Aptamers as Insights for Targeting SARS-CoV-2
Abstract
:1. Introduction
2. Aptamers
2.1. Artificial Intelligence: An Emerging Approach to Aptamer Selection
2.2. Aptamers against the Virus: Therapeutic and Diagnostic Applications
| Aptamer Type | Aptamer Name | Molecular Target | Organism | Effect | Reference |
|---|---|---|---|---|---|
| Phosphorothioate guanosine-quartets oligonucleotide | T2G4T2 | V3 region of the gp120 protein | HIV | Blocking the virus–cell membrane fusion | [44] |
| 6-mer sequence d(TGGGAG) | R-95288 | V3 and CD4-binding site of gp120 | HIV | Anti-virus activity in the range of micromolar and low toxicity | [45,46,47,48,49,50] |
| RNA | RNA-Tat | TAT | HIV | Even in the presence of a significant abundance of HIV TAR in cell culture, exhibits strong binding to Tat | [41] |
| RNA | B40 | Gp120 | HIV | Blockage of the interaction between gp120 and C-C chemokine receptor type 5 | [43] |
| DNA | T30695 | Integrase | HIV | HIV-1 integrase inhibition | [61] |
| 2′-DEOXY-2′-FLUOROARABINONUCLEOTIDE (FANA) | FA1 | RT | HIV RT inhibition | [62] | |
| RNA | G6-16 | N53 protein | HCV | N53 inhibition | [63] |
| DNA | ZE2 | E2 | HCV | Inhibition of HCV in vitro | [51] |
| DNA | C4 | Core protein | HCV | Inhibition of HCV | [64] |
| DNA | E10 | HA | Influenza A virus (H5N1) | Receptor-binding inhibition | [54] |
| DNA | HA12-16 | Glycosylated HA | Influenza A virus (H5N2) | Prevention of influenza virus infection | [55] |
| RNA | S9 | Truncated P protein | HBV | Inhibition of P protein binding | [65] |
| DNA | S15 | Envelope protein domain III | DENV-2 | Inhibition of proliferation | [58] |
| DNA | GE54 | Glycoprotein | RABV | Inhibition of viral replication | [59] |
| RNA | G5 α3N.4 | E7 protein | Human papillomavirus 16 | E7 Inhibition | [66] |
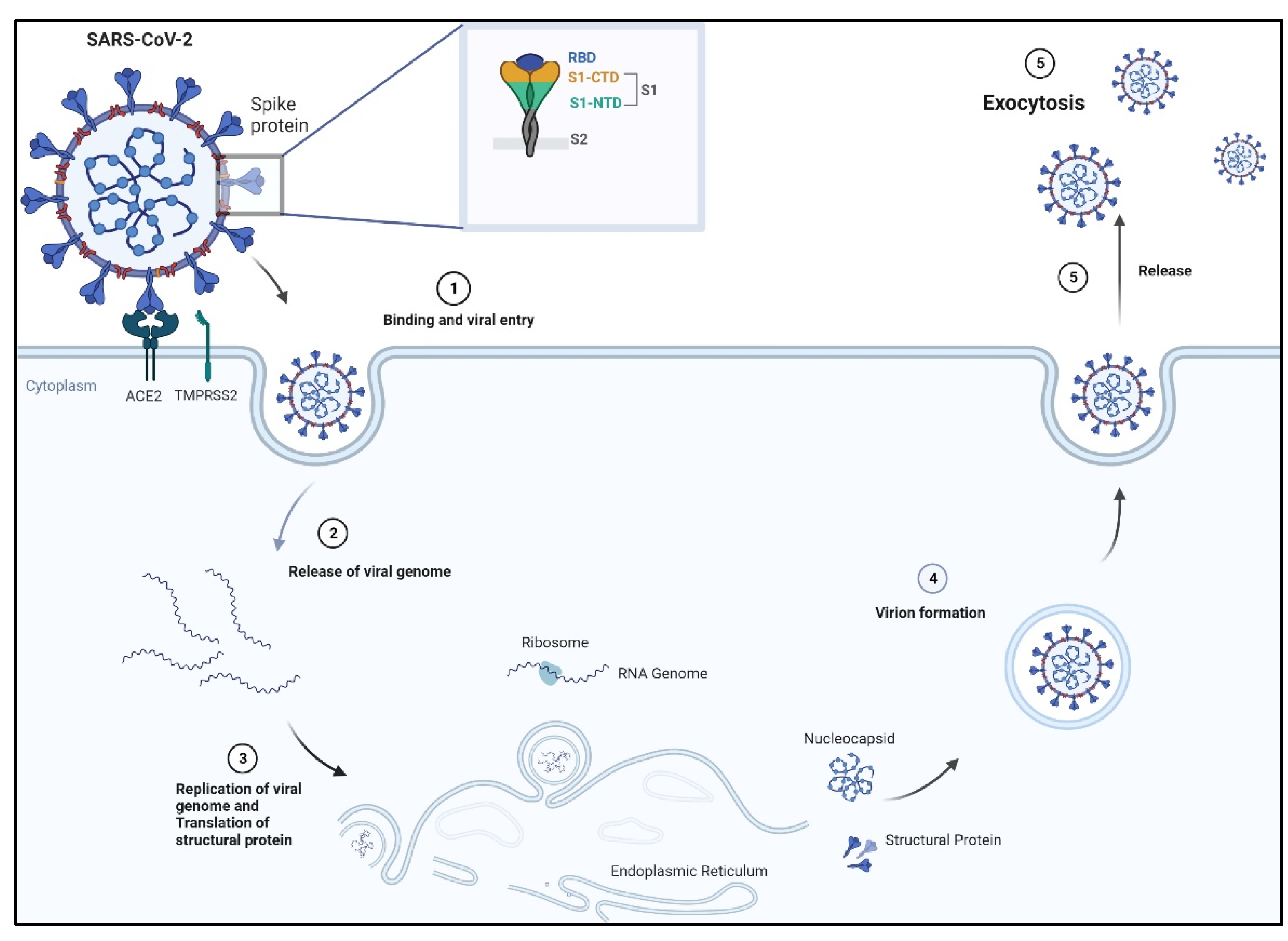
3. Severe Acute Respiratory Syndrome Coronavirus SARS-CoV-2
3.1. Spike Receptor: Its Structure and Mechanism of Binding
3.2. Aptamers against Spike Receptors
| Aptamer Type | Aptamer Name | Molecular Target | SELEX Method | Reference |
|---|---|---|---|---|
| DNA | CoV2-RBD-1C CoV2-RBD-4C | RBD | Bead-based | [89] |
| DNA | CoV2-6C3 cb-CoV2-6C3 | RBD | Bead-based | [90] |
| DNA | Aptamer-1 Aptamer-2 | RBD | Bead-based | [92] |
| DNA | nCoV-S1-Apt1 | RBD, S1 protein | Capillary electrophoresis-based | [8] |
| DNA | MSA1 MSA5 | RBD, S1 protein, trimeric S protein | EMSA (electrophoretic mobility shift assay and magnetic bead-based) | [95] |
| RNA | RBD-PB6 | RBD | Bead-based | [91] |
| DNA | XN-268s | S1 protein | Magnetic bead-based | [96] |
| DNA | ST-6 ST-6-1 ST-6-2 | Trimeric S protein | Bead-based | [93] |
| DNA | S2A2C1 S1B6C3 | S2 protein, RBD | Bead-based | [97] |
| DNA | MSA52 | S1 protein | Electrophoretic mobility shift assay, EMSA-based | [94] |
3.3. Aptamers as Diagnostics for SARS-CoV-2
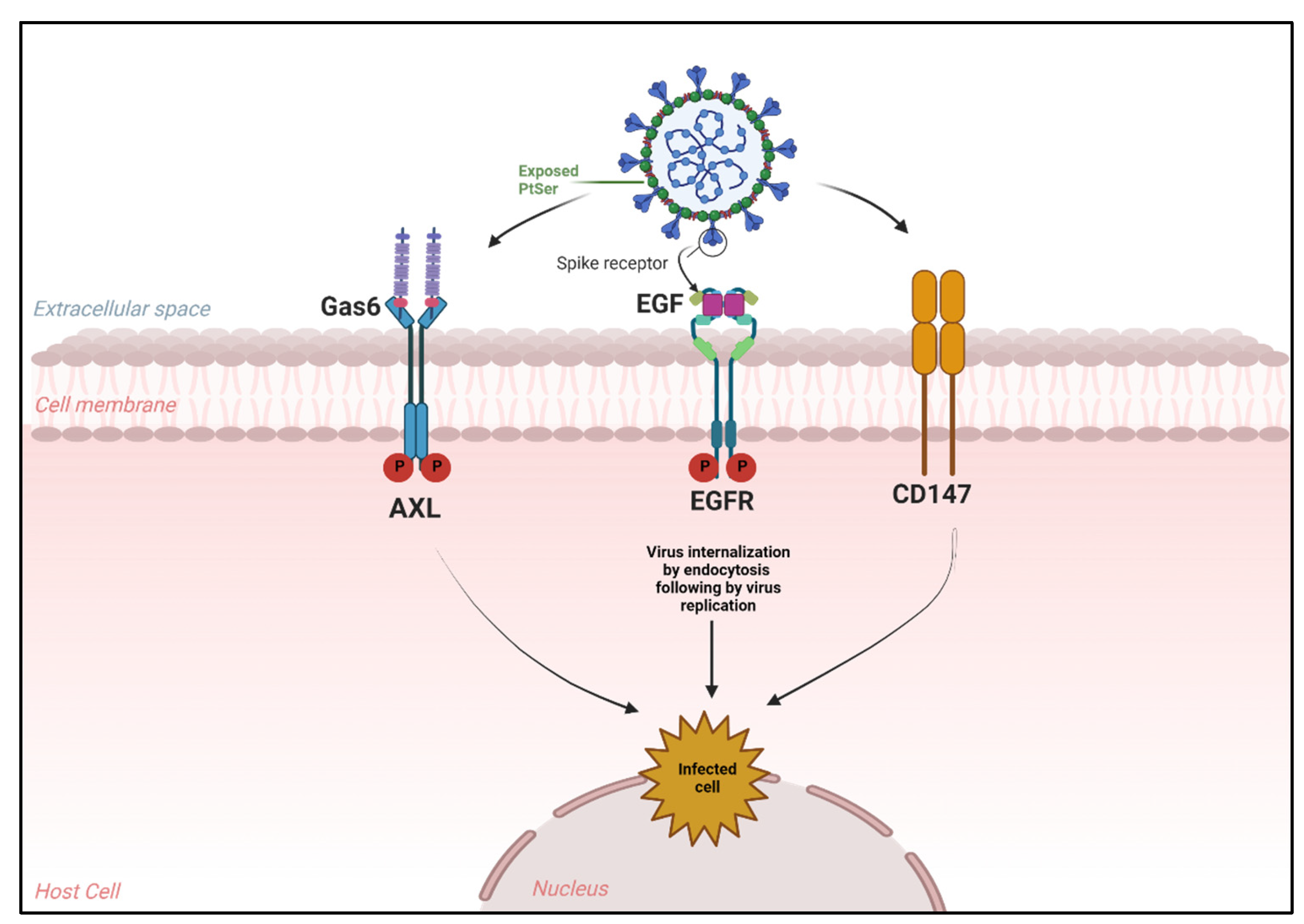
4. Novel Receptors Promote SARS-CoV-2 Entry into Host Cells
4.1. Alternative Receptors That Mediate SARS-CoV-2 Entry
4.2. Aptamers against Those Receptors
| Aptamer Type | Aptamer Name | Molecular Target | SELEX-Method | References |
|---|---|---|---|---|
| RNA | GL21.T | AXL | Bead-based | [125] |
| DNA | AXL-APTAMER | AXL | Bead-Based | [17] |
| DNA | AXL-APTAMER | AXL | Bead-based | [16] |
| DNA | AXL-APTAMER | AXL | Bead-based | [129] |
| DNA | EGFR APTAMER | EGFR | Bead-based | [126] |
| RNA | CL4 Aptamer | EGFR | Bead-based | [130,131] |
5. Single-Cell Transcriptomics Analysis as a Potential Tool to Define Novel Targets of SARS-CoV-2
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| SARS-CoV-2 | Severe Acute Respiratory Syndrome coronavirus |
| COVID-19 | Coronavirus disease 2019 |
| MERS-CoV | Middle East Respiratory Syndrome coronavirus |
| WHO | World Health Organization |
| S | Spike |
| ACE 2 | Angiotensin-converting enzyme 2 |
| AXL | Anexelekto |
| AI | Artificial Intelligence |
| SELEX | System evolution of ligand by exponential enrichment |
| CoVs | Coronaviruses |
| KREMEN1 | Kringle Containing Transmembrane Protein 1 |
| HCV | Hepatitis C Virus |
| HIV | Human Immunodeficiency Virus |
| TMPRSS2 | Type 2 TM serine protease TM protease serine 2 |
| S-RBD | Receptor-binding domain |
| TM | Transmembrane |
| FS | Fusion Peptide |
| HR1 | Heptapeptide repeat sequence 1 |
| HR2 | Heptapeptide repeat sequence 2 |
| RTK | Receptor Tyrosine Kinase |
| Gas6 | Growth Arrest Protein 6 |
| TAM | TYRO3, AXL, and MERTK Family |
| EGFR | Epidermal Growth Factor Receptor |
| LDLR | Low-Density Lipoprotein Receptor |
| CD147 | Cluster of Differentiation 147 |
| ASGR1 | Asialoglycoprotein receptor 1 |
| LDLRAD3 | Low-Density Lipoprotein Receptor Class A Domain Containing 3 |
| TMEM30A | Transmembrane Protein 30A |
| CD209 | Cluster of Differentiation 209 |
| CD209L | Cluster of Differentiation 209 Ligand |
| L-SIGN | Liver/lymph node-specific intercellular adhesion molecule-3-grabbing integrin |
| DC-SIGN | Dendritic Cell-Specific Intercellular Adhesion molecule-3-Grabbing Non-integrin |
| CCR5 | C–C chemokine receptor type 5 |
