Efficient Elimination of mtDNA from Mammalian Cells with 2′,3′-Dideoxycytidine
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines and Culture Conditions
2.2. Determination of mtDNA Copy Number (mtCN) by Direct Digital Droplet PCR
2.3. Clonal Isolation of ρ0 Cells and Confirmation of the ρ0 Status
3. Results
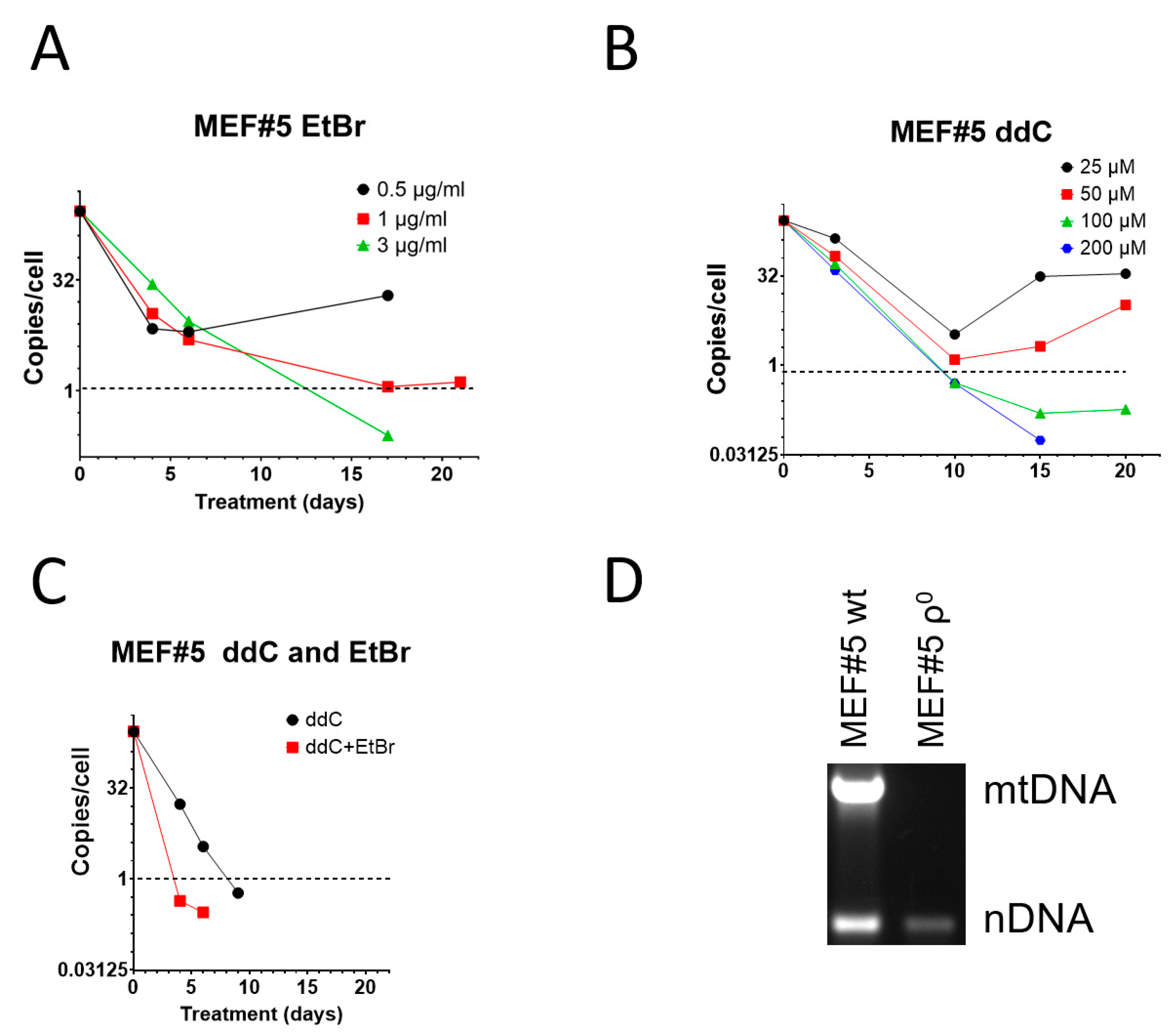
3.1. ddC Is More Effective than EtBr in mtDNA Elimination from Immortalized Mouse Embryonic Fibroblasts
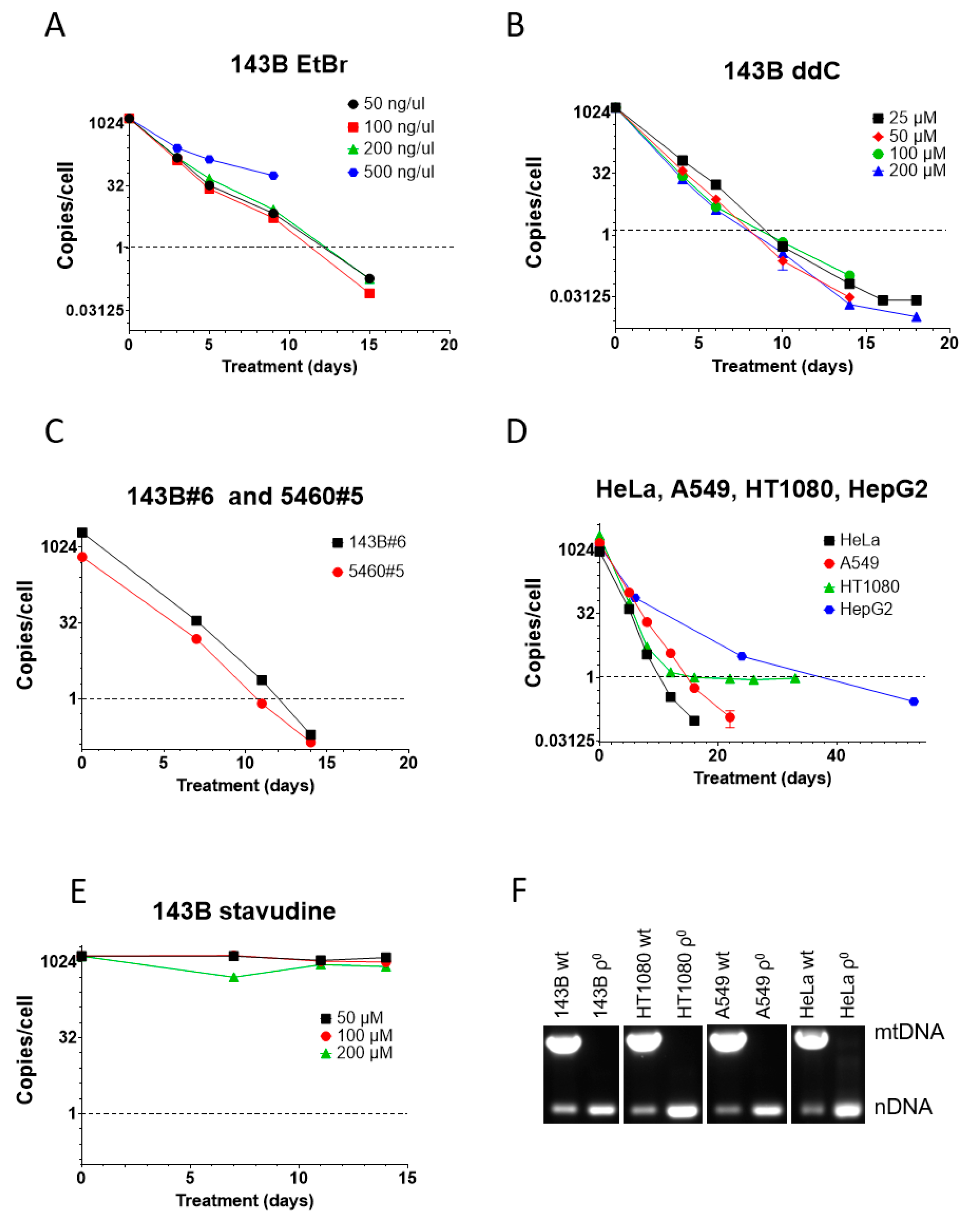
3.2. ddC Is More Effective than EtBr in mtDNA Elimination from Human Cells
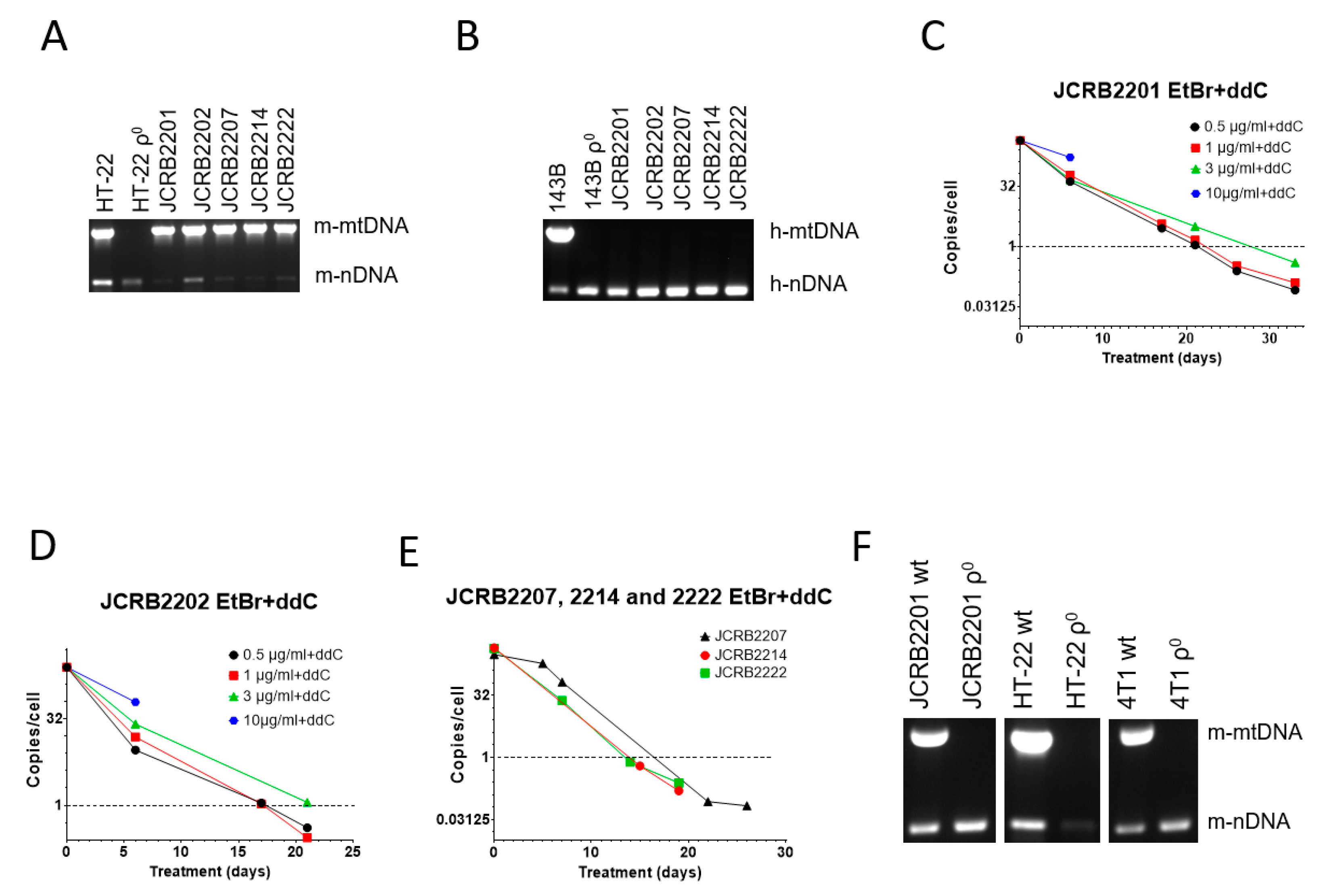
3.3. A Combination of EtBr and ddC Is Effective for mtDNA Elimination from Mouse/Human Somatic Cell Hybrids
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kozhukhar, N.; Spadafora, D.; Rodriguez, Y.A.R.; Alexeyev, M.F. A Method for In Situ Reverse Genetic Analysis of Proteins Involved mtDNA Replication. Cells 2022, 11, 2168. [Google Scholar] [CrossRef]
- Larsson, N.G.; Wang, J.; Wilhelmsson, H.; Oldfors, A.; Rustin, P.; Lewandoski, M.; Barsh, G.S.; Clayton, D.A. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat. Genet. 1998, 18, 231–236. [Google Scholar] [CrossRef]
- Humble, M.M.; Young, M.J.; Foley, J.F.; Pandiri, A.R.; Travlos, G.S.; Copeland, W.C. Polg2 is essential for mammalian embryogenesis and is required for mtDNA maintenance. Hum. Mol. Genet. 2012, 22, 1017–1025. [Google Scholar] [CrossRef]
- Hance, N.; Ekstrand, M.I.; Trifunovic, A. Mitochondrial DNA polymerase gamma is essential for mammalian embryogenesis. Hum. Mol. Genet. 2005, 14, 1775–1783. [Google Scholar] [CrossRef]
- Kuhl, I.; Miranda, M.; Posse, V.; Milenkovic, D.; Mourier, A.; Siira, S.J.; Bonekamp, N.A.; Neumann, U.; Filipovska, A.; Polosa, P.L.; et al. POLRMT regulates the switch between replication primer formation and gene expression of mammalian mtDNA. Sci. Adv. 2016, 2, e1600963. [Google Scholar] [CrossRef]
