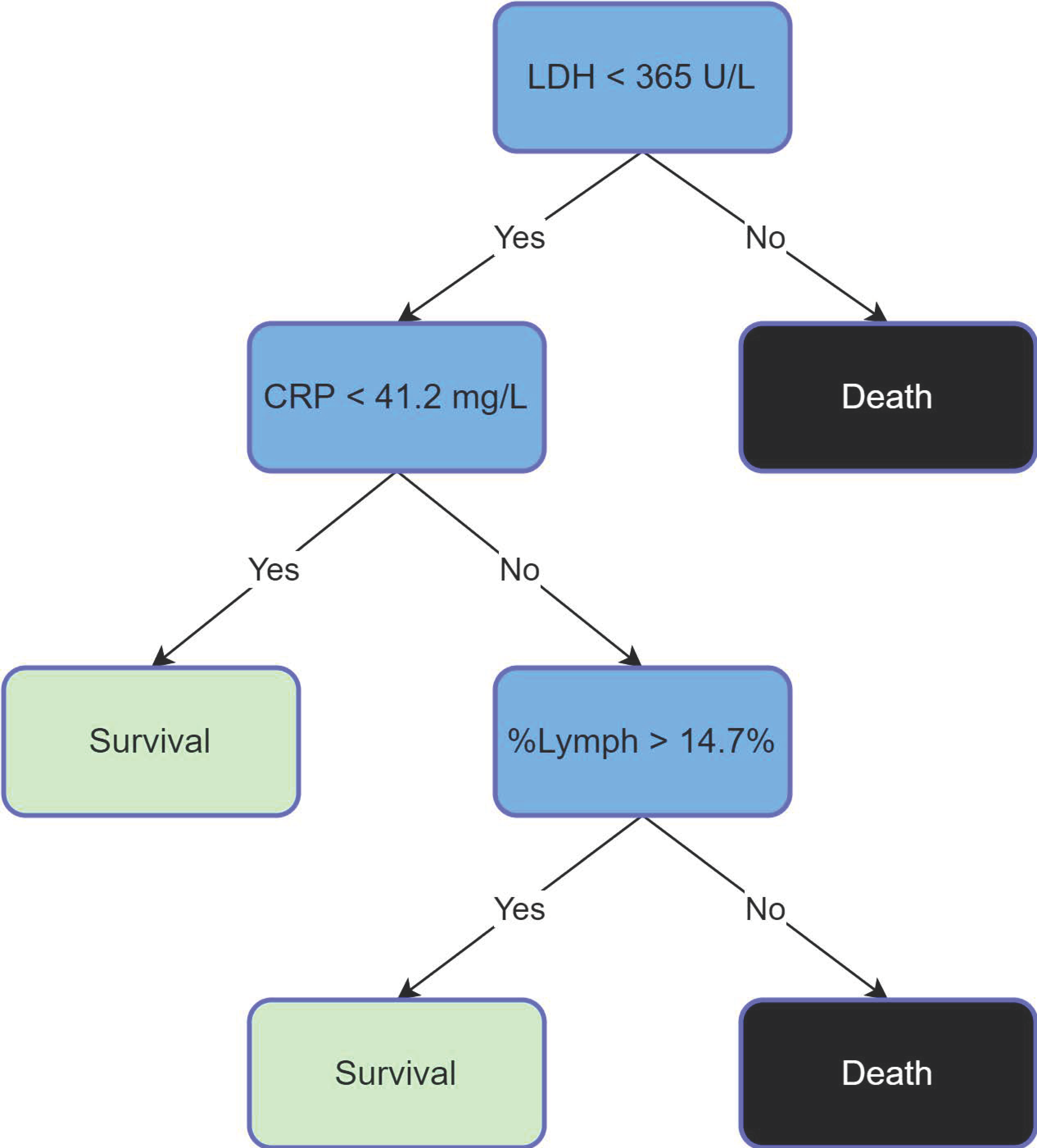
2.5. Mortality Prediction Model Building
2.5.1. Minimal Model (MM)
2.5.2. Generalizable Model (GM)
2.5.3. Model Comparison
3. Results
3.1. Systematic Search for Publicly Available Data
3.2. Patient Characteristics
3.3. Univariate Analysis
3.4. Model Building and Comparison
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- WHO Coronavirus (COVID-19) Dashboard. Available online: https://covid19.who.int/ (accessed on 21 September 2021).
