Expression of Cowpea VuWRKY21 and VuWRKY87 Genes in Arabidopsis thaliana Confers Plant Tolerance to Salt Stress
Abstract
:1. Introduction
2. Materials and Methods
2.1. Gene Cloning and Plant Transformation
2.2. Subcellular Location, Molecular Weight (kDa), and Isoelectric Point of VuWRKY21 and VuWRKY87 Proteins
2.3. Establishment and Generation Advance of Transformed A. thaliana
2.4. Germination and Root Elongation under Salt Stress
2.5. Exposure to Salt Stress
2.6. Hydrogen Peroxide (H2O2) and Malondialdehyde (MDA) Quantification
2.7. Enzymatic Tests and Quantification of Soluble Proteins
2.8. RNA Extraction, cDNA Synthesis, and Analysis of the Differential Expression by qPCR
2.9. Statistical Analyses of the Relative Gene Expression
3. Results
3.1. Subcellular Location, Molecular Weight (kDa), and Isoelectric Point of VuWRKY21 and VuWRKY87 Proteins
3.2. Seed Germination and Root Elongation Tests
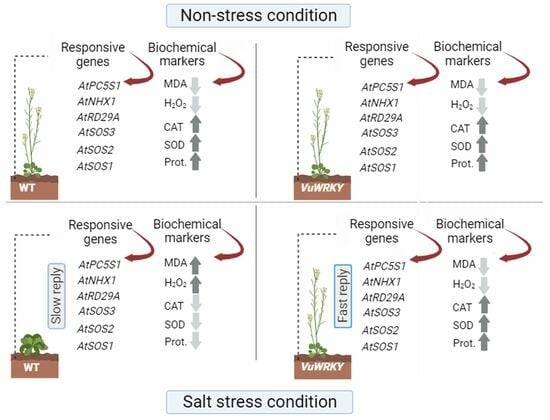
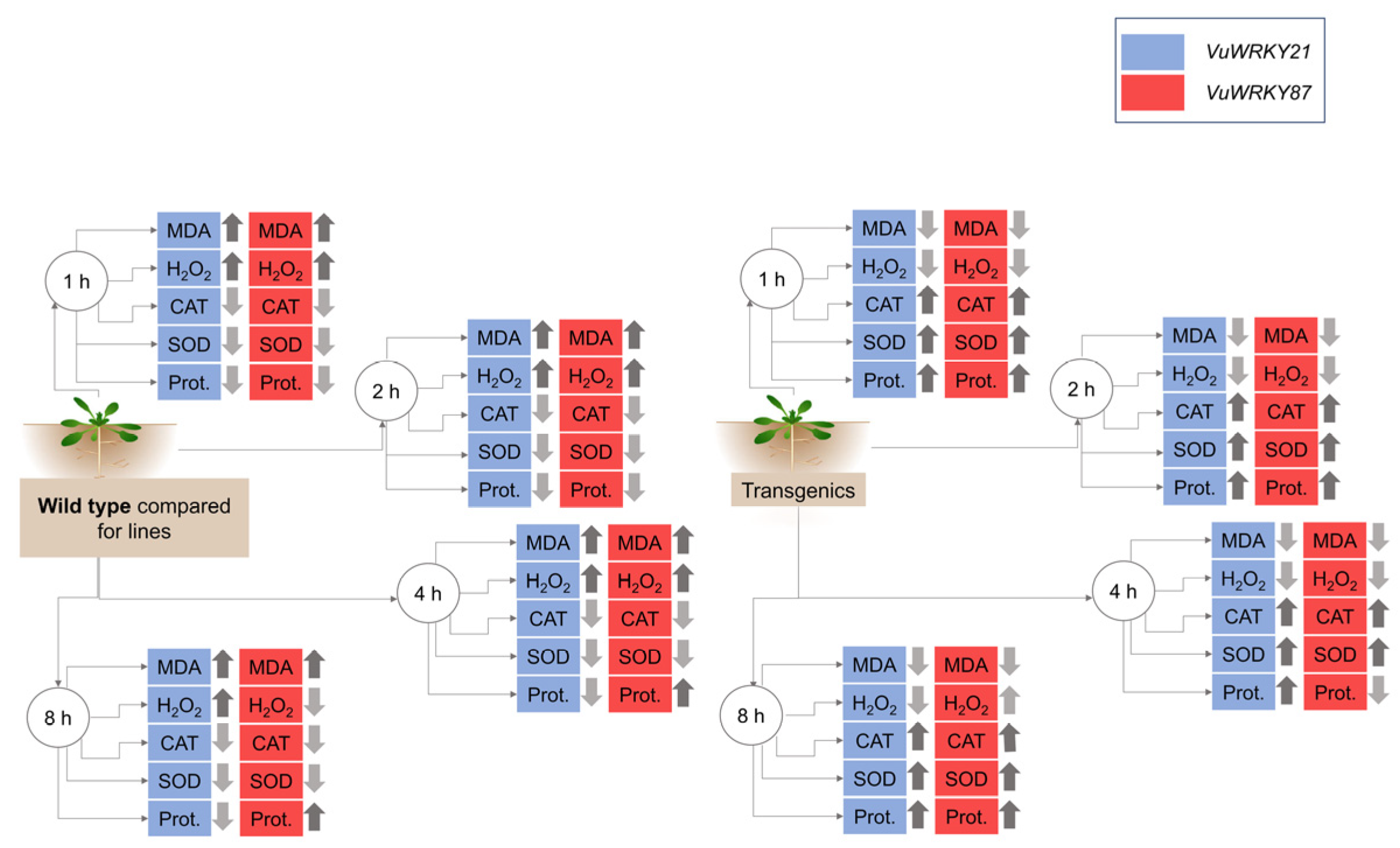
3.3. Biochemical Responses
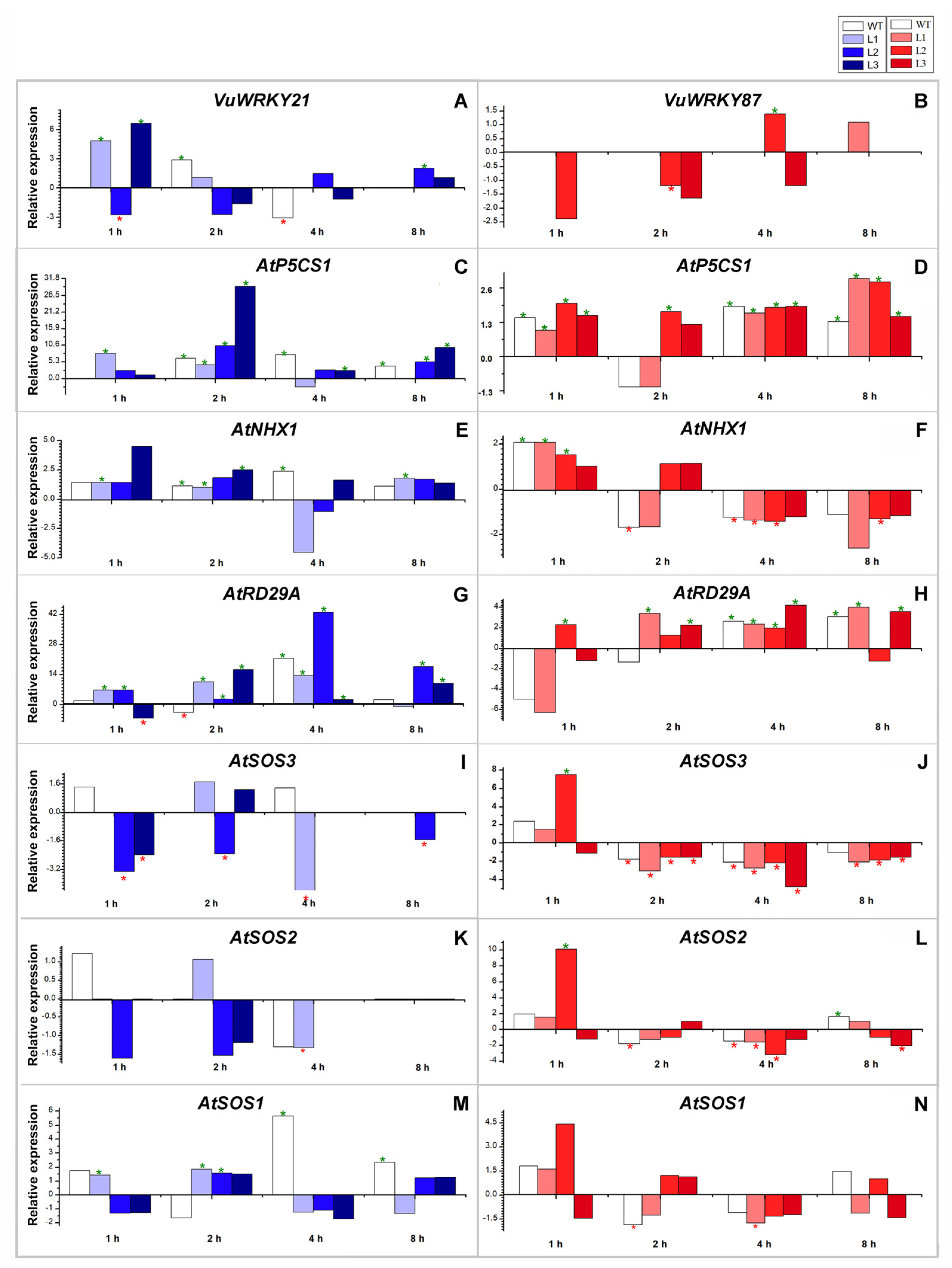
3.4. Effects of VuWRKY21 and VuWRKY87 Genes in Arabidopsis under Salt Stress
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Abiala, M.A.; Abdelrahman, M.; Burritt, D.J.; Tran, L.P. Salt stress tolerance mechanisms and potential applications of legumes for sustainable reclamation of salt-degraded soils. Land Degrad. Dev. 2018, 29, 3812–3822. [Google Scholar] [CrossRef]
- Brozynska, M.; Furtado, A.; Henry, R.J. Genomics of crop wild relatives: Expanding the gene pool for crop improvement. Plant Biotechnol. J. 2016, 14, 1070–1085. [Google Scholar] [CrossRef]
- Ravelombola, W.; Shi, A.; Weng, Y.; Mou, B.; Motes, D.; Clark, J.; Chen, P.; Srivastava, V.; Qin, J.; Dong, L.; et al. Association analysis of salt tolerance in cowpea (Vigna unguiculata (L.) Walp) at germination and seedling stages. Theor. Appl. Genet. 2018, 131, 79–91. [Google Scholar] [CrossRef]
- Merwad, A.-R.M.A.; Desoky, E.-S.M.; Rady, M.M. Response of water deficit-stressed Vigna unguiculata performances to silicon, proline or methionine foliar application. Sci. Hortic. 2018, 228, 132–144. [Google Scholar] [CrossRef]
- Joshi, R.; Wani, S.H.; Singh, B.; Bohra, A.; Dar, Z.A.; Lone, A.A.; Pareek, A.; Singla-Pareek, S.L. Transcription factors and plants response to drought stress: Current understanding and future directions. Front. Plant Sci. 2016, 7, 1029. [Google Scholar] [CrossRef] [PubMed]
- Maruyama, K.; Urano, K.; Yoshiwara, K.; Morishita, Y.; Sakurai, N.; Suzuki, H.; Kojima, M.; Sakakibara, H.; Shibata, D.; Saito, K.; et al. Integrated analysis of the effects of cold and dehydration on rice metabolites, phytohormones, and gene transcripts. Plant Physiol. 2014, 164, 1759–1771. [Google Scholar] [CrossRef]