- Jiang, M.; Xie, X.; Zhu, X.; Jiang, S.; Milenkovic, D.; Misic, J.; Shi, Y.; Tandukar, N.; Li, X.; Atanassov, I.; et al. The mitochondrial single-stranded DNA binding protein is essential for initiation of mtDNA replication. Sci. Adv. 2021, 7, eabf8631. [Google Scholar] [CrossRef]
- Milenkovic, D.; Matic, S.; Kuhl, I.; Ruzzenente, B.; Freyer, C.; Jemt, E.; Park, C.B.; Falkenberg, M.; Larsson, N.G. TWINKLE is an essential mitochondrial helicase required for synthesis of nascent D-loop strands and complete mtDNA replication. Hum. Mol. Genet. 2013, 22, 1983–1993. [Google Scholar] [CrossRef]
- King, M.P.; Attardi, G. Human cells lacking mtDNA: Repopulation with exogenous mitochondria by complementation. Science 1989, 246, 500–503. [Google Scholar] [CrossRef]
- King, M.P.; Attardi, G. Isolation of human cell lines lacking mitochondrial DNA. Methods Enzymol. 1996, 264, 304–313. [Google Scholar]
- Morais, R.; Gregoire, M.; Jeannotte, L.; Gravel, D. Chick embryo cells rendered respiration-deficient by chloramphenicol and ethidium bromide are auxotrophic for pyrimidines. Biochem. Biophys. Res. Commun. 1980, 94, 71–77. [Google Scholar] [CrossRef]
- Wilkins, H.M.; Carl, S.M.; Swerdlow, R.H. Cytoplasmic hybrid (cybrid) cell lines as a practical model for mitochondriopathies. Redox Biol. 2014, 2C, 619–631. [Google Scholar] [CrossRef] [PubMed]
- Ziegler, M.L. Phenotypic expression of malignancy in hybrid and cybrid mouse cells. Somatic Cell Genet. 1978, 4, 477–489. [Google Scholar] [CrossRef]
- Giles, R.E.; Stroynowski, I.; Wallace, D.C. Characterization of mitochondrial DNA in chloramphenicol-resistant interspecific hybrids and a cybrid. Somatic Cell Genet. 1980, 6, 543–554. [Google Scholar] [CrossRef]
- Hayashi, J.; Gotoh, O.; Tagashira, Y.; Tosu, M.; Sekiguchi, T. Identification of mitochondrial DNA species in interspecific cybrids and reconstituted cells using restriction endonuclease. FEBS Lett. 1980, 117, 59–62. [Google Scholar] [CrossRef]
