Carbon Dioxide Fluxes Associated with Prokaryotic and Eukaryotic Communities in Ice-Free Areas on King George Island, Maritime Antarctica
Abstract
1. Introduction
2. Methods
2.1. Study Areas
2.2. Soil Sampling
2.3. DNA Extraction, Data Analyses, and Organism Identification
2.4. Diversity Indices
2.5. Carbon Dioxide Flux
3. Results
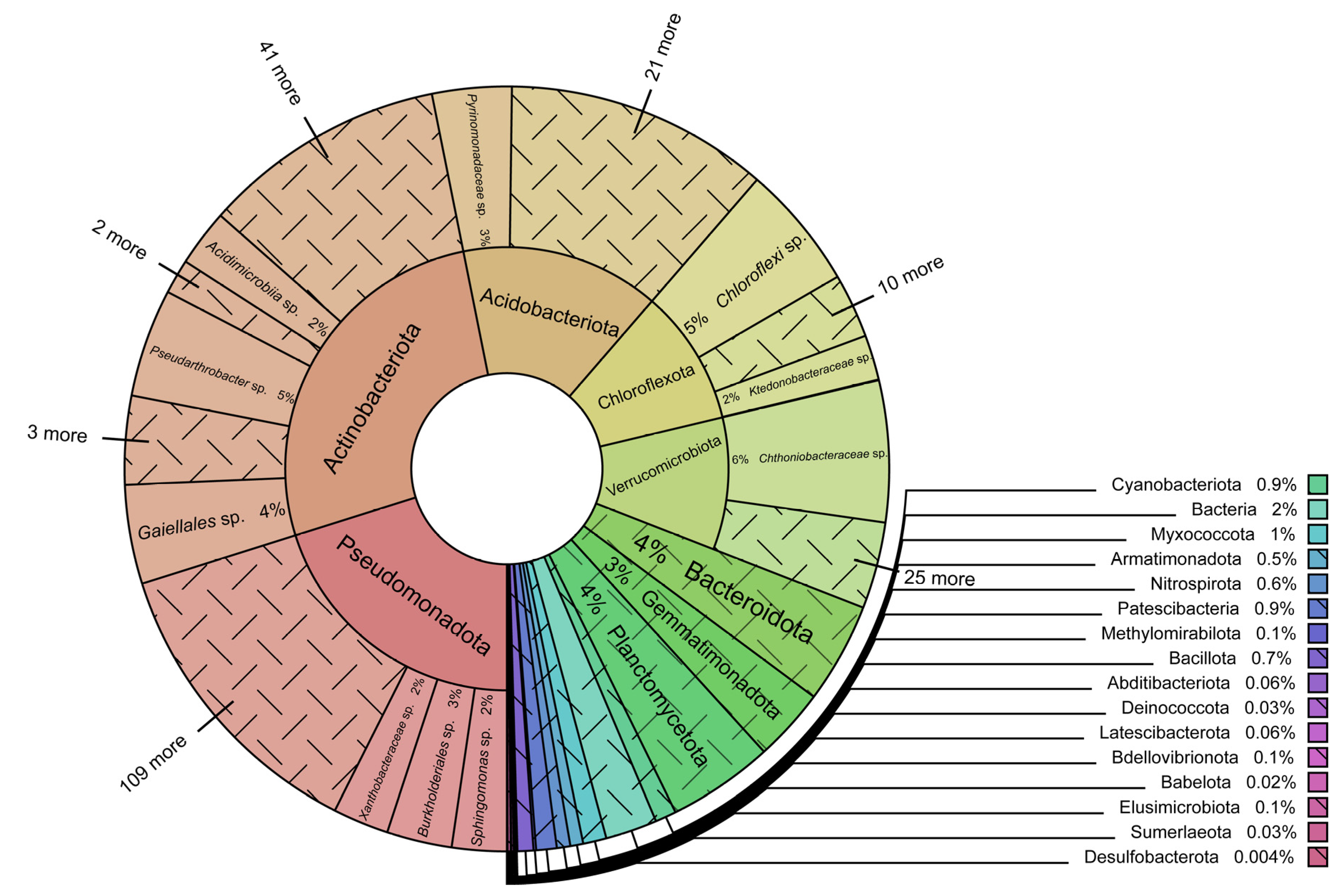
3.1. Prokaryotic Taxonomy and Abundance
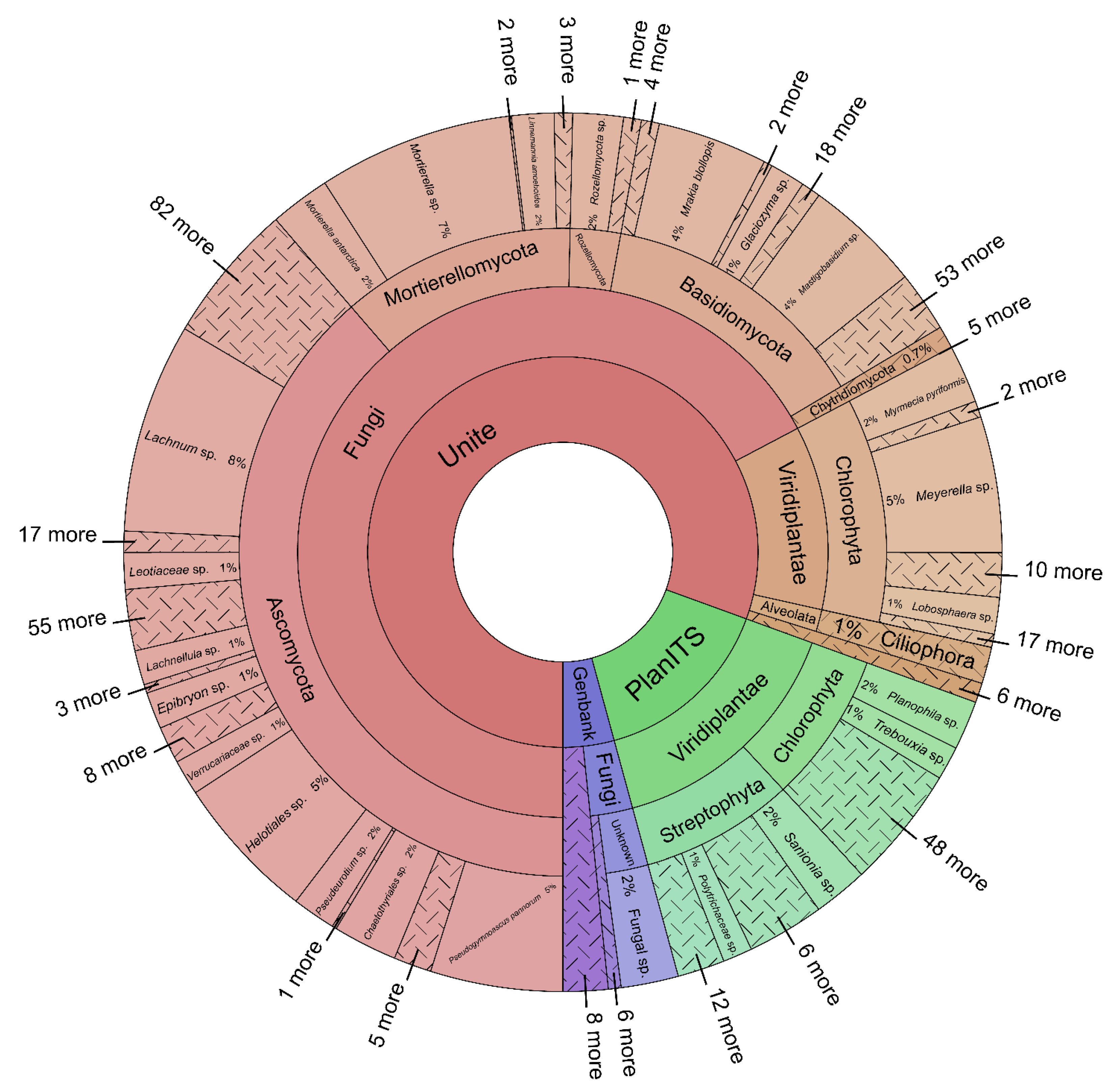
3.2. Eukaryotic Taxonomy and Abundance
3.3. Fungi
3.4. Viridiplantae
3.5. Stramenopila, Chromista, Protozoa, Holozoa, and Metazoa
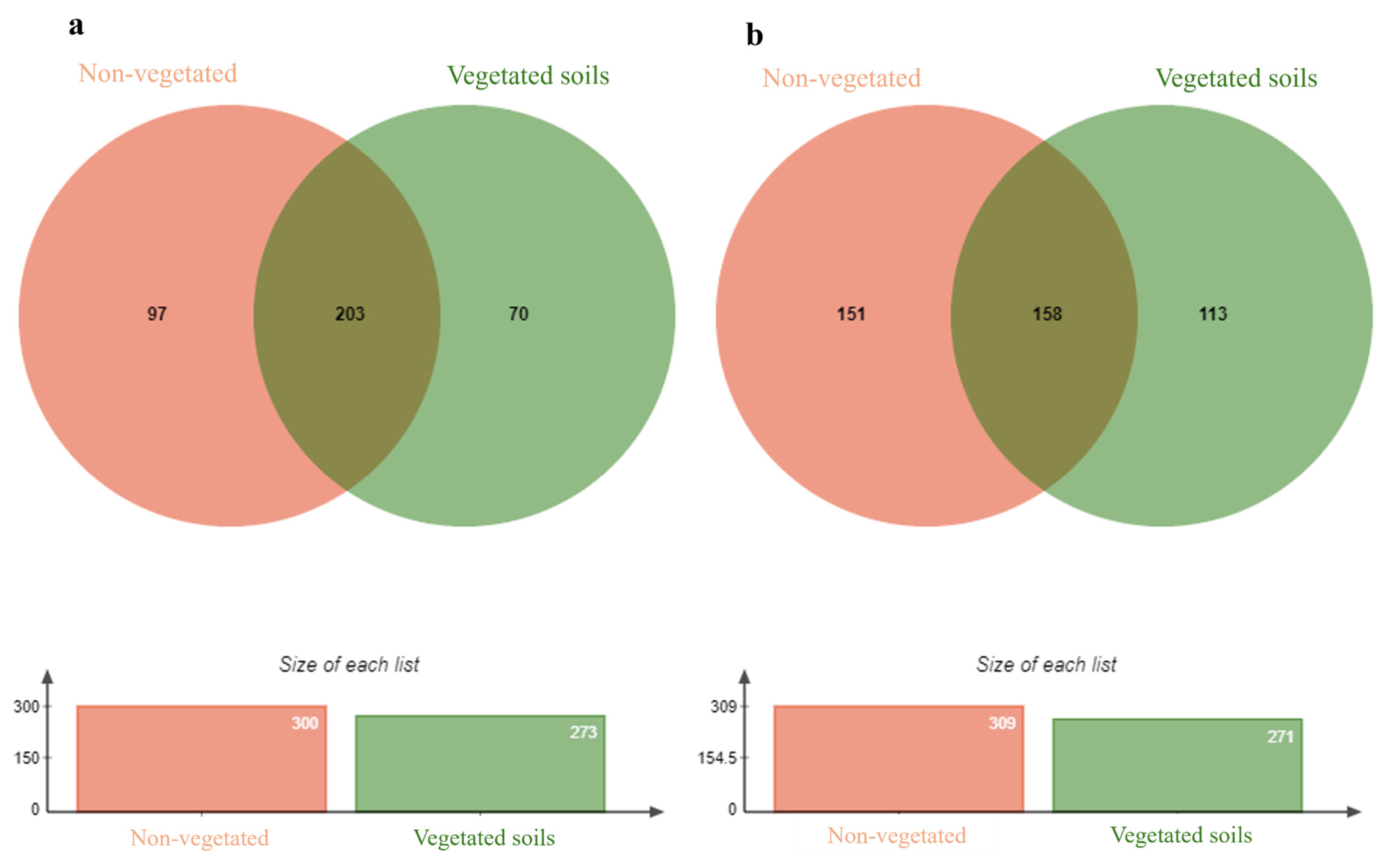
3.6. Prokaryotic and Eukaryotic Diversity and Distribution
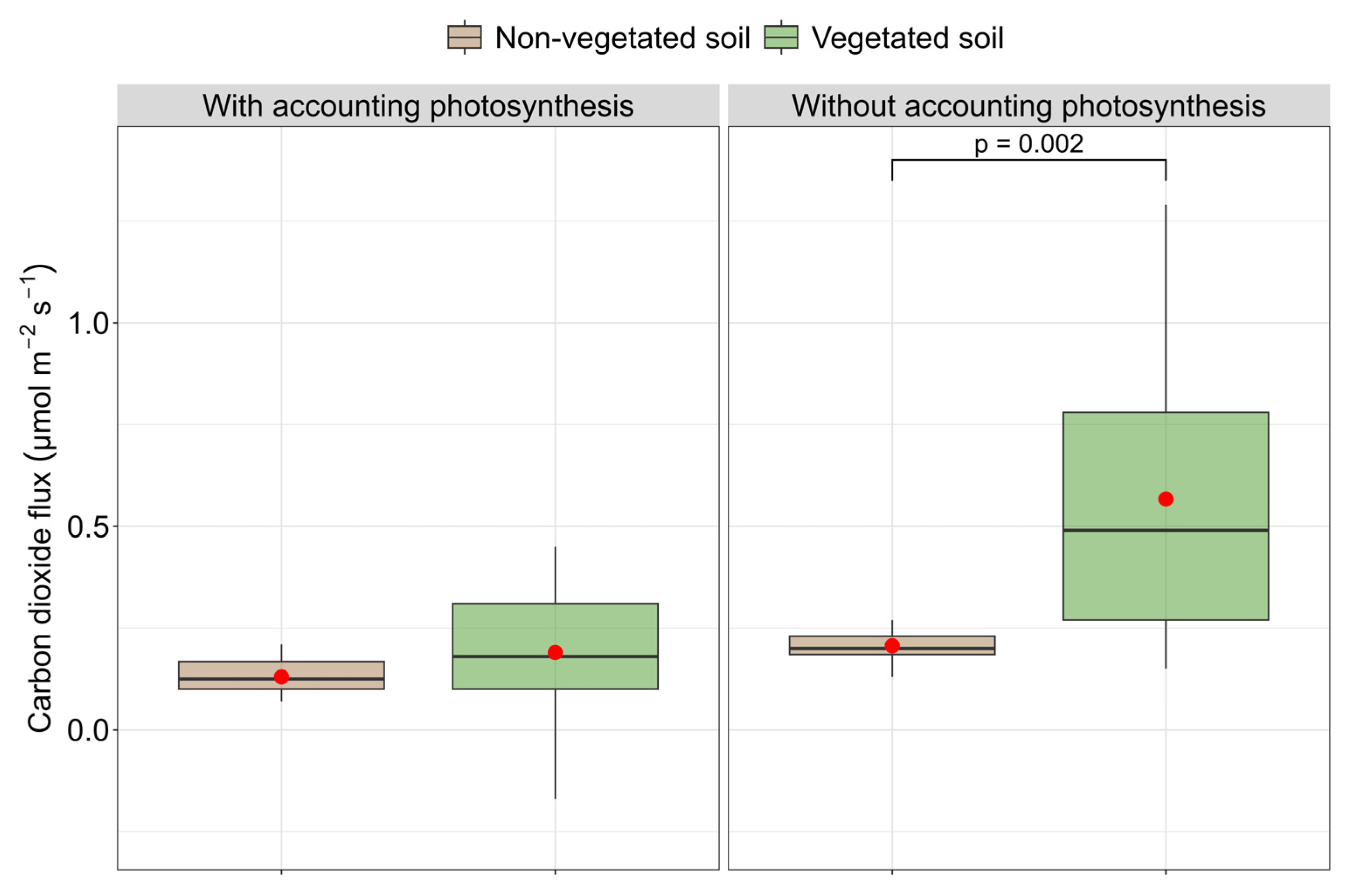
3.7. CO2 Flux and Prokaryotic and Eukaryotic Diversity
4. Discussion
4.1. Prokaryotic Taxonomy
4.2. Eukaryotic Taxonomy