- Impact of COVID-19 on People’s Livelihoods, Their Health and Our Food Systems. Available online: https://www.who.int/news/item/13-10-2020-impact-of-covid-19-on-people’s-livelihoods-their-health-and-our-food-systems (accessed on 21 September 2021).
- Saladino, V.; Algeri, D.; Auriemma, V. The psychological and social impact of Covid-19: New perspectives of well-being. Front. Psychol. 2020, 11, 2550. [Google Scholar] [CrossRef] [PubMed]
- Cascella, M.; Rajnik, M.; Aleem, A.; Dulebohn, S.; Di Napoli, R. Features, evaluation, and treatment of coronavirus (COVID-19). StatPearls 2021. Available online: https://www.statpearls.com/ArticleLibrary/viewarticle/52171 (accessed on 21 September 2021).
- Heustess, A.M.; Allard, M.A.; Thompson, D.K.; Fasinu, P.S. Clinical Management of COVID-19: A Review of Pharmacological Treatment Options. Pharmaceuticals 2021, 14, 520. [Google Scholar] [CrossRef] [PubMed]
- Wynants, L.; Van Calster, B.; Collins, G.S.; Riley, R.D.; Heinze, G.; Schuit, E.; Bonten, M.M.; Dahly, D.L.; Damen, J.A.; Debray, T.P.; et al. Prediction models for diagnosis and prognosis of COVID-19: Systematic review and critical appraisal. BMJ 2020, 369, m1328. [Google Scholar] [CrossRef] [Green Version]
- Knight, S.R.; Ho, A.; Pius, R.; Buchan, I.; Carson, G.; Drake, T.M.; Dunning, J.; Fairfield, C.J.; Gamble, C.; Green, C.A.; et al. Risk stratification of patients admitted to hospital with covid-19 using the ISARIC WHO Clinical Characterisation Protocol: Development and validation of the 4C Mortality Score. BMJ 2020, 370, m3339. [Google Scholar] [CrossRef] [PubMed]
- Bonanad, C.; García-Blas, S.; Tarazona-Santabalbina, F.; Sanchis, J.; Bertomeu-González, V.; Fácila, L.; Ariza, A.; Núñez, J.; Cordero, A. The effect of age on mortality in patients with COVID-19: A meta-analysis with 611,583 subjects. J. Am. Med. Dir. Assoc. 2020, 21, 915–918. [Google Scholar] [CrossRef]
- Chatterjee, A.; Wu, G.; Primakov, S.; Oberije, C.; Woodruff, H.; Kubben, P.; Henry, R.; Aries, M.J.; Beudel, M.; Noordzij, P.G.; et al. Can predicting COVID-19 mortality in a European cohort using only demographic and comorbidity data surpass age-based prediction: An externally validated study. PLoS ONE 2021, 16, e0249920. [Google Scholar] [CrossRef]
- Magro, B.; Zuccaro, V.; Novelli, L.; Zileri, L.; Celsa, C.; Raimondi, F.; Gori, M.; Cammà, G.; Battaglia, S.; Genova, V.G.; et al. Predicting in-hospital mortality from Coronavirus Disease 2019: A simple validated app for clinical use. PLoS ONE 2021, 16, e0245281. [Google Scholar] [CrossRef]
- Stekhoven, D.J.; Bühlmann, P. MissForest—non-parametric missing value imputation for mixed-type data. Bioinformatics 2012, 28, 112–118. [Google Scholar] [CrossRef] [Green Version]
- Waljee, A.K.; Mukherjee, A.; Singal, A.G.; Zhang, Y.; Warren, J.; Balis, U.; Marrero, J.; Zhu, J.; Higgins, P.D. Comparison of imputation methods for missing laboratory data in medicine. BMJ Open 2013, 3, e002847. [Google Scholar] [CrossRef]