- Dunbar, D.R.; Moonie, P.A.; Zeviani, M.; Holt, I.J. Complex I deficiency is associated with 3243G:C mitochondrial DNA in osteosarcoma cell cybrids. Hum. Mol. Genet. 1996, 5, 123–129. [Google Scholar] [CrossRef] [PubMed]
- Trounce, I.; Wallace, D.C. Production of transmitochondrial mouse cell lines by cybrid rescue of rhodamine-6G pre-treated L-cells. Somat. Cell Mol. Genet. 1996, 22, 81–85. [Google Scholar] [CrossRef] [PubMed]
- Barrientos, A.; Kenyon, L.; Moraes, C.T. Human xenomitochondrial cybrids. Cellular models of mitochondrial complex I deficiency. J. Biol. Chem. 1998, 273, 14210–14217. [Google Scholar] [CrossRef]
- Pye, D.; Kyriakouli, D.S.; Taylor, G.A.; Johnson, R.; Elstner, M.; Meunier, B.; Chrzanowska-Lightowlers, Z.M.; Taylor, R.W.; Turnbull, D.M.; Lightowlers, R.N. Production of transmitochondrial cybrids containing naturally occurring pathogenic mtDNA variants. Nucleic Acids Res. 2006, 34, e95. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, K.; Hayashi, J. Generation of mtDNA-exchanged cybrids for determination of the effects of mtDNA mutations on tumor phenotypes. Methods Enzymol. 2009, 457, 335–346. [Google Scholar] [CrossRef]
- Ishikawa, K.; Takenaga, K.; Akimoto, M.; Koshikawa, N.; Yamaguchi, A.; Imanishi, H.; Nakada, K.; Honma, Y.; Hayashi, J.I. ROS-Generating Mitochondrial DNA Mutations Can Regulate Tumor Cell Metastasis. Science 2008, 320, 661–664. [Google Scholar] [CrossRef]
- Hashizume, O.; Shimizu, A.; Yokota, M.; Sugiyama, A.; Nakada, K.; Miyoshi, H.; Itami, M.; Ohira, M.; Nagase, H.; Takenaga, K.; et al. Specific mitochondrial DNA mutation in mice regulates diabetes and lymphoma development. Proc. Natl. Acad. Sci. USA 2012, 109, 10528–10533. [Google Scholar] [CrossRef]
- Hashizume, O.; Yamanashi, H.; Taketo, M.M.; Nakada, K.; Hayashi, J. A Specific Nuclear DNA Background Is Required for High Frequency Lymphoma Development in Transmitochondrial Mice with G13997A mtDNA. PLoS ONE 2015, 10, e0118561. [Google Scholar] [CrossRef] [PubMed]