| SCAP | Spherical cocktail aptamers–gold nanoparticles |
| SERS | Surface-enhanced Raman Spectroscopy |
References
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef] [PubMed]
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.; Vijayan, R. Dynamics of the ACE2–SARS-CoV-2/SARS-CoV spike protein interface reveal unique mechanisms. Sci. Rep. 2020, 10, 14214. [Google Scholar] [CrossRef]
- Wang, S.; Qiu, Z.; Hou, Y.; Deng, X.; Xu, W.; Zheng, T.; Wu, P.; Xie, S.; Bian, W.; Zhang, C.; et al. AXL is a candidate receptor for SARS-CoV-2 that promotes infection of pulmonary and bronchial epithelial cells. Cell Res. 2021, 31, 126–140. [Google Scholar] [CrossRef]
- Wang, S.; Qiu, Z.; Hou, Y.; Deng, X.; Zheng, T.; Yan, R.; Wu, P.; Xie, S.; Zhou, Q.; Huang, J.; et al. AXL Promotes SARS-CoV-2 Infection of Pulmonary and Bronchial Epithelial Cells. Res. Sq. 2021. [Google Scholar] [CrossRef]
- Dai, M.; Liu, D.; Liu, M.; Zhou, F.; Li, G.; Chen, Z.; Zhang, Z.; You, H.; Wu, M.; Zheng, Q.; et al. Patients with Cancer Appear More Vulnerable to SARS-CoV-2: A Multicenter Study during the COVID-19 Outbreak. Cancer Discov. 2020, 10, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Mehta, V.; Goel, S.; Kabarriti, R.; Cole, D.; Goldfinger, M.; Acuna-Villaorduna, A.; Pradhan, K.; Thota, R.; Reissman, S.; Sparano, J.A.; et al. Case Fatality Rate of Cancer Patients with COVID-19 in a New York Hospital System. Cancer Discov. 2020, 10, 935–941. [Google Scholar] [CrossRef]
- Yang, G.; Li, Z.; Mohammed, I.; Zhao, L.; Wei, W.; Xiao, H.; Guo, W.; Zhao, Y.; Qu, F.; Huang, Y. Identification of SARS-CoV-2-against aptamer with high neutralization activity by blocking the RBD domain of spike protein 1. Signal Transduct. Target. Ther. 2021, 6, 227. [Google Scholar] [CrossRef] [PubMed]
- Thiel, K.W.; Giangrande, P.H. Therapeutic applications of DNA and RNA aptamers. Oligonucleotides 2009, 19, 209–222. [Google Scholar] [CrossRef]
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef]
- Schneider, D.; Tuerk, C.; Gold, L. Selection of high affinity RNA ligands to the bacteriophage R17 coat protein. J. Mol. Biol. 1992, 228, 862–869. [Google Scholar] [CrossRef] [PubMed]
- Torres-Chavolla, E.; Alocilja, E.C. Aptasensors for detection of microbial and viral pathogens. Biosens. Bioelectron. 2009, 24, 3175–3182. [Google Scholar] [CrossRef] [PubMed]
- Ku, T.H.; Zhang, T.; Luo, H.; Yen, T.M.; Chen, P.W.; Han, Y.; Lo, Y.H. Nucleic Acid Aptamers: An Emerging Tool for Biotechnology and Biomedical Sensing. Sensors 2015, 15, 16281–16313. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Zhang, X.; Zhou, S.; Shen, J.; Yang, D.; Wu, J.; Li, X.; Li, M.; Huang, X.; Sealy, J.E.; et al. A DNA aptamer efficiently inhibits the infectivity of Bovine herpesvirus 1 by blocking viral entry. Sci. Rep. 2017, 7, 11796. [Google Scholar] [CrossRef]
- Bouchard, P.R.; Hutabarat, R.M.; Thompson, K.M. Discovery and development of therapeutic aptamers. Annu. Rev. Pharmacol. Toxicol. 2010, 50, 237–257. [Google Scholar] [CrossRef]
- Amero, P.; Lokesh, G.L.R.; Chaudhari, R.R.; Cardenas-Zuniga, R.; Schubert, T.; Attia, Y.M.; Montalvo-Gonzalez, E.; Elsayed, A.M.; Ivan, C.; Wang, Z.; et al. Conversion of RNA Aptamer into Modified DNA Aptamers Provides for Prolonged Stability and Enhanced Antitumor Activity. J. Am. Chem. Soc. 2021, 143, 7655–7670. [Google Scholar] [CrossRef]
- Kanlikilicer, P.; Ozpolat, B.; Aslan, B.; Bayraktar, R.; Gurbuz, N.; Rodriguez-Aguayo, C.; Bayraktar, E.; Denizli, M.; Gonzalez-Villasana, V.; Ivan, C.; et al. Therapeutic Targeting of AXL Receptor Tyrosine Kinase Inhibits Tumor Growth and Intraperitoneal Metastasis in Ovarian Cancer Models. Mol. Ther. Nucleic Acids 2017, 9, 251–262. [Google Scholar] [CrossRef]
- Morita, Y.; Leslie, M.; Kameyama, H.; Volk, D.E.; Tanaka, T. Aptamer Therapeutics in Cancer: Current and Future. Cancers 2018, 10, 80. [Google Scholar] [CrossRef]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef]
- Reinemann, C.; Strehlitz, B. Aptamer-modified nanoparticles and their use in cancer diagnostics and treatment. Swiss Med. Wkly. 2014, 144, w13908. [Google Scholar] [CrossRef]
- Zhou, J.; Rossi, J. Aptamers as targeted therapeutics: Current potential and challenges. Nat. Rev. Drug Discov. 2017, 16, 181–202. [Google Scholar] [CrossRef] [PubMed]
- Hori, S.I.; Herrera, A.; Rossi, J.J.; Zhou, J. Current Advances in Aptamers for Cancer Diagnosis and Therapy. Cancers 2018, 10, 9. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.; Rossi, J.J. Aptamers: Uptake mechanisms and intracellular applications. Adv. Drug Deliv. Rev. 2018, 134, 22–35. [Google Scholar] [CrossRef] [PubMed]
- Tatiparti, K.; Sau, S.; Kashaw, S.K.; Iyer, A.K. siRNA Delivery Strategies: A Comprehensive Review of Recent Developments. Nanomaterials 2017, 7, 77. [Google Scholar] [CrossRef]
- Catuogno, S.; Esposito, C.L.; de Franciscis, V. Aptamer-Mediated Targeted Delivery of Therapeutics: An Update. Pharmaceuticals 2016, 9, 69. [Google Scholar] [CrossRef]
- Chen, Z.; Hu, L.; Zhang, B.T.; Lu, A.; Wang, Y.; Yu, Y.; Zhang, G. Artificial Intelligence in Aptamer-Target Binding Prediction. Int. J. Mol. Sci. 2021, 22, 3605. [Google Scholar] [CrossRef]