- Dong, J.; Chen, C.; Chen, Z. Expression profiles of the Arabidopsis WRKY gene superfamily during plant defense response. Plant Mol. Biol. 2003, 51, 21–37. [Google Scholar] [CrossRef]
- Dong, Q.; Zheng, W.; Duan, D.; Huang, D.; Wang, Q.; Liu, C.; Li, C.; Gong, X.; Li, C.; Mao, K.; et al. MdWRKY30, a group IIa WRKY gene from apple, confers tolerance to salinity and osmotic stresses in transgenic apple callus and Arabidopsis seedlings. Plant Sci. 2020, 299, 110611. [Google Scholar] [CrossRef]
- Msanne, J.; Lin, J.; Stone, J.M.; Awada, T. Characterization of abiotic stress-responsive Arabidopsis thaliana RD29A and RD29B genes and evaluation of transgenes. Planta 2011, 234, 97–107. [Google Scholar] [CrossRef] [PubMed]
- Ullah, A.; Sun, H.; Hakim; Yang, X.; Zhang, X. A novel cotton WRKY gene, GhWRKY6-like, improves salt tolerance by activating the ABA signaling pathway and scavenging of reactive oxygen species. Physiol. Plant. 2018, 162, 439–454. [Google Scholar] [CrossRef]
- Li, X.; Tang, Y.; Zhou, C.; Zhang, L.; Lv, J. A wheat WRKY transcription factor TaWRKY46 enhances tolerance to osmotic stress in transgenic Arabidopsis plants. Int. J. Mol. Sci. 2020, 21, 1321. [Google Scholar] [CrossRef] [PubMed]
- Amin, I.; Rasool, S.; Mir, M.A.; Wani, W.; Masoodi, K.Z.; Ahmad, P. Ion homeostasis for salinity tolerance in plants: A molecular approach. Physiol. Plant. 2021, 171, 578–594. [Google Scholar] [CrossRef] [PubMed]
- Dai, W.; Wang, M.; Gong, X.; Liu, J. The transcription factor FcWRKY 40 of Fortunella crassifolia functions positively in salt tolerance through modulation of ion homeostasis and proline biosynthesis by directly regulating SOS2 and P5CS1 homologs. New Phytol. 2018, 219, 972–989. [Google Scholar] [CrossRef] [PubMed]
- Brady, S.M.; Orlando, D.A.; Lee, J.-Y.; Wang, J.Y.; Koch, J.; Dinneny, J.R.; Mace, D.; Ohler, U.; Benfey, P.N. A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 2007, 318, 801–806. [Google Scholar] [CrossRef]
- Tang, R.-J.; Yang, Y.; Yang, L.; Liu, H.; Wang, C.-T.; Yu, M.-M.; Gao, X.-S.; Zhang, H.-X. Poplar calcineurin B-like proteins PtCBL10A and PtCBL10B regulate shoot salt tolerance through interaction with PtSOS2 in the vacuolar membrane. Plant Cell Environ. 2014, 37, 573–588. [Google Scholar] [CrossRef]
- Lv, B.; Wu, Q.; Wang, A.; Li, Q.; Dong, Q.; Yang, J.; Zhao, H.; Wang, X.; Chen, H.; Li, C. A WRKY transcription factor, FtWRKY46, from Tartary buckwheat improves salt tolerance in transgenic Arabidopsis thaliana. Plant Physiol. Biochem. 2020, 147, 43–53. [Google Scholar] [CrossRef]