4.3. Prokaryotic and Eukaryotic Diversity and CO2 Flux
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Godinho, V.M.; Gonçalves, V.N.; Santiago, I.F.; Figueredo, H.M.; Vitoreli, G.A.; Schaefer, C.E.G.R.; Barbosa, E.C.; Oliveira, J.G.; Alves, T.M.A.; Zani, C.L.; et al. Diversity and bioprospection of fungal community present in oligotrophic soil of continental Antarctica. Extremophiles 2015, 19, 585–596. [Google Scholar] [CrossRef]
- Bokhorst, S.; Convey, P.; Aerts, R. Nitrogen inputs by marine vertebrates drive abundance and richness in Antarctic terrestrial ecosystems. Curr. Biol. 2019, 29, 1721–1727. [Google Scholar] [CrossRef] [PubMed]
- Fell, J.W.; Scorzetti, G.; Connell, L.; Craig, S. Biodiversity of microeukaryotes in Antarctic Dry Valley soils with >5% soil moisture. Soil Biol. Biochem. 2006, 38, 3107–3119. [Google Scholar] [CrossRef]
- Ruisi, S.; Barreca, D.; Selbmann, L.; Zucconi, L.; Onofri, S. Fungi in Antarctica. Rev. Environ. Sci. Biotechnol. 2007, 6, 127–141. [Google Scholar] [CrossRef]
- Cowan, D.A.; Chown, S.L.; Convey, P.; Tuffin, M.; Hughes, K.; Pointing, S.; Vincent, W.F. Non-indigenous microorganisms in the Antarctic: Assessing the risks. Trends Microbiol. 2011, 19, 540–548. [Google Scholar] [CrossRef]
- Cavicchioli, R. Microbial ecology of Antarctic aquatic systems. Nat. Rev. Microbiol. 2015, 13, 691–706. [Google Scholar] [CrossRef] [PubMed]
- Rosa, L.H.; Zani, C.L.; Cantrell, C.L.; Duke, S.O.; Dijck, P.V.; Desideri, A.; Rosa, C.A. Fungi in Antarctica: Diversity, ecology, effects of climate change, and bioprospection for bioactive compounds. In Fungi of Antarctica: Diversity, Ecology and Biotechnological Applications; Springer: Cham, Switzerland, 2019; pp. 1–18. [Google Scholar] [CrossRef]