- Kinghorn, A.B.; Fraser, L.A.; Lang, S.; Shiu, S.C.C.; Tanner, J.A. Aptamer Bioinformatics. Int. J. Mol. Sci. 2017, 18, 2516. [Google Scholar] [CrossRef]
- Buglak, A.A.; Samokhvalov, A.V.; Zherdev, A.V.; Dzantiev, B.B. Methods and Applications of In Silico Aptamer Design and Modeling. Int. J. Mol. Sci. 2020, 21, 8420. [Google Scholar] [CrossRef]
- Chushak, Y.; Stone, M.O. In silico selection of RNA aptamers. Nucleic Acids Res. 2009, 37, e87. [Google Scholar] [CrossRef]
- Hofacker, I.L. Vienna RNA secondary structure server. Nucleic Acids Res. 2003, 31, 3429–3431. [Google Scholar] [CrossRef]
- Ahirwar, R.; Nahar, S.; Aggarwal, S.; Ramachandran, S.; Maiti, S.; Nahar, P. In silico selection of an aptamer to estrogen receptor alpha using computational docking employing estrogen response elements as aptamer-alike molecules. Sci. Rep. 2016, 6, 21285. [Google Scholar] [CrossRef] [PubMed]
- Thafar, M.; Raies, A.B.; Albaradei, S.; Essack, M.; Bajic, V.B. Comparison Study of Computational Prediction Tools for Drug-Target Binding Affinities. Front. Chem. 2019, 7, 782. [Google Scholar] [CrossRef]
- Ain, Q.U.; Aleksandrova, A.; Roessler, F.D.; Ballester, P.J. Machine-learning scoring functions to improve structure-based binding affinity prediction and virtual screening. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2015, 5, 405–424. [Google Scholar] [CrossRef] [PubMed]
- Zhuo, Z.; Wan, Y.; Guan, D.; Ni, S.; Wang, L.; Zhang, Z.; Liu, J.; Liang, C.; Yu, Y.; Lu, A.; et al. A Loop-Based and AGO-Incorporated Virtual Screening Model Targeting AGO-Mediated miRNA-mRNA Interactions for Drug Discovery to Rescue Bone Phenotype in Genetically Modified Mice. Adv. Sci. 2020, 7, 1903451. [Google Scholar] [CrossRef] [PubMed]
- Eilers, A.; Witt, S.; Walter, J. Aptamer-Modified Nanoparticles in Medical Applications. Adv. Biochem. Eng. Biotechnol. 2020, 174, 161–193. [Google Scholar] [CrossRef] [PubMed]
- Zou, X.; Wu, J.; Gu, J.; Shen, L.; Mao, L. Application of Aptamers in Virus Detection and Antiviral Therapy. Front. Microbiol. 2019, 10, 1462. [Google Scholar] [CrossRef]
- Nimjee, S.M.; Rusconi, C.P.; Sullenger, B.A. Aptamers: An emerging class of therapeutics. Annu. Rev. Med. 2005, 56, 555–583. [Google Scholar] [CrossRef]
- Parekh, P.; Tang, Z.; Turner, P.C.; Moyer, R.W.; Tan, W. Aptamers recognizing glycosylated hemagglutinin expressed on the surface of vaccinia virus-infected cells. Anal. Chem. 2010, 82, 8642–8649. [Google Scholar] [CrossRef]
- Pan, Q.; Luo, F.; Liu, M.; Zhang, X.L. Oligonucleotide aptamers: Promising and powerful diagnostic and therapeutic tools for infectious diseases. J. Infect. 2018, 77, 83–98. [Google Scholar] [CrossRef]
- Chakraborty, B.; Das, S.; Gupta, A.; Xiong, Y.; T-V, V.; Kizer, M.E.; Duan, J.; Chandrasekaran, A.R.; Wang, X. Aptamers for Viral Detection and Inhibition. ACS Infect. Dis. 2022, 8, 667–692. [Google Scholar] [CrossRef]
- Yamamoto, R.; Katahira, M.; Nishikawa, S.; Baba, T.; Taira, K.; Kumar, P.K.R. A novel RNA motif that binds efficiently and specifically to the Tat protein of HIV and inhibits the trans-activation by Tat of transcription in vitro and in vivo. Genes Cells 2000, 5, 371–388. [Google Scholar] [CrossRef] [PubMed]
- Minunni, M.; Tombelli, S.; Gullotto, A.; Luzi, E.; Mascini, M. Development of biosensors with aptamers as bio-recognition element: The case of HIV-1 Tat protein. Biosens. Bioelectron. 2004, 20, 1149–1156. [Google Scholar] [CrossRef] [PubMed]
- Dey, A.K.; Griffiths, C.; Lea, S.M.; James, W. Structural characterization of an anti-gp120 RNA aptamer that neutralizes R5 strains of HIV-1. RNA 2005, 11, 873–884. [Google Scholar] [CrossRef] [PubMed]
- Wyatt, J.R.; Vickers, T.A.; Roberson, J.L.; Buckheit, R.W., Jr.; Klimkait, T.; DeBaets, E.; Davis, P.W.; Rayner, B.; Imbach, J.L.; Ecker, D.J. Combinatorially selected guanosine-quartet structure is a potent inhibitor of human immunodeficiency virus envelope-mediated cell fusion. Proc. Natl. Acad. Sci. USA 1994, 91, 1356–1360. [Google Scholar] [CrossRef]
- Hotoda, H.; Koizumi, M.; Koga, R.; Kaneko, M.; Momota, K.; Ohmine, T.; Furukawa, H.; Agatsuma, T.; Nishigaki, T.; Sone, J.; et al. Biologically Active Oligodeoxyribonucleotides. 5. 5′-End-Substituted d(TGGGAG) Possesses Anti-Human Immunodeficiency Virus Type 1 Activity by Forming a G-Quadruplex Structure. J. Med. Chem. 1998, 41, 3655–3663. [Google Scholar] [CrossRef]
- D’Onofrio, J.; Petraccone, L.; Erra, E.; Martino, L.; Di Fabio, G.; De Napoli, L.; Giancola, C.; Montesarchio, D. 5′-Modified G-Quadruplex Forming Oligonucleotides Endowed with Anti-HIV Activity: Synthesis and Biophysical Properties. Bioconjug. Chem. 2007, 18, 1194–1204. [Google Scholar] [CrossRef]
- Oliviero, G.; Amato, J.; Borbone, N.; D’Errico, S.; Galeone, A.; Mayol, L.; Haider, S.; Olubiyi, O.; Hoorelbeke, B.; Balzarini, J.; et al. Tetra-end-linked oligonucleotides forming DNA G-quadruplexes: A new class of aptamers showing anti-HIV activity. Chem. Commun. 2010, 46, 8971–8973. [Google Scholar] [CrossRef]