- Chatterjee, A.; Woodruff, H.; Wu, G.; Lambin, P. Limitations of Only Reporting the Odds Ratio in the Age of Precision Medicine: A Deterministic Simulation Study. Front. Med. 2021, 8, 640854. [Google Scholar] [CrossRef] [PubMed]
- Fawcett, T. ROC graphs: Notes and practical considerations for researchers. Mach. Learn. 2004, 31, 1–38. [Google Scholar]
- De Long, E.R.; DeLong, D.M.; Clarke-Pearson, D.L. Comparing the areas under two or more correlated receiver operating characteristic curves: A nonparametric approach. Biometrics 1988, 44, 837–845. [Google Scholar] [CrossRef]
- Yan, L.; Zhang, H.T.; Goncalves, J.; Xiao, Y.; Wang, M.; Guo, Y.; Sun, C.; Tang, X.; Jing, L.; Zhang, M.; et al. An interpretable mortality prediction model for COVID-19 patients. Nat. Mach. Intell. 2020, 2, 283–288. [Google Scholar] [CrossRef]
- Quanjel, M.J.; Van Holten, T.C.; Gunst-van der Vliet, P.C.; Wielaard, J.; Karakaya, B.; Söhne, M.; Moeniralam, H.S.; Grutters, J.C. Replication of a mortality prediction model in Dutch patients with COVID-19. Nat. Mach. Intell. 2021, 3, 23–24. [Google Scholar] [CrossRef]
- Dupuis, C.; De Montmollin, E.; Neuville, M.; Mourvillier, B.; Ruckly, S.; Timsit, J.F. Limited applicability of a COVID-19 specific mortality prediction rule to the intensive care setting. Nat. Mach. Intell. 2021, 3, 20–22. [Google Scholar] [CrossRef]
- Wilkinson, M.D.; Dumontier, M.; Aalbersberg, I.J.; Appleton, G.; Axton, M.; Baak, A.; Blomberg, N.; Boiten, J.W.; Da Silva Santos, L.B.; Bourne, P.E.; et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data 2016, 3, 1–9. [Google Scholar] [CrossRef] [Green Version]
- The Cancer Imaging Archive. Available online: https://www.cancerimagingarchive.net/ (accessed on 21 September 2021).
- Levin, A.T.; Hanage, W.P.; Owusu-Boaitey, N.; Cochran, K.B.; Walsh, S.P.; Meyerowitz-Katz, G. Assessing the age specificity of infection fatality rates for COVID-19: Systematic review, meta-analysis, and public policy implications. Eur. J. Epidemiol. 2020, 35, 1123–1138. [Google Scholar] [CrossRef]
- Liu, Y.; Mao, B.; Liang, S.; Yang, J.W.; Lu, H.W.; Chai, Y.H.; Wang, L.; Zhang, L.; Li, Q.H.; Zhao, L.; et al. Association between age and clinical characteristics and outcomes of COVID-19. Eur. Respir. J. 2020, 55, 2001112. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Hou, J.; Ma, F.Z.; Li, J.; Xue, S.; Xu, Z.G. The common risk factors for progression and mortality in COVID-19 patients: A meta-analysis. Arch. Virol. 2021, 166, 2071–2087. [Google Scholar] [CrossRef]
- Dai, Z.; Zeng, D.; Cui, D.; Wang, D.; Feng, Y.; Shi, Y.; Zhao, L.; Xu, J.; Guo, W.; Yang, Y.; et al. Prediction of COVID-19 patients at high risk of progression to severe disease. Front. Public Health 2020, 8, 574915. [Google Scholar] [CrossRef]