- Qin, Y.; Tian, Y.; Liu, X. A wheat salinity-induced WRKY transcription factor TaWRKY93 confers multiple abiotic stress tolerance in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 2015, 464, 428–433. [Google Scholar] [CrossRef]
- Zhu, L.; Li, S.; Ouyang, M.; Yang, L.; Sun, S.; Wang, Y.; Cai, X.; Wu, G.; Li, Y. Overexpression of watermelon ClWRKY20 in transgenic Arabidopsis improves salt and low-temperature tolerance. Sci. Hortic. 2022, 295, 110848. [Google Scholar] [CrossRef]
- Wu, M.; Zhang, K.; Xu, Y.; Wang, L.; Liu, H.; Qin, Z.; Xiang, Y. The moso bamboo WRKY transcription factor, PheWRKY86, regulates drought tolerance in transgenic plants. Plant Physiol. Biochem. 2022, 170, 180–191. [Google Scholar] [CrossRef]
- Zhu, H.; Jiang, Y.; Guo, Y.; Huang, J.; Zhou, M.; Tang, Y.; Sui, J.; Wang, J.; Qiao, L. A novel salt inducible WRKY transcription factor gene, AhWRKY75, confers salt tolerance in transgenic peanut. Plant Physiol. Biochem. 2021, 160, 175–183. [Google Scholar] [CrossRef]
- Kang, G.; Yan, D.; Chen, X.; Yang, L.; Zeng, R. HbWRKY82, a novel IIc WRKY transcription factor from Hevea brasiliensis associated with abiotic stress tolerance and leaf senescence in Arabidopsis. Physiol. Plant. 2021, 171, 151–160. [Google Scholar] [CrossRef]
- Baptista, A.; Pinho, O.; Pinto, E.; Casal, S.; Mota, C.; Ferreira, I.M.P.L.V.O. Characterization of protein and fat composition of seeds from common beans (Phaseolus vulgaris L.), cowpea (Vigna unguiculata L. Walp) and bambara groundnuts (Vigna subterranea L. Verdc) from Mozambique. J. Food Meas. Charact. 2017, 11, 442–450. [Google Scholar] [CrossRef]
- Amorim, M.R.; Mendes, L.W.; Antunes, J.E.L.; Oliveira, L.M.d.S.; Melo, V.M.M.; Oliveira, F.A.S.; Aquino, J.P.A.; Rocha, S.M.B.; Pereira, A.P.d.A.; da Costa, A.F.; et al. Cowpea nodules host a similar bacterial community regardless of soil properties. Appl. Soil Ecol. 2022, 172, 104354. [Google Scholar] [CrossRef]
- Masete, F.M.; Munjonji, L.; Ayisi, K.K.; Mopape-Mabapa, M.P. Cowpea growth and nitrogen fixation performance under different mulch treatments. Agriculture 2022, 12, 1144. [Google Scholar] [CrossRef]
- Opoku, V.A.; Yawson, D.O.; Asare, P.A.; Afutu, E.; Kotochi, M.C.; Amoah, K.K.; Adu, M.O. Root hair and rhizosheath traits contribute to genetic variation and phosphorus use efficiency in cowpea (Vigna unguiculata (L.) Walp). Rhizosphere 2022, 21, 100463. [Google Scholar] [CrossRef]
- Abdelrahman, M.; Jogaiah, S.; Burritt, D.J.; Tran, L.-S.P. Legume genetic resources and transcriptome dynamics under abiotic stress conditions. Plant Cell Environ. 2018, 41, 1972–1983. [Google Scholar] [CrossRef] [PubMed]