- D’Onofrio, J.; Petraccone, L.; Martino, L.; Fabio, G.D.; Iadonisi, A.; Balzarini, J.; Giancola, C.; Montesarchio, D. Synthesis, Biophysical Characterization, and Anti-HIV Activity of Glyco-Conjugated G-Quadruplex-Forming Oligonucleotides. Bioconjug. Chem. 2008, 19, 607–616. [Google Scholar] [CrossRef]
- Di Fabio, G.; D’Onofrio, J.; Chiapparelli, M.; Hoorelbeke, B.; Montesarchio, D.; Balzarini, J.; De Napoli, L. Discovery of novel anti-HIV active G-quadruplex-forming oligonucleotides. Chem. Commun. 2011, 47, 2363–2365. [Google Scholar] [CrossRef]
- D’Atri, V.; Oliviero, G.; Amato, J.; Borbone, N.; D’Errico, S.; Mayol, L.; Piccialli, V.; Haider, S.; Hoorelbeke, B.; Balzarini, J.; et al. New anti-HIV aptamers based on tetra-end-linked DNA G-quadruplexes: Effect of the base sequence on anti-HIV activity. Chem. Commun. 2012, 48, 9516–9518. [Google Scholar] [CrossRef]
- Chen, F.; Hu, Y.L.; Li, D.Q.; Chen, H.D.; Zhang, X.L. CS-SELEX Generates High-Affinity ssDNA Aptamers as Molecular Probes for Hepatitis C Virus Envelope Glycoprotein E2. PLoS ONE 2009, 4, e8142. [Google Scholar] [CrossRef] [PubMed]
- Romero-Lopez, C.; Lahlali, T.; Berzal-Herranz, B.; Berzal-Herranz, A. Development of Optimized Inhibitor RNAs Allowing Multisite-Targeting of the HCV Genome. Molecules 2017, 22, 861. [Google Scholar] [CrossRef] [PubMed]
- Hung, L.Y.; Huang, T.B.; Tsai, Y.C.; Yeh, C.S.; Lei, H.Y.; Lee, G.B. A microfluidic immunomagnetic bead-based system for the rapid detection of influenza infections: From purified virus particles to clinical specimens. Biomed. Microdevices 2013, 15, 539–551. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.S.; Dong, J.; Yao, L.H.; Chen, A.J.; Jia, R.Q.; Huan, L.F.; Guo, J.L.; Shu, Y.L.; Zhang, Z.Q. Potent inhibition of human influenza H5N1 virus by oligonucleotides derived by SELEX. Biochem. Biophys. Res. Commun. 2008, 366, 670–674. [Google Scholar] [CrossRef] [PubMed]
- Kwon, H.M.; Lee, K.H.; Han, B.W.; Han, M.R.; Kim, D.H.; Kim, D.E. An RNA Aptamer That Specifically Binds to the Glycosylated Hemagglutinin of Avian Influenza Virus and Suppresses Viral Infection in Cells. PLoS ONE 2014, 9, e97574. [Google Scholar] [CrossRef]
- Wang, R.; Zhao, J.; Jiang, T.; Kwon, Y.M.; Lu, H.; Jiao, P.; Liao, M.; Li, Y. Selection and characterization of DNA aptamers for use in detection of avian influenza virus H5N1. J. Virol. Methods 2013, 189, 362–369. [Google Scholar] [CrossRef]
- Liu, J.; Yang, Y.; Hu, B.; Ma, Z.Y.; Huang, H.P.; Yu, Y.; Liu, S.P.; Lu, M.J.; Yang, D.L. Development of HBsAg-binding aptamers that bind HepG2.2.15 cells via HBV surface antigen. Virol. Sin. 2010, 25, 27–35. [Google Scholar] [CrossRef]
- Chen, H.L.; Hsiao, W.H.; Lee, H.C.; Wu, S.C.; Cheng, J.W. Selection and Characterization of DNA Aptamers Targeting All Four Serotypes of Dengue Viruses. PLoS ONE 2015, 10, e0131240. [Google Scholar] [CrossRef]
- Liang, H.R.; Hu, G.Q.; Li, L.; Gao, Y.W.; Yang, S.T.; Xia, X.Z. Aptamers targeting rabies virus-infected cells inhibit street rabies virus in vivo. Int. Immunopharmacol. 2014, 21, 432–438. [Google Scholar] [CrossRef]
- Liang, H.R.; Hu, G.Q.; Xue, X.H.; Li, L.; Zheng, X.X.; Gao, Y.W.; Yang, S.T.; Xia, X.Z. Selection of an aptamer against rabies virus: A new class of molecules with antiviral activity. Virus Res. 2014, 184, 7–13. [Google Scholar] [CrossRef]
- Jing, N.J.; Hogan, M.E. Structure-activity of tetrad-forming oligonucleotides as a potent anti-HIV therapeutic drug. J. Biol. Chem. 1998, 273, 34992–34999. [Google Scholar] [CrossRef] [PubMed]
- Ferreira-Bravo, I.A.; Cozens, C.; Holliger, P.; DeStefano, J.J. Selection of 2′-deoxy-2′-fluoroarabinonucleotide (FANA) aptamers that bind HIV-1 reverse transcriptase with picomolar affinity. Nucleic Acids Res. 2015, 43, 9587–9599. [Google Scholar] [CrossRef]
- Kumar, P.K.R.; Machida, K.; Urvil, P.T.; Kakiuchi, N.; Vishnuvardhan, D.; Shimotohno, K.; Taira, K.; Nishikawa, S. Isolation of RNA aptamers specific to the NS3 protein of hepatitis C virus from a pool of completely random RNA. Virology 1997, 237, 270–282. [Google Scholar] [CrossRef] [PubMed]
- Shi, S.L.; Yu, X.Y.; Gao, Y.M.; Xue, B.B.; Wu, X.J.; Wang, X.H.; Yang, D.R.; Zhu, H.Z. Inhibition of Hepatitis C Virus Production by Aptamers against the Core Protein. J. Virol. 2014, 88, 1990–1999. [Google Scholar] [CrossRef] [PubMed]
- Feng, H.; Beck, J.; Nassal, M.; Hu, K.H. A SELEX-Screened Aptamer of Human Hepatitis B Virus RNA Encapsidation Signal Suppresses Viral Replication. PLoS ONE 2011, 6, e27862. [Google Scholar] [CrossRef]
- Toscano-Garibay, J.D.; Benitez-Hess, M.L.; Alvarez-Salas, L.M. Isolation and Characterization of an RNA Aptamer for the HPV-16 E7 Oncoprotein. Arch. Med. Res. 2011, 42, 88–96. [Google Scholar] [CrossRef]
- Siu, Y.L.; Teoh, K.T.; Lo, J.; Chan, C.M.; Kien, F.; Escriou, N.; Tsao, S.W.; Nicholls, J.M.; Altmeyer, R.; Peiris, J.S.; et al. The M, E, and N structural proteins of the severe acute respiratory syndrome coronavirus are required for efficient assembly, trafficking, and release of virus-like particles. J. Virol. 2008, 82, 11318–11330. [Google Scholar] [CrossRef]