- Khozhukhar, N.; Spadafora, D.; Rodriguez, Y.; Alexeyev, M. Elimination of Mitochondrial DNA from Mammalian Cells. Curr. Protoc. Cell Biol. 2018, 78, 20.11.1–20.11.14. [Google Scholar] [CrossRef] [PubMed]
- Nass, M.M. Differential effects of ethidium bromide on mitochondrial and nuclear DNA synthesis in vivo in cultured mammalian cells. Exp. Cell Res. 1972, 72, 211–222. [Google Scholar] [CrossRef] [PubMed]
- Wiseman, A.; Attardi, G. Reversible tenfod reduction in mitochondria DNA content of human cells treated with ethidium bromide. Mol. Gen. Genet. 1978, 167, 51–63. [Google Scholar] [CrossRef] [PubMed]
- Desjardins, P.; de Muys, J.M.; Morais, R. An established avian fibroblast cell line without mitochondrial DNA. Somat. Cell Mol. Genet. 1986, 12, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Inoue, K.; Ito, S.; Takai, D.; Soejima, A.; Shisa, H.; LePecq, J.B.; Segal-Bendirdjian, E.; Kagawa, Y.; Hayashi, J.I. Isolation of mitochondrial DNA-less mouse cell lines and their application for trapping mouse synaptosomal mitochondrial DNA with deletion mutations. J. Biol. Chem. 1997, 272, 15510–15515. [Google Scholar] [CrossRef] [PubMed]
- Inoue, K.; Takai, D.; Hosaka, H.; Ito, S.; Shitara, H.; Isobe, K.; LePecq, J.B.; Segal-Bendirdjian, E.; Hayashi, J. Isolation and characterization of mitochondrial DNA-less lines from various mammalian cell lines by application of an anticancer drug, ditercalinium. Biochem. Biophys. Res. Commun. 1997, 239, 257–260. [Google Scholar] [CrossRef] [PubMed]
- Okamaoto, M.; Ohsato, T.; Nakada, K.; Isobe, K.; Spelbrink, J.N.; Hayashi, J.; Hamasaki, N.; Kang, D. Ditercalinium chloride, a pro-anticancer drug, intimately associates with mammalian mitochondrial DNA and inhibits its replication. Curr. Genet. 2003, 43, 364–370. [Google Scholar] [CrossRef]