- Matos, M.K.d.S.; Benko-Iseppon, A.M.; Bezerra-Neto, J.P.; Ferreira-Neto, J.R.C.; Wang, Y.; Liu, H.; Pandolfi, V.; Amorim, L.L.B.; Willadino, L.; do Vale Amorim, T.C.; et al. The WRKY transcription factor family in cowpea: Genomic characterization and transcriptomic profiling under root dehydration. Gene 2022, 823, 146377. [Google Scholar] [CrossRef]
- Singh, D.; Debnath, P.; Sane, A.P.; Sane, V.A. Tomato (Solanum lycopersicum) WRKY23 enhances salt and osmotic stress tolerance by modulating the ethylene and auxin pathways in transgenic Arabidopsis. Plant Physiol. Biochem. 2023, 195, 330–340. [Google Scholar] [CrossRef]
- Nakagawa, T.; Suzuki, T.; Murata, S.; Nakamura, S.; Hino, T.; Maeo, K.; Tabata, R.; Kawai, T.; Tanaka, K.; Niwa, Y.; et al. Improved Gateway binary vectors: High-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci. Biotechnol. Biochem. 2007, 71, 2095–2100. [Google Scholar] [CrossRef] [PubMed]
- Bechtold, N.; Bouchez, D. In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. In Gene Transfer to Plants; Springer: Berlin/Heidelberg, Germany, 1995; pp. 19–23. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Chou, K.-C.; Shen, H.-B. Cell-PLoc 2.0: An improved package of web-servers for predicting subcellular localization of proteins in various organisms. Nat. Sci. 2010, 2, 1090–1103. [Google Scholar] [CrossRef]
- Hiller, K.; Grote, A.; Maneck, M.; Münch, R.; Jahn, D. JVirGel 2.0: Computational prediction of proteomes separated via two-dimensional gel electrophoresis under consideration of membrane and secreted proteins. Bioinformatics 2006, 22, 2441–2443. [Google Scholar] [CrossRef] [PubMed]
- Murashige, T.; Skoog, F. A Revised Medium for Rapid Growth and Bio Assays with Tobacco Tissue Cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Alexieva, V.; Sergiev, I.; Mapelli, S.; Karanov, E. The effect of drought and ultraviolet radiation on growth and stress markers in pea and wheat. Plant Cell Environ. 2001, 24, 1337–1344. [Google Scholar] [CrossRef]
- Heath, R.L.; Packer, L. Photoperoxidation in isolated chloroplasts. Arch. Biochem. Biophys. 1968, 125, 189–198. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Havir, E.A.; McHale, N.A. Biochemical and developmental characterization of multiple forms of catalase in tobacco leaves. Plant Physiol. 1987, 84, 450–455. [Google Scholar] [CrossRef]
- Giannopolitis, C.N.; Ries, S.K. Superoxide Dismutases: I. Occurrence in higher plants. Plant Physiol. 1977, 59, 309–314. [Google Scholar] [CrossRef]
- Zhao, L.; Ding, Q.; Zeng, J.; Wang, F.-R.; Zhang, J.; Fan, S.-J.; He, X.-Q. An improved CTAB-ammonium acetate method for total RNA isolation from cotton. Phytochem. Anal. 2012, 23, 647–650. [Google Scholar] [CrossRef]