- Kirchdoerfer, R.N.; Cottrell, C.A.; Wang, N.; Pallesen, J.; Yassine, H.M.; Turner, H.L.; Corbett, K.S.; Graham, B.S.; McLellan, J.S.; Ward, A.B. Pre-fusion structure of a human coronavirus spike protein. Nature 2016, 531, 118–121. [Google Scholar] [CrossRef]
- Grunewald, M.E.; Fehr, A.R.; Athmer, J.; Perlman, S. The coronavirus nucleocapsid protein is ADP-ribosylated. Virology 2018, 517, 62–68. [Google Scholar] [CrossRef]
- Kuhn, J.H.; Li, W.; Choe, H.; Farzan, M. Angiotensin-converting enzyme 2: A functional receptor for SARS coronavirus. Cell Mol. Life Sci. 2004, 61, 2738–2743. [Google Scholar] [CrossRef]
- Der Sarkissian, S.; Huentelman, M.J.; Stewart, J.; Katovich, M.J.; Raizada, M.K. ACE2: A novel therapeutic target for cardiovascular diseases. Prog. Biophys. Mol. Biol. 2006, 91, 163–198. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Kruger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280. [Google Scholar] [CrossRef] [PubMed]
- Yuan, M.; Wu, N.C.; Zhu, X.; Lee, C.D.; So, R.T.Y.; Lv, H.; Mok, C.K.P.; Wilson, I.A. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science 2020, 368, 630–633. [Google Scholar] [CrossRef] [PubMed]
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef]
- Chen, L.; Liu, W.; Zhang, Q.; Xu, K.; Ye, G.; Wu, W.; Sun, Z.; Liu, F.; Wu, K.; Zhong, B.; et al. RNA based mNGS approach identifies a novel human coronavirus from two individual pneumonia cases in 2019 Wuhan outbreak. Emerg. Microbes Infect. 2020, 9, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.F.; Kok, K.H.; Zhu, Z.; Chu, H.; To, K.K.; Yuan, S.; Yuen, K.Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020, 9, 221–236. [Google Scholar] [CrossRef] [PubMed]
- Letko, M.; Marzi, A.; Munster, V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020, 5, 562–569. [Google Scholar] [CrossRef] [PubMed]
- Bosch, B.J.; van der Zee, R.; de Haan, C.A.M.; Rottier, P.J.M. The coronavirus spike protein is a class I virus fusion protein: Structural and functional characterization of the fusion core complex. J. Virol. 2003, 77, 8801–8811. [Google Scholar] [CrossRef]
- Xia, S.; Zhu, Y.; Liu, M.Q.; Lan, Q.H.; Xu, W.; Wu, Y.L.; Ying, T.L.; Liu, S.W.; Shi, Z.L.; Jiang, S.B.; et al. Fusion mechanism of 2019-nCoV and fusion inhibitors targeting HR1 domain in spike protein. Cell Mol. Immunol. 2020, 17, 765–767. [Google Scholar] [CrossRef]
- Tang, T.; Bidon, M.; Jaimes, J.A.; Whittaker, G.R.; Daniel, S. Coronavirus membrane fusion mechanism offers a potential target for antiviral development. Antiviral. Res. 2020, 178, 104792. [Google Scholar] [CrossRef]
- Fehr, A.R.; Perlman, S. Coronaviruses: An Overview of Their Replication and Pathogenesis. Methods Mol. Biol. 2015, 1282, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Walls, A.C.; Park, Y.J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein (vol 180, 281.e1, 2020). Cell 2020, 183, 1735. [Google Scholar] [CrossRef] [PubMed]
- Lan, J.; Ge, J.W.; Yu, J.F.; Shan, S.S.; Zhou, H.; Fan, S.L.; Zhang, Q.; Shi, X.L.; Wang, Q.S.; Zhang, L.Q.; et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020, 581, 215–220. [Google Scholar] [CrossRef] [PubMed]
- Yan, R.H.; Zhang, Y.Y.; Li, Y.N.; Xia, L.; Guo, Y.Y.; Zhou, Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 2020, 367, 1444–1448. [Google Scholar] [CrossRef]
- Huang, Y.; Yang, C.; Xu, X.F.; Xu, W.; Liu, S.W. Structural and functional properties of SARS-CoV-2 spike protein: Potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 2020, 41, 1141–1149. [Google Scholar] [CrossRef]
- Dai, L.P.; Gao, G.F. Viral targets for vaccines against COVID-19. Nat. Rev. Immunol. 2021, 21, 73–82. [Google Scholar] [CrossRef]
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020, 367, 1260–1263. [Google Scholar] [CrossRef]
- Amini, R.; Zhang, Z.; Li, J.; Gu, J.; Brennan, J.D.; Li, Y. Aptamers for SARS-CoV-2: Isolation, Characterization, and Diagnostic and Therapeutic Developments. Anal. Sens. 2022, 2, e202200012. [Google Scholar] [CrossRef]
- Song, Y.; Song, J.; Wei, X.; Huang, M.; Sun, M.; Zhu, L.; Lin, B.; Shen, H.; Zhu, Z.; Yang, C. Discovery of Aptamers Targeting the Receptor-Binding Domain of the SARS-CoV-2 Spike Glycoprotein. Anal. Chem. 2020, 92, 9895–9900. [Google Scholar] [CrossRef]
- Sun, M.; Liu, S.; Wei, X.; Wan, S.; Huang, M.; Song, T.; Lu, Y.; Weng, X.; Lin, Z.; Chen, H.; et al. Aptamer Blocking Strategy Inhibits SARS-CoV-2 Virus Infection. Angew. Chem. Int. Ed. Engl. 2021, 60, 10266–10272. [Google Scholar] [CrossRef]