- Segal-Bendirdjian, E.; Coulaud, D.; Roques, B.P.; Le Pecq, J.B. Selective loss of mitochondrial DNA after treatment of cells with ditercalinium (NSC 335153), an antitumor bis-intercalating agent. Cancer Res. 1988, 48, 4982–4992. [Google Scholar]
- Shokolenko, I.N.; Wilson, G.L.; Alexeyev, M.F. Persistent damage induces mitochondrial DNA degradation. DNA Repair 2013, 12, 488–499. [Google Scholar] [CrossRef] [PubMed]
- Spadafora, D.; Kozhukhar, N.; Chouljenko, V.N.; Kousoulas, K.G.; Alexeyev, M.F. Methods for Efficient Elimination of Mitochondrial DNA from Cultured Cells. PLoS ONE 2016, 11, e0154684. [Google Scholar] [CrossRef] [PubMed]
- Khozhukhar, N.; Spadafora, D.; Rodriguez, Y.A.R.; Fayzulin, R.; Alexeyev, M. Generation of Mammalian Cells Devoid of Mitochondrial DNA (rho(0) cells). Curr. Protoc. 2023, 3, e679. [Google Scholar] [CrossRef] [PubMed]
- Kukat, A.; Kukat, C.; Brocher, J.; Schafer, I.; Krohne, G.; Trounce, I.A.; Villani, G.; Seibel, P. Generation of rho0 cells utilizing a mitochondrially targeted restriction endonuclease and comparative analyses. Nucleic Acids Res. 2008, 36, e44. [Google Scholar] [CrossRef] [PubMed]
- Adkins, J.C.; Peters, D.H.; Faulds, D. Zalcitabine. An update of its pharmacodynamic and pharmacokinetic properties and clinical efficacy in the management of HIV infection. Drugs 1997, 53, 1054–1080. [Google Scholar] [CrossRef] [PubMed]
- Walker, U.A.; Bauerle, J.; Laguno, M.; Murillas, J.; Mauss, S.; Schmutz, G.; Setzer, B.; Miquel, R.; Gatell, J.M.; Mallolas, J. Depletion of mitochondrial DNA in liver under antiretroviral therapy with didanosine, stavudine, or zalcitabine. Hepatology 2004, 39, 311–317. [Google Scholar] [CrossRef] [PubMed]
- Walker, U.A.; Setzer, B.; Venhoff, N. Increased long-term mitochondrial toxicity in combinations of nucleoside analogue reverse-transcriptase inhibitors. AIDS 2002, 16, 2165–2173. [Google Scholar] [CrossRef] [PubMed]