- Raso, A.; Biassoni, R. Twenty Years of qPCR: A Mature Technology? Springer: Berlin/Heidelberg, Germany, 2014; pp. 1–3. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Cruz, C.D. Genes: A software package for analysis in experimental statistics and quantitative genetics. Acta Sci. Agron. 2013, 35, 271–276. [Google Scholar] [CrossRef]
- Pfaffl, M.W. Relative expression software tool (REST(C)) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002, 30, e36. [Google Scholar] [CrossRef] [PubMed]
- Schmid, M.; Davison, T.S.; Henz, S.R.; Pape, U.J.; Demar, M.; Vingron, M.; Scholkopf, B.; Weigel, D.; Lohmann, J.U. A gene expression map of Arabidopsis thaliana development. Nat. Genet. 2005, 37, 501–506. [Google Scholar] [CrossRef] [PubMed]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [PubMed]
- Bandeoğlu, E.; Eyidoğan, F.; Yücel, M.; Avni Öktem, H. Antioxidant responses of shoots and roots of lentil to NaCl-salinity stress. Plant Growth Regul. 2004, 42, 69–77. [Google Scholar] [CrossRef]
- Shafiq, S.; Akram, N.A.; Ashraf, M. Assessment of physio-biochemical indicators for drought tolerance in different cultivars of maize (Zea mays L.). Pak. J. Bot. 2019, 51, 1241–1247. [Google Scholar] [CrossRef]
- Zhou, Q.-Y.; Tian, A.-G.; Zou, H.-F.; Xie, Z.-M.; Lei, G.; Huang, J.; Wang, C.-M.; Wang, H.-W.; Zhang, J.-S.; Chen, S.-Y. Soybean WRKY-type transcription factor genes, GmWRKY13, GmWRKY21, and GmWRKY54, confer differential tolerance to abiotic stresses in transgenic Arabidopsis plants. Plant Biotechnol. J. 2008, 6, 486–503. [Google Scholar] [CrossRef]
- Anwar, K.; Joshi, R.; Dhankher, O.P.; Singla-Pareek, S.L.; Pareek, A. Elucidating the response of crop plants towards individual, combined and sequentially occurring abiotic stresses. Int. J. Mol. Sci. 2021, 22, 6119. [Google Scholar] [CrossRef]
- Silva, P.C.C.; Azevedo Neto, A.D.; Gheyi, H.R.; Ribas, R.F.; Silva, C.R.R.; Cova, A.M.W. Seed priming with H2O2 improves photosynthetic efficiency and biomass production in sunflower plants under salt stress. Arid. Land Res. Manag. 2022, 36, 283–297. [Google Scholar] [CrossRef]
- Mansoor, S.; Ali Wani, O.; Lone, J.K.; Manhas, S.; Kour, N.; Alam, P.; Ahmad, A.; Ahmad, P. Reactive oxygen species in plants: From source to sink. Antioxidants 2022, 11, 225. [Google Scholar] [CrossRef]
- Foyer, C.H.; Noctor, G. Redox regulation in photosynthetic organisms: Signaling, acclimation, and practical implications. Antioxid. Redox Signal. 2009, 11, 861–905. [Google Scholar] [CrossRef] [PubMed]
- Foyer, C.H.; Noctor, G. Redox homeostasis and antioxidant signaling: A metabolic interface between stress perception and physiological responses. Plant Cell 2005, 17, 1866–1875. [Google Scholar] [CrossRef]