- Peckham, H.; De Gruijter, N.M.; Raine, C.; Radziszewska, A.; Ciurtin, C.; Wedderburn, L.R.; Rosser, E.C.; Webb, K.; Deakin, C.T. Male sex identified by global COVID-19 meta-analysis as a risk factor for death and ITU admission. Nat. Commun. 2020, 11, 6317. [Google Scholar] [CrossRef]
- Kelada, M.; Anto, A.; Dave, K.; Saleh, S.N. The role of sex in the risk of mortality from COVID-19 amongst adult patients: A systematic review. Cureus 2020, 12, e10114. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Ye, J.; Chen, Q.; Hu, W.; Wang, L.; Fan, Y.; Lu, Z.; Chen, J.; Chen, Z.; Chen, S.; et al. Elevated lactate dehydrogenase (LDH) level as an independent risk factor for the severity and mortality of COVID-19. Aging (Albany NY) 2020, 12, 15670. [Google Scholar] [CrossRef]
- Han, Y.; Zhang, H.; Mu, S.; Wei, W.; Jin, C.; Tong, C.; Song, Z.; Zha, Y.; Xue, Y.; Gu, G. Lactate dehydrogenase, an independent risk factor of severe COVID-19 patients: A retrospective and observational study. Aging (Albany NY) 2020, 12, 11245. [Google Scholar] [CrossRef]
- Zhao, Q.; Meng, M.; Kumar, R.; Wu, Y.; Huang, J.; Deng, Y.; Weng, Z.; Yang, L. Lymphopenia is associated with severe coronavirus disease 2019 (COVID-19) infections: A systemic review and meta-analysis. Int. J. Infect. Dis. 2020, 96, 131–135. [Google Scholar] [CrossRef] [PubMed]
- Somasekar, J.; Kumar, P.P.; Sharma, A.; Ramesh, G. Machine learning and image analysis applications in the fight against COVID-19 pandemic: Datasets, research directions, challenges and opportunities. Mater. Today Proc. 2020. Available online: https://www.sciencedirect.com/science/article/pii/S2214785320370620 (accessed on 21 October 2021).
- Noor, F.M.; Islam, M.M. Prevalence and associated risk factors of mortality among COVID-19 patients: A meta-analysis. J. Community Health 2020, 45, 1270–1282. [Google Scholar] [CrossRef]
- Najera, H.; Ortega-Avila, A.G. Health and Institutional Risk Factors of COVID-19 Mortality in Mexico, 2020. Am. J. Prev. Med. 2021, 60, 471–477. [Google Scholar] [CrossRef]
- Halilaj, I.; Chatterjee, A.; Van Wijk, Y.; Wu, G.; Van Eeckhout, B.; Oberije, C.; Lambin, P. Covid19Risk.ai: An Open Source Repository and Online Calculator of Prediction Models for Early Diagnosis and Prognosis of COVID-19. BioMed 2021, 1, 41–49. [Google Scholar] [CrossRef]
- Collins, G.S.; Reitsma, J.B.; Altman, D.G.; Moons, K.G. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD) the TRIPOD statement. Circulation 2015, 131, 211–219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhaskaran, K.; Smeeth, L. What is the difference between missing completely at random and missing at random? Int. J. Epidemiol. 2014, 43, 1336–1339. [Google Scholar] [CrossRef] [PubMed]


| Mortality Group | Missing Values | Non-Mortality Group | Missing Values | p-Value | |