- Valero, J.; Civit, L.; Dupont, D.M.; Selnihhin, D.; Reinert, L.S.; Idorn, M.; Israels, B.A.; Bednarz, A.M.; Bus, C.; Asbach, B.; et al. A serum-stable RNA aptamer specific for SARS-CoV-2 neutralizes viral entry. Proc. Natl. Acad. Sci. USA 2021, 118, e2112942118. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wang, Y.L.; Wu, J.; Qi, J.; Zeng, Z.; Wan, Q.; Chen, Z.; Manandhar, P.; Cavener, V.S.; Boyle, N.R.; et al. Neutralizing Aptamers Block S/RBD-ACE2 Interactions and Prevent Host Cell Infection. Angew. Chem. Weinheim. Bergstr. Ger. 2021, 133, 10361–10366. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Zhang, S.; Wang, Y.; Li, L.; Li, Y.; Yuan, D.; Luo, F.; Zhao, J.; Song, X.; Zhao, Y. Aptamer blocking S-TLR4 interaction selectively inhibits SARS-CoV-2 induced inflammation. Signal Transduct. Target Ther. 2022, 7, 120. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.J.; Li, J.X.; Gu, J.; Amini, R.; Stacey, H.D.; Ang, J.C.; White, D.; Filipe, C.D.M.; Mossman, K.; Miller, M.S.; et al. A Universal DNA Aptamer that Recognizes Spike Proteins of Diverse SARS-CoV-2 Variants of Concern. Chem.-Eur. J. 2022, 28, e202200078. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhang, Z.; Gu, J.; Stacey, H.D.; Ang, J.C.; Capretta, A.; Filipe, C.D.M.; Mossman, K.L.; Balion, C.; Salena, B.J.; et al. Diverse high-affinity DNA aptamers for wild-type and B.1.1.7 SARS-CoV-2 spike proteins from a pre-structured DNA library. Nucleic Acids Res. 2021, 49, 7267–7279. [Google Scholar] [CrossRef]
- Shi, L.; Wang, L.; Ma, X.; Fang, X.; Xiang, L.; Yi, Y.; Li, J.; Luo, Z.; Li, G. Aptamer-Functionalized Nanochannels for One-Step Detection of SARS-CoV-2 in Samples from COVID-19 Patients. Anal. Chem. 2021, 93, 16646–16654. [Google Scholar] [CrossRef]
- Silwal, A.P.; Thennakoon, S.K.S.; Arya, S.P.; Postema, R.M.; Jahan, R.; Phuoc, C.M.T.; Tan, X. DNA aptamers inhibit SARS-CoV-2 spike-protein binding to hACE2 by an RBD- independent or dependent approach. Theranostics 2022, 12, 5522–5536. [Google Scholar] [CrossRef]
- Singh, B.; Datta, B.; Ashish, A.; Dutta, G. A comprehensive review on current COVID-19 detection methods: From lab care to point of care diagnosis. Sens. Int. 2021, 2, 100119. [Google Scholar] [CrossRef]
- Jung, Y.; Park, G.-S.; Moon, J.H.; Ku, K.; Beak, S.-H.; Lee, C.-S.; Kim, S.; Park, E.C.; Park, D.; Lee, J.-H.; et al. Comparative Analysis of Primer–Probe Sets for RT-qPCR of COVID-19 Causative Virus (SARS-CoV-2). ACS Infect. Dis. 2020, 6, 2513–2523. [Google Scholar] [CrossRef]
- Yüce, M.; Filiztekin, E.; Özkaya, K.G. COVID-19 diagnosis—A review of current methods. Biosens. Bioelectron. 2021, 172, 112752. [Google Scholar] [CrossRef]
- Wandtke, T.; Wędrowska, E.; Szczur, M.; Przybylski, G.; Libura, M.; Kopiński, P. Aptamers-Diagnostic and Therapeutic Solution in SARS-CoV-2. Int. J. Mol. Sci. 2022, 23, 1412. [Google Scholar] [CrossRef] [PubMed]
- Binny, R.N.; Priest, P.; French, N.P.; Parry, M.; Lustig, A.; Hendy, S.C.; Maclaren, O.J.; Ridings, K.M.; Steyn, N.; Vattiato, G.; et al. Sensitivity of Reverse Transcription Polymerase Chain Reaction Tests for Severe Acute Respiratory Syndrome Coronavirus 2 Through Time. J. Infect. Dis. 2022, 227, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Teymouri, M.; Mollazadeh, S.; Mortazavi, H.; Naderi Ghale-Noie, Z.; Keyvani, V.; Aghababaei, F.; Hamblin, M.R.; Abbaszadeh-Goudarzi, G.; Pourghadamyari, H.; Hashemian, S.M.R.; et al. Recent advances and challenges of RT-PCR tests for the diagnosis of COVID-19. Pathol. Res. Pract. 2021, 221, 153443. [Google Scholar] [CrossRef] [PubMed]
- Kundu, D.; Gautam, P.; Dayanand, D.; Gunasekaran, K.; Manesh, A.; Sebastian, M.; Abhilash, K.P.P.; Zachariah, A.; George, T.; Sathyendra, S.; et al. The role and diagnostic accuracy of serology for COVID-19. BMC Infect. Dis. 2022, 22, 390. [Google Scholar] [CrossRef] [PubMed]
- Gong, F.; Wei, H.X.; Li, Q.; Liu, L.; Li, B. Evaluation and Comparison of Serological Methods for COVID-19 Diagnosis. Front. Mol. Biosci. 2021, 8, 682405. [Google Scholar] [CrossRef] [PubMed]
- Kyosei, Y.; Yamura, S.; Namba, M.; Yoshimura, T.; Watabe, S.; Ito, E. Antigen tests for COVID-19. Biophys. Physicobiol. 2021, 18, 28–39. [Google Scholar] [CrossRef] [PubMed]
- Pendergrast, P.S.; Marsh, H.N.; Grate, D.; Healy, J.M.; Stanton, M. Nucleic acid aptamers for target validation and therapeutic applications. J. Biomol. Tech. 2005, 16, 224–234. [Google Scholar]
- Dzuvor, C.K.O.; Tettey, E.L.; Danquah, M.K. Aptamers as promising nanotheranostic tools in the COVID-19 pandemic era. WIREs Nanomed. Nanobiotechnol. 2022, 14, e1785. [Google Scholar] [CrossRef]
- Bruno, J.G. A Review of Therapeutic Aptamer Conjugates with Emphasis on New Approaches. Pharmaceuticals 2013, 6, 340–357. [Google Scholar] [CrossRef]
- Zhang, Y.; Juhas, M.; Kwok, C.K. Aptamers targeting SARS-CoV-2: A promising tool to fight against COVID-19. Trends Biotechnol. 2023, 41, 528–544. [Google Scholar] [CrossRef]
- Gupta, A.; Anand, A.; Jain, N.; Goswami, S.; Anantharaj, A.; Patil, S.; Singh, R.; Kumar, A.; Shrivastava, T.; Bhatnagar, S.; et al. A novel G-quadruplex aptamer-based spike trimeric antigen test for the detection of SARS-CoV-2. Mol. Ther. Nucleic Acids 2021, 26, 321–332. [Google Scholar] [CrossRef] [PubMed]
- Gu, M.-M.; Guan, P.-C.; Xu, S.-S.; Li, H.-M.; Kou, Y.-C.; Lin, X.-D.; Kathiresan, M.; Song, Y.; Zhang, Y.-J.; Jin, S.-Z.; et al. Ultrasensitive detection of SARS-CoV-2 S protein with aptamers biosensor based on surface-enhanced Raman scattering. J. Chem. Phys. 2023, 158, 024203. [Google Scholar] [CrossRef] [PubMed]