- Cavalcante Granja, M.M.; Lacerda e Medeiros, M.J.; Medeiros de Araújo Silva, M.; Camara, T.; Willadino, L.; Ulisses, C. Response to in vitro salt stress in sugarcane is conditioned by concentration and condition of exposure to NaCl. Acta Biológica Colomb. 2018, 23, 30–38. [Google Scholar] [CrossRef]
- Ahanger, M.A.; Tomar, N.S.; Tittal, M.; Argal, S.; Agarwal, R.M. Plant growth under water/salt stress: ROS production; antioxidants and significance of added potassium under such conditions. Physiol. Mol. Biol. Plants 2017, 23, 731–744. [Google Scholar] [CrossRef] [PubMed]
- Willadino, L.; Camara, T.R.; Ribeiro, M.B.; Amaral, D.O.J.D.; Suassuna, F.; Silva, M.V.D. Mechanisms of tolerance to salinity in banana: Physiological, biochemical, and molecular aspects. Rev. Bras. Frutic. 2017, 39, e-723. [Google Scholar] [CrossRef]
- Zulfiqar, F.; Ashraf, M. Bioregulators: Unlocking their potential role in regulation of the plant oxidative defense system. Plant Mol. Biol. 2021, 105, 11–41. [Google Scholar] [CrossRef]
- Singh, R.; Misra, A.N.; Sharma, P. Genome–wide transcriptional response of contrasting genotypes of industrial crop castor to As(V) stress: Identification of genes and mechanisms associated with As(V) tolerance. Ind. Crops Prod. 2022, 179, 114678. [Google Scholar] [CrossRef]
- Akbari, B.; Baghaei-Yazdi, N.; Bahmaie, M.; Mahdavi Abhari, F. The role of plant-derived natural antioxidants in reduction of oxidative stress. BioFactors 2022, 48, 611–633. [Google Scholar] [CrossRef]
- Di Carli, M.; Benvenuto, E.; Donini, M. Recent insights into plant–virus interactions through proteomic analysis. J. Proteome Res. 2012, 11, 4765–4780. [Google Scholar] [CrossRef]
- Das, P.; Majumder, A.L. Transcriptome analysis of grapevine under salinity and identification of key genes responsible for salt tolerance. Funct. Integr. Genom. 2019, 19, 61–73. [Google Scholar] [CrossRef]
- Joshi, R.; Sahoo, K.K.; Tripathi, A.K.; Kumar, R.; Gupta, B.K.; Pareek, A.; Singla-Pareek, S.L. Knockdown of an inflorescence meristem-specific cytokinin oxidase–OsCKX2 in rice reduces yield penalty under salinity stress condition. Plant Cell Environ. 2018, 41, 936–946. [Google Scholar] [CrossRef] [PubMed]
- Wani, S.H.; Anand, S.; Singh, B.; Bohra, A.; Joshi, R. WRKY transcription factors and plant defense responses: Latest discoveries and future prospects. Plant Cell Rep. 2021, 40, 1071–1085. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Li, H.; Yang, Y.; Wang, Y.; Mo, Y.; Zhang, R.; Zhang, Y.; Ma, J.; Wei, C.; Zhang, X. Identification and expression analyses of WRKY genes reveal their involvement in growth and abiotic stress response in watermelon (Citrullus lanatus). PLoS ONE 2018, 13, e0191308. [Google Scholar] [CrossRef] [PubMed]