| Total | 372 (27%) | NA | 987 (73%) | NA | NA |
| Age | 76 (72, 82) | 0 | 63 (53, 73) | 0 | <0.01 |
| Male | 271 (72.8%) | 0 | 682 (69.1%) | 0 | 0.18 |
| Diabetes | 83 (23.3%) | 16 (4.3%) | 141 (14.8%) | 36 (3.6%) | <0.01 |
| COPD | 29 (8.1%) | 16 (4.3%) | 49 (5.2%) | 36 (3.6%) | 0.05 |
| Tumor | 17 (4.8%) | 16 (4.3%) | 30 (3.2%) | 37 (3.7%) | 0.18 |
| CHD | 48 (13.5%) | 17 (4.6%) | 59 (6.2%) | 36 (3.6%) | <0.01 |
| CLD | 14 (3.9%) | 17 (4.6%) | 17 (1.8%) | 37 (3.7%) | 0.04 |
| GPT, U/L | 37 (24, 57) | 118 (32%) | 36 (24, 60) | 342 (35%) | 0.98 |
| CRP, mg/dL | 13 (9, 18) | 88 (24%) | 8 (4, 15) | 265 (27%) | <0.01 |
| LDH, U/L | 438 (345, 587) | 31 (8%) | 365 (291, 486) | 123 (12%) | <0.01 |
| Platelets, ×109 per L | 180 (126, 234) | 52 (14%) | 191 (135, 262) | 128 (13%) | 0.02 |
| Mortality Group | Non-Mortality Group | p-Value | |||
| Total | 372 (27%) | 987 (73%) | NA | ||
| Age, years | 76 (72, 82) | 63 (53, 73) | <0.01 | ||
| Male | 271 (72.8%) | 682 (69.1%) | 0.18 | ||
| Diabetes | 85 (22.8%) | 141 (14.3%) | <0.01 | ||
| COPD | 29 (7.8%) | 49 (5.0%) | 0.05 | ||
| Tumor | 17 (4.6%) | 30 (3.0%) | 0.18 | ||
| CHD | 49 (13.2%) | 59 (6.0%) | <0.01 | ||
| CLD | 14 (3.8%) | 17 (1.7%) | 0.04 | ||
| GPT, U/L | 42 (30, 59) | 41 (29, 61) | 0.48 | ||
| CRP, mg/dL | 14 (10, 18) | 10 (5, 15) | <0.01 | ||
| LDH, U/L | 462 (356, 609) | 372 (299, 500) | <0.01 | ||
| Platelets, ×109 per L | 182 (135, 229) | 191 (141, 252) | 0.02 | ||
| Italian Dataset [10] | Mortality Group | Non-Mortality Group | p-Value |
| Total | 41 (12%) | 315 (88%) | NA |
| Age, years | 84 (78, 88) | 66 (54, 76) | <0.01 |
| Male | 27 (65.9%) | 200 (63.5%) | 0.86 |
| Diabetes | 14 (34.1%) | 46 (14.6%) | <0.01 |
| COPD | 9 (22.0%) | 33 (10.5%) | 0.04 |
| Tumor | 8 (19.5%) | 22 (7.0%) | 0.01 |
| CHD | 11 (26.8%) | 29 (9.2%) | <0.01 |
| CLD | 1 (2.4%) | 2 (0.6%) | 0.31 |
| GPT, U/L | 18 (14, 35) | 29 (16, 49) | 0.03 |
| CRP, mg/dL | 14 (9, 19) | 7 (3, 15) | <0.01 |
| LDH, U/L | 370 (300, 460) | 290 (235, 402) | <0.01 |
| Platelets, ×109 per L | 188 (129, 243) | 216 (169, 284) | <0.01 |
| Chinese Dataset [16] | Mortality Group | Non-Mortality Group | p-Value |
| Total | 154 (45%) | 190 (55%) | NA |
| Age, years | 70 (63, 77) | 51 (37, 62) | <0.01 |
| Male | 113 (73.4%) | 90 (47.4%) | <0.01 |
| CRP, mg/L | 113 (61, 165) | 19 (4, 50) | <0.01 |
| LDH, U/L | 558 (420, 719) | 251 (201, 312) | <0.01 |
| % Lymphocytes | 6 (3, 10) | 24 (17, 34) | <0.01 |
| Dutch Dataset [17] | Mortality Group | Non-Mortality Group | p-Value |
| Total | 61 (20%) | 244 (80%) | NA |
| Age, years | 75 (69, 78) | 60 (50, 73) | <0.01 |
| Male | 39 (63.9%) | 149 (61.1%) | 0.77 |
| CRP, mg/L | 107 (51, 166) | 66 (31, 116) | <0.01 |
| LDH, U/L | 443 (351, 555) | 314 (247, 433) | <0.01 |
| % Lymphocytes | 11 (6, 16) | 15 (9, 22) | <0.01 |
| French Dataset [18] | Mortality Group | Non-Mortality Group | p-Value |