- Chariot, P.; Drogou, I.; de Lacroix-Szmania, I.; Eliezer-Vanerot, M.C.; Chazaud, B.; Lombes, A.; Schaeffer, A.; Zafrani, E.S. Zidovudine-induced mitochondrial disorder with massive liver steatosis, myopathy, lactic acidosis, and mitochondrial DNA depletion. J. Hepatol. 1999, 30, 156–160. [Google Scholar] [CrossRef] [PubMed]
- Brivet, F.G.; Nion, I.; Megarbane, B.; Slama, A.; Brivet, M.; Rustin, P.; Munnich, A. Fatal lactic acidosis and liver steatosis associated with didanosine and stavudine treatment: A respiratory chain dysfunction? J. Hepatol. 2000, 32, 364–365. [Google Scholar] [CrossRef]
- Gerschenson, M.; Nguyen, V.T.; St Claire, M.C.; Harbaugh, S.W.; Harbaugh, J.W.; Proia, L.A.; Poirier, M.C. Chronic stavudine exposure induces hepatic mitochondrial toxicity in adult Erythrocebus patas monkeys. J. Hum. Virol. 2001, 4, 335–342. [Google Scholar]
- Gaou, I.; Malliti, M.; Guimont, M.C.; Letteron, P.; Demeilliers, C.; Peytavin, G.; Degott, C.; Pessayre, D.; Fromenty, B. Effect of stavudine on mitochondrial genome and fatty acid oxidation in lean and obese mice. J. Pharmacol. Exp. Ther. 2001, 297, 516–523. [Google Scholar] [PubMed]
- Kakuda, T.N. Pharmacology of nucleoside and nucleotide reverse transcriptase inhibitor-induced mitochondrial toxicity. Clin. Ther. 2000, 22, 685–708. [Google Scholar] [CrossRef] [PubMed]
- Sterky, F.H.; Lee, S.; Wibom, R.; Olson, L.; Larsson, N.G. Impaired mitochondrial transport and Parkin-independent degeneration of respiratory chain-deficient dopamine neurons in vivo. Proc. Natl. Acad. Sci. USA 2011, 108, 12937–12942. [Google Scholar] [CrossRef] [PubMed]
- Koi, M.; Morita, H.; Shimizu, M.; Oshimura, M. Construction of mouse A9 clones containing a single human chromosome (X/autosome translocation) via micro-cell fusion. Jpn. J. Cancer Res. 1989, 80, 122–125. [Google Scholar] [CrossRef] [PubMed]