- Shang, Y.; Yan, L.; Liu, Z.-Q.; Cao, Z.; Mei, C.; Xin, Q.; Wu, F.-Q.; Wang, X.-F.; Du, S.-Y.; Jiang, T.; et al. The Mg-Chelatase H subunit of Arabidopsis antagonizes a group of WRKY transcription repressors to relieve ABA-responsive genes of inhibition. Plant Cell 2010, 22, 1909–1935. [Google Scholar] [CrossRef]
- Turchetto-Zolet, A.C.; Margis-Pinheiro, M.; Margis, R. The evolution of pyrroline-5-carboxylate synthase in plants: A key enzyme in proline synthesis. Mol. Genet. Genom. 2009, 281, 87–97. [Google Scholar] [CrossRef]
- Chen, W.; Zheng, C.; Yao, M.; Chen, L. The tea plant CsWRKY26 promotes drought tolerance in transgenic Arabidopsis plants. Beverage Plant Res. 2021, 1, 3. [Google Scholar] [CrossRef]
- Garg, G.; Neha, P. Plant transcription factors networking of pyrroline-5-carboxylate (p5c) enzyme under stress condition: A review. Plant Arch. 2019, 19, 562–569. [Google Scholar]
- Furlan, A.L.; Bianucci, E.; Giordano, W.; Castro, S.; Becker, D.F. Proline metabolic dynamics and implications in drought tolerance of peanut plants. Plant Physiol. Biochem. 2020, 151, 566–578. [Google Scholar] [CrossRef]
- Zhu, D.; Hou, L.; Xiao, P.; Guo, Y.; Deyholos, M.K.; Liu, X. VvWRKY30, a grape WRKY transcription factor, plays a positive regulatory role under salinity stress. Plant Sci. 2019, 280, 132–142. [Google Scholar] [CrossRef]
- Thiery, L.; Leprince, A.-S.; Lefebvre, D.; Ghars, M.A.; Debarbieux, E.; Savouré, A. Phospholipase D is a negative regulator of proline biosynthesis in Arabidopsis thaliana. J. Biol. Chem. 2004, 279, 14812–14818. [Google Scholar] [CrossRef]
- Li, P.; Song, A.; Gao, C.; Wang, L.; Wang, Y.; Sun, J.; Jiang, J.; Chen, F.; Chen, S. Chrysanthemum WRKY gene CmWRKY17 negatively regulates salt stress tolerance in transgenic Chrysanthemum and Arabidopsis plants. Plant Cell Rep. 2015, 34, 1365–1378. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Wang, N.-N.; Gong, S.-Y.; Lu, R.; Li, Y.; Li, X.-B. Overexpression of a cotton (Gossypium hirsutum) WRKY gene, GhWRKY34, in Arabidopsis enhances salt-tolerance of the transgenic plants. Plant Physiol. Biochem. 2015, 96, 311–320. [Google Scholar] [CrossRef] [PubMed]
- Julkowska, M.M.; Testerink, C. Tuning plant signaling and growth to survive salt. Trends Plant Sci. 2015, 20, 586–594. [Google Scholar] [CrossRef] [PubMed]
- Ismail, A.M.; Horie, T. Genomics, physiology, and molecular breeding approaches for improving salt tolerance. Annu. Rev. Plant Biol. 2017, 68, 405–434. [Google Scholar] [CrossRef] [PubMed]
- Hussain, S.; Ali, B.; Ren, X.; Chen, X.; Li, Q.; Saqib, M.; Ahmad, N. Recent progress in understanding salinity tolerance in plants: Story of Na+/K+ balance and beyond. Plant Physiol. Biochem. 2021, 160, 239–256. [Google Scholar] [CrossRef] [PubMed]