| Total | 42 (37%) | 74 (63%) | NA |
| Age, years | 64 (54, 71) | 58 (50, 66) | <0.01 |
| Male | 35 (83%) | 59 (80%) | 0.81 |
| CRP, mg/L | 146 (70, 226) | 136 (78, 189) | 0.65 |
| LDH, U/L | 493 (418, 623) | 389 (324, 515) | <0.01 |
| % Lymphocytes | 9 (4, 15) | 9 (7, 14) | 0.65 |
| Internal Validation | Sensitivity | Specificity | B. Accuracy | AUC | AU-PRC | p-Value |
| Web model | 0.836 | 0.750 | 0.793 | 0.857 | 0.653 | NA |
| Minimal Model | 0.847 | 0.737 | 0.792 | 0.851 | 0.656 | 0.353 |
| Generalizable Model | 0.836 | 0.750 | 0.793 | 0.849 | 0.637 | 0.420 |
| Age-only Model | 0.608 | 0.842 | 0.725 | 0.790 | 0.535 | <0.001 |
| Italian External | Sensitivity | Specificity | B. Accuracy | AUC | AU-PRC | p-Value |
| Web model | 0.714 | 0.927 | 0.821 | 0.873 | 0.499 | NA |
| Minimal Model | 0.727 | 0.902 | 0.815 | 0.869 | 0.503 | 0.267 |
| Generalizable Model | 0.844 | 0.829 | 0.837 | 0.890 | 0.494 | 0.104 |
| Age-only Model | 0.803 | 0.805 | 0.804 | 0.853 | 0.381 | 0.157 |
| Chinese External | Sensitivity | Specificity | B. Accuracy | AUC | AU-PRC | p-Value |
| Age-only Model | 0.726 | 0.792 | 0.759 | 0.833 | 0.785 | NA |
| Generalizable Model | 0.847 | 0.831 | 0.839 | 0.909 | 0.886 | <0.001 |
| Yan’s Rule | 0.938 | 0.791 | 0.865 | 0.868 | NA | 0.197 |
| Dutch External | Sensitivity | Specificity | B. Accuracy | AUC | AU-PRC | p-Value |
| Age-only Model | 0.582 | 0.918 | 0.750 | 0.775 | 0.387 | NA |
| Generalizable Model | 0.598 | 0.918 | 0.758 | 0.806 | 0.454 | 0.037 |
| Yan’s Rule | 0.924 | 0.265 | 0.594 | 0.633 | NA | <0.001 |
| French External | Sensitivity | Specificity | B. Accuracy | AUC | AU-PRC | p-Value |
| Age-only Model | 0.581 | 0.714 | 0.648 | 0.645 | 0.512 | NA |
| Generalizable Model | 0.608 | 0.714 | 0.661 | 0.664 | 0.473 | 0.574 |
| Yan’s Rule | 0.813 | 0.390 | 0.601 | 0.552 | NA | 0.135 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chatterjee, A.; Wilmink, G.; Woodruff, H.; Lambin, P. Improving and Externally Validating Mortality Prediction Models for COVID-19 Using Publicly Available Data. BioMed 2022, 2, 13-26. https://doi.org/10.3390/biomed2010002
Chatterjee A, Wilmink G, Woodruff H, Lambin P. Improving and Externally Validating Mortality Prediction Models for COVID-19 Using Publicly Available Data. BioMed. 2022; 2(1):13-26. https://doi.org/10.3390/biomed2010002
Chicago/Turabian StyleChatterjee, Avishek, Guus Wilmink, Henry Woodruff, and Philippe Lambin. 2022. "Improving and Externally Validating Mortality Prediction Models for COVID-19 Using Publicly Available Data" BioMed 2, no. 1: 13-26. https://doi.org/10.3390/biomed2010002
APA StyleChatterjee, A., Wilmink, G., Woodruff, H., & Lambin, P. (2022). Improving and Externally Validating Mortality Prediction Models for COVID-19 Using Publicly Available Data. BioMed, 2(1), 13-26. https://doi.org/10.3390/biomed2010002