- Minhas, N.; Gurav, Y.K.; Sambhare, S.; Potdar, V.; Choudhary, M.L.; Bhardwaj, S.D.; Abraham, P. Cost-analysis of real time RT-PCR test performed for COVID-19 diagnosis at India’s national reference laboratory during the early stages of pandemic mitigation. PLoS ONE 2023, 18, e0277867. [Google Scholar] [CrossRef] [PubMed]
- Lukassen, S.; Chua, R.L.; Trefzer, T.; Kahn, N.C.; Schneider, M.A.; Muley, T.; Winter, H.; Meister, M.; Veith, C.; Boots, A.W.; et al. SARS-CoV-2 receptor ACE2 and TMPRSS2 are primarily expressed in bronchial transient secretory cells. EMBO J. 2020, 39, e105114. [Google Scholar] [CrossRef] [PubMed]
- Jackson, C.B.; Farzan, M.; Chen, B.; Choe, H. Mechanisms of SARS-CoV-2 entry into cells. Nat. Rev. Mol. Cell Biol. 2022, 23, 3–20. [Google Scholar] [CrossRef]
- Graham, D.K.; DeRyckere, D.; Davies, K.D.; Earp, H.S. The TAM family: Phosphatidylserine sensing receptor tyrosine kinases gone awry in cancer. Nat. Rev. Cancer 2014, 14, 769–785. [Google Scholar] [CrossRef]
- Varnum, B.C.; Young, C.; Elliott, G.; Garcia, A.; Bartley, T.D.; Fridell, Y.W.; Hunt, R.W.; Trail, G.; Clogston, C.; Toso, R.J.; et al. Axl receptor tyrosine kinase stimulated by the vitamin K-dependent protein encoded by growth-arrest-specific gene 6. Nature 1995, 373, 623–626. [Google Scholar] [CrossRef]
- Shieh, Y.S.; Lai, C.Y.; Kao, Y.R.; Shiah, S.G.; Chu, Y.W.; Lee, H.S.; Wu, C.W. Expression of axl in lung adenocarcinoma and correlation with tumor progression. Neoplasia 2005, 7, 1058–1064. [Google Scholar] [CrossRef]
- Bohan, D.; Van Ert, H.; Ruggio, N.; Rogers, K.J.; Badreddine, M.; Aguilar Briseno, J.A.; Elliff, J.M.; Rojas Chavez, R.A.; Gao, B.; Stokowy, T.; et al. Phosphatidylserine receptors enhance SARS-CoV-2 infection. PLoS Pathog. 2021, 17, e1009743. [Google Scholar] [CrossRef]
- Boytz, R.; Slabicki, M.; Ramaswamy, S.; Patten, J.J.; Zou, C.; Meng, C.; Hurst, B.L.; Wang, J.; Nowak, R.P.; Yang, P.L.; et al. Anti-SARS-CoV-2 activity of targeted kinase inhibitors: Repurposing clinically available drugs for COVID-19 therapy. J. Med. Virol. 2022, 95, e28157. [Google Scholar] [CrossRef]
- Wang, K.; Chen, W.; Zhang, Z.; Deng, Y.Q.; Lian, J.Q.; Du, P.; Wei, D.; Zhang, Y.; Sun, X.X.; Gong, L.; et al. CD147-spike protein is a novel route for SARS-CoV-2 infection to host cells. Signal Transduct. Target. Ther. 2020, 5, 283. [Google Scholar] [CrossRef] [PubMed]
- Shilts, J.; Crozier, T.W.M.; Greenwood, E.J.D.; Lehner, P.J.; Wright, G.J. No evidence for basigin/CD147 as a direct SARS-CoV-2 spike binding receptor. Sci. Rep. 2021, 11, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Rahimi, N. C-type Lectin CD209L/L-SIGN and CD209/DC-SIGN: Cell Adhesion Molecules Turned to Pathogen Recognition Receptors. Biology 2020, 10, 1. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Cao, J.; Zhang, X.; Gao, H.; Wang, Y.; Wang, J.; He, J.; Jiang, X.; Zhang, J.; Shen, G.; et al. Receptome profiling identifies KREMEN1 and ASGR1 as alternative functional receptors of SARS-CoV-2. Cell Res. 2021, 32, 24–37. [Google Scholar] [CrossRef]
- Cerchia, L.; Esposito, C.L.; Camorani, S.; Rienzo, A.; Stasio, L.; Insabato, L.; Affuso, A.; de Franciscis, V. Targeting Axl with an high-affinity inhibitory aptamer. Mol. Ther. 2012, 20, 2291–2303. [Google Scholar] [CrossRef]
- Li, N.; Nguyen, H.H.; Byrom, M.; Ellington, A.D. Inhibition of cell proliferation by an anti-EGFR aptamer. PLoS ONE 2011, 6, e20299. [Google Scholar] [CrossRef]
- Darabi, F.; Saidijam, M.; Nouri, F.; Mahjub, R.; Soleimani, M. Anti-CD44 and EGFR Dual-Targeted Solid Lipid Nanoparticles for Delivery of Doxorubicin to Triple-Negative Breast Cancer Cell Line: Preparation, Statistical Optimization, and In Vitro Characterization. Biomed. Res. Int. 2022, 2022, 6253978. [Google Scholar] [CrossRef]
- Passariello, M.; Camorani, S.; Vetrei, C.; Ricci, S.; Cerchia, L.; De Lorenzo, C. Ipilimumab and Its Derived EGFR Aptamer-Based Conjugate Induce Efficient NK Cell Activation against Cancer Cells. Cancers 2020, 12, 331. [Google Scholar] [CrossRef]
- Hwang, J.A.; Hur, J.Y.; Kim, Y.; Im, J.H.; Jin, S.H.; Ryu, S.H.; Choi, C.M. Efficacy of newly discovered DNA aptamers targeting AXL in a lung cancer cell with acquired resistance to Erlotinib. Transl. Cancer Res. 2021, 10, 1025–1033. [Google Scholar] [CrossRef]
- Esposito, C.L.; Passaro, D.; Longobardo, I.; Condorelli, G.; Marotta, P.; Affuso, A.; de Franciscis, V.; Cerchia, L. A neutralizing RNA aptamer against EGFR causes selective apoptotic cell death. PLoS ONE 2011, 6, e24071. [Google Scholar] [CrossRef]
- Camorani, S.; Crescenzi, E.; Gramanzini, M.; Fedele, M.; Zannetti, A.; Cerchia, L. Aptamer-mediated impairment of EGFR-integrin alphavbeta3 complex inhibits vasculogenic mimicry and growth of triple-negative breast cancers. Sci. Rep. 2017, 7, 46659. [Google Scholar] [CrossRef] [PubMed]
- Booth, A.; Reed, A.B.; Ponzo, S.; Yassaee, A.; Aral, M.; Plans, D.; Labrique, A.; Mohan, D. Population risk factors for severe disease and mortality in COVID-19: A global systematic review and meta-analysis. PLoS ONE 2021, 16, e0247461. [Google Scholar] [CrossRef] [PubMed]