- Pabuayon, I.C.M.; Jiang, J.; Qian, H.; Chung, J.-S.; Shi, H. Gain-of-function mutations of AtNHX1 suppress sos1 salt sensitivity and improve salt tolerance in Arabidopsis. Stress Biol. 2021, 1, 14. [Google Scholar] [CrossRef]
- Köster, P.; Wallrad, L.; Edel, K.H.; Faisal, M.; Alatar, A.A.; Kudla, J. The battle of two ions: Ca2+ signalling against Na+ stress. Plant Biol. 2019, 21, 39–48. [Google Scholar] [CrossRef]
- Yousefirad, S.; Soltanloo, H.; Ramezanpour, S.S.; Zaynali Nezhad, K.; Shariati, V. The RNA-seq transcriptomic analysis reveals genes mediating salt tolerance through rapid triggering of ion transporters in a mutant barley. PLoS ONE 2020, 15, e0229513. [Google Scholar] [CrossRef]
- Procino, G.; Gerbino, A.; Milano, S.; Nicoletti, M.C.; Mastrofrancesco, L.; Carmosino, M.; Svelto, M. Rosiglitazone promotes AQP2 plasma membrane expression in renal cells via a Ca2+-dependent/cAMP-independent mechanism. Cell Physiol. Biochem. 2015, 35, 1070–1085. [Google Scholar] [CrossRef]
- Zhu, J.-K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002, 53, 247–273. [Google Scholar] [CrossRef]
- Goyal, V.; Jhanghel, D.; Mehrotra, S. Emerging warriors against salinity in plants: Nitric oxide and hydrogen sulphide. Physiol. Plant 2021, 171, 896–908. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Crispim, J.G.; Souza, E.d.S.; Antunes, M.F.K.; Liu, H.; Pandolfi, V.; Morais, M.B.d.; Sun, L.; Ulisses, C.; Rabara, R.C.; Ferreira-Neto, J.R.C.; et al. Expression of Cowpea VuWRKY21 and VuWRKY87 Genes in Arabidopsis thaliana Confers Plant Tolerance to Salt Stress. DNA 2023, 3, 168-185. https://doi.org/10.3390/dna3040014
Crispim JG, Souza EdS, Antunes MFK, Liu H, Pandolfi V, Morais MBd, Sun L, Ulisses C, Rabara RC, Ferreira-Neto JRC, et al. Expression of Cowpea VuWRKY21 and VuWRKY87 Genes in Arabidopsis thaliana Confers Plant Tolerance to Salt Stress. DNA. 2023; 3(4):168-185. https://doi.org/10.3390/dna3040014
Chicago/Turabian StyleCrispim, Joelson Germano, Elenilson dos Santos Souza, Marina Ferreira Kitazono Antunes, Hai Liu, Valesca Pandolfi, Marciana Bizerra de Morais, Lili Sun, Cláudia Ulisses, Roel Collamat Rabara, José Ribamar Costa Ferreira-Neto, and et al. 2023. "Expression of Cowpea VuWRKY21 and VuWRKY87 Genes in Arabidopsis thaliana Confers Plant Tolerance to Salt Stress" DNA 3, no. 4: 168-185. https://doi.org/10.3390/dna3040014
APA StyleCrispim, J. G., Souza, E. d. S., Antunes, M. F. K., Liu, H., Pandolfi, V., Morais, M. B. d., Sun, L., Ulisses, C., Rabara, R. C., Ferreira-Neto, J. R. C., Benko-Iseppon, A. M., Timko, M. P., & Brasileiro-Vidal, A. C. (2023). Expression of Cowpea VuWRKY21 and VuWRKY87 Genes in Arabidopsis thaliana Confers Plant Tolerance to Salt Stress. DNA, 3(4), 168-185. https://doi.org/10.3390/dna3040014