- Kozhukhar, N.; Fant, A.; Alexeyev, M.F. Quantification of mtDNA content in cultured cells by direct droplet digital PCR. Mitochondrion 2021, 61, 102–113. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, J.; Tanaka, M.; Sato, W.; Ozawa, T.; Yonekawa, H.; Kagawa, Y.; Ohta, S. Effects of ethidium bromide treatment of mouse cells on expression and assembly of nuclear-coded subunits of complexes involved in the oxidative phosphorylation. Biochem. Biophys. Res. Commun. 1990, 167, 216–221. [Google Scholar] [CrossRef] [PubMed]
- Fayzulin, R.Z.; Perez, M.; Kozhukhar, N.; Spadafora, D.; Wilson, G.L.; Alexeyev, M.F. A method for mutagenesis of mouse mtDNA and a resource of mouse mtDNA mutations for modeling human pathological conditions. Nucleic Acids Res. 2015, 43, e62. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fernandez-Moreno, M.; Hermida-Gomez, T.; Gallardo, M.E.; Dalmao-Fernandez, A.; Rego-Perez, I.; Garesse, R.; Blanco, F.J. Generating Rho-0 Cells Using Mesenchymal Stem Cell Lines. PLoS ONE 2016, 11, e0164199. [Google Scholar] [CrossRef] [PubMed]
- Clayton, D.A.; Teplitz, R.L.; Nabholz, M.; Dovey, H.; Bodmer, W. Mitochondrial DNA of human-mouse cell hybrids. Nature 1971, 234, 560–562. [Google Scholar] [CrossRef]
- Attardi, B.; Attardi, G. Fate of mitochondrial DNA in human-mouse somatic cell hybrids (density gradient centrifugation-ethidium bromide-karyotype). Proc. Natl. Acad. Sci. USA 1972, 69, 129–133. [Google Scholar] [CrossRef]
- Kirsanov, K.I.; Lesovaya, E.A.; Yakubovskaya, M.G.; Belitsky, G.A. SYBR Gold and SYBR Green II are not mutagenic in the Ames test. Mutat. Res. 2010, 699, 1–4. [Google Scholar] [CrossRef]
- Singer, V.L.; Lawlor, T.E.; Yue, S. Comparison of SYBR Green I nucleic acid gel stain mutagenicity and ethidium bromide mutagenicity in the Salmonella/mammalian microsome reverse mutation assay (Ames test). Mutat. Res. 1999, 439, 37–47. [Google Scholar] [CrossRef]
- Singh, K.K.; Kulawiec, M.; Still, I.; Desouki, M.M.; Geradts, J.; Matsui, S. Inter-genomic cross talk between mitochondria and the nucleus plays an important role in tumorigenesis. Gene 2005, 354, 140–146. [Google Scholar] [CrossRef]