- Shaath, H.; Vishnubalaji, R.; Elkord, E.; Alajez, N.M. Single-Cell Transcriptome Analysis Highlights a Role for Neutrophils and Inflammatory Macrophages in the Pathogenesis of Severe COVID-19. Cells 2020, 9, 2374. [Google Scholar] [CrossRef] [PubMed]
- Brendel, M.; Su, C.; Bai, Z.; Zhang, H.; Elemento, O.; Wang, F. Application of Deep Learning on Single-cell RNA Sequencing Data Analysis: A Review. Genom. Proteom. Bioinform. 2022, 20, 814–835. [Google Scholar] [CrossRef]
- Zhang, J.-Y.; Wang, X.-M.; Xing, X.; Xu, Z.; Zhang, C.; Song, J.-W.; Fan, X.; Xia, P.; Fu, J.-L.; Wang, S.-Y.; et al. Single-cell landscape of immunological responses in patients with COVID-19. Nat. Immunol. 2020, 21, 1107–1118. [Google Scholar] [CrossRef]
- Lee, T.; Kim, Y.; Kim, H.J.; Ha, N.Y.; Lee, S.; Chin, B.; Cho, N.H. Acute Surge of Atypical Memory and Plasma B-Cell Subsets Driven by an Extrafollicular Response in Severe COVID-19. Front. Cell Infect. Microbiol. 2022, 12, 909218. [Google Scholar] [CrossRef]
- Joshee, S.; Vatti, N.; Chang, C. Long-Term Effects of COVID-19. Mayo Clin. Proc. 2022, 97, 579–599. [Google Scholar] [CrossRef]
- Pathak, E.; Atri, N.; Mishra, R. Single-Cell Transcriptome Analysis Reveals the Role of Pancreatic Secretome in COVID-19 Associated Multi-organ Dysfunctions. Interdiscip. Sci. 2022, 14, 863–878. [Google Scholar] [CrossRef]
- Guo, C.; Li, B.; Ma, H.; Wang, X.; Cai, P.; Yu, Q.; Zhu, L.; Jin, L.; Jiang, C.; Fang, J.; et al. Single-cell analysis of two severe COVID-19 patients reveals a monocyte-associated and tocilizumab-responding cytokine storm. Nat. Commun. 2020, 11, 3924. [Google Scholar] [CrossRef]
- Xiang, J.; Lu, M.; Shi, M.; Cheng, X.; Kwakwa, K.A.; Davis, J.L.; Su, X.; Bakewell, S.J.; Zhang, Y.; Fontana, F.; et al. Heparanase Blockade as a Novel Dual-Targeting Therapy for COVID-19. J. Virol. 2022, 96, e0005722. [Google Scholar] [CrossRef]
- Zhang, Z.; Cui, F.; Cao, C.; Wang, Q.; Zou, Q. Single-cell RNA analysis reveals the potential risk of organ-specific cell types vulnerable to SARS-CoV-2 infections. Comput. Biol. Med. 2021, 140, 105092. [Google Scholar] [CrossRef] [PubMed]

| Assay | Mechanism | Specificity and Sensitivity | Advantages | Disadvantages | References |
|---|---|---|---|---|---|
| RT-PCR | Based on the polymerase chain reaction | Sensitivity: 92.7% (4–5 days after infection), 88% (5–14 days after infection), and specificity ~100% | The best available diagnostic test enables the qualitative and quantitative determination of the COVID-19 virus in the downloaded material | -Expensive -Risk of false negative or false positive results -Requires special equipment -Not fast as antigen tests | [102,103] |
| Antibody | Monoclonal antibodies detect the host’s antibodies (usually directed against two virus proteins, S and N) | Depends on the detected antibody; average specificity: 75–90% and sensitivity: 77–90% | Helps to detect who previously had COVID-19 or evaluate how the vaccine affected the immune response | -Gives information only about the number of antibodies in the patient’s blood -Does not provide information about how strong the infection is -Does not detect antibodies in early stages of the disease -The level of antibodies does not correspond to the stage of the disease -Risk of cross-reactions and similarity to the SARS-CoV-2 virus sequence to other viruses | [101,104,105] |
| Antigen | Detect SARS-CoV-2 proteins | Specificity: 55–100% Sensitivity: 30–100% | Fast time to obtain results, does not require expensive equipment, and is cheap | -False-positive results in patients without symptoms -Accuracy of tests is unknown -These tests are not approved by the WHO for diagnosis | [106] |
| Aptamers | Recognize S and N proteins of SARS-CoV-2 | Depending on the aptamer, sensitivity, and specificity are comparable with RT-PCR | Cheaper than RT_PCR, detects virus in early stages, detects small amounts of the virus in the sample, does not require extraction and amplification of the virus’s genetic material, and low cost of tests | -Expensive cost of synthesis aptamers on a big scale (kilograms) | [107,108,109] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karadeniz Saygılı, S.; Szymanowska, A.; Lopez-Berestein, G.; Rodriguez-Aguayo, C.; Amero, P. Aptamers as Insights for Targeting SARS-CoV-2. Biologics 2023, 3, 116-137. https://doi.org/10.3390/biologics3020007
Karadeniz Saygılı S, Szymanowska A, Lopez-Berestein G, Rodriguez-Aguayo C, Amero P. Aptamers as Insights for Targeting SARS-CoV-2. Biologics. 2023; 3(2):116-137. https://doi.org/10.3390/biologics3020007
Chicago/Turabian StyleKaradeniz Saygılı, Suna, Anna Szymanowska, Gabriel Lopez-Berestein, Cristian Rodriguez-Aguayo, and Paola Amero. 2023. "Aptamers as Insights for Targeting SARS-CoV-2" Biologics 3, no. 2: 116-137. https://doi.org/10.3390/biologics3020007
APA StyleKaradeniz Saygılı, S., Szymanowska, A., Lopez-Berestein, G., Rodriguez-Aguayo, C., & Amero, P. (2023). Aptamers as Insights for Targeting SARS-CoV-2. Biologics, 3(2), 116-137. https://doi.org/10.3390/biologics3020007