- Fan, H.; Penman, S. Mitochondrial RNA synthesis during mitosis. Science 1970, 168, 135–138. [Google Scholar] [CrossRef]
| Purpose | Name | Sequence | Amplicon, bp |
|---|---|---|---|
| Genotyping of the ρ0 state in human cells | hMitF | AATGTCTGCACAGCCACTTTCCAC | 901 |
| hMitR | TCGTAGTGTTCTGGCGAGCAGTTT | ||
| hNucF | CGGACAGGATTGACAGATTGA | 389 | |
| hNucR | AGCTTATGACCCGCACTTAC | ||
| Genotyping the ρ0 state in murine cells | mMitF | AAAGCATCTGGCCTACACCCAGAA | 1041 |
| mMitR | ACCCTCGTTTAGCCGTTCATGCTA | ||
| mNucF | CCACGTGCTCTGTATGAGATT | 636 | |
| mNucR | ATGCTGGCTTATCTGTTCCTT | ||
| mtDNA copy number determination in human cells by dddPCR | NucF | AACTTGTAAGTGGTAGTGCATAGA | N/A |
| NucR | GTAGGAGGACATTTGAGGAGTG | ||
| NucProbe | FAM-TCAGGCAGACTGACACTAGAGTTCACA-BHQ1 | ||
| MitF | CTGATCAGGGTGAGCATCAAA | ||
| MitR | GAATGATGGCTAGGGTGACTTC | ||
| MitProbe | Hex-TGCGAGCAGTAGCCCAAACAATCT-BHQ1 | ||
| mtDNA copy number determination in mouse cells by dddPCR | NucF | CCTGGGCTTTGAACTTGTCTA | N/A |
| NucR | TGAGGGCATTGGAGATTGTG | ||
| NucProbe | FAM-TGGTCCTGCTATCAGAGATGCAACG-BHQ1 | ||
| MitF | GGCCTATTAATCGCAGCTACAG | ||
| MitR | GTAGTGCTGAAACTGGTGTAGG | ||
| MitProbe | FAM-ATTTGGCCTCCACCCATGACTACC-BHQ1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kozhukhar, N.; Alexeyev, M.F. Efficient Elimination of mtDNA from Mammalian Cells with 2′,3′-Dideoxycytidine. DNA 2024, 4, 201-211. https://doi.org/10.3390/dna4030013
Kozhukhar N, Alexeyev MF. Efficient Elimination of mtDNA from Mammalian Cells with 2′,3′-Dideoxycytidine. DNA. 2024; 4(3):201-211. https://doi.org/10.3390/dna4030013
Chicago/Turabian StyleKozhukhar, Natalya, and Mikhail F. Alexeyev. 2024. "Efficient Elimination of mtDNA from Mammalian Cells with 2′,3′-Dideoxycytidine" DNA 4, no. 3: 201-211. https://doi.org/10.3390/dna4030013
APA StyleKozhukhar, N., & Alexeyev, M. F. (2024). Efficient Elimination of mtDNA from Mammalian Cells with 2′,3′-Dideoxycytidine. DNA, 4(3), 201-211. https://doi.org/10.3390/dna4030013