- Lopes, D.V.; Schaefer, C.E.G.R.; Souza, J.J.L.; de Oliveira, F.S.; Simas, F.N.B.; Daher, M.; Gjorup, D.F. Concretionary horizons, unusual pedogenetic processes and features of sulfate affected soils from Antarctica. Geoderma 2019, 347, 13–24. [Google Scholar] [CrossRef]
- Porto, B.A.; da Silva, T.H.; Machado, M.R.; de Oliveira, F.S.; Rosa, C.A.; Rosa, L.H. Diversity and distribution of cultivable fungi present in acid sulphate soils in chronosequence under para-periglacial conditions in King George Island, Antarctica. Extremophiles 2020, 24, 797–807. [Google Scholar] [CrossRef]
- Simas, F.N.B.; Schaefer, C.G.R.; Filho, M.R.A.; Francelino, M.R.; Fernandes Filho, E.I.; da Costa, L.M. Genesis, properties and classification of Cryosols from Admiralty Bay, Maritime Antarctica. Geoderma 2008, 144, 116–122. [Google Scholar] [CrossRef]
- Cao, J.J.; Li, Y.K.; Jiang, T.; Hu, G. Sulphur-containing particles emitted by concealed sulfide ore deposits: An unknown source of sulphur-containing particles in the atmosphere. Atmos. Chem. Phys. 2015, 12, 6959–6969. [Google Scholar] [CrossRef]
- Raich, J.W.; Schlesinger, W.H. The global carbon-dioxide flux in soil respiration and its relationship to vegetation and climate. Tellus Ser. B 1992, 44, 81–99. [Google Scholar] [CrossRef]
- La Scala, N.; de Sá Mendonça, E.; de Souza, J.V.; Panosso, A.R.; Simas, F.N.; Schaefer, C.E. Spatial and temporal variability in soil CO2-C emissions and relation to soil temperature at King George Island, Maritime Antarctica. Polar Sci. 2010, 4, 479–487. [Google Scholar] [CrossRef]
- Reichstein, M.; Rey, A.; Freibauer, A.; Tenhunen, J.; Valentini, R.; Banza, J.; Casals, P.; Cheng, Y.; Grunzweig, J.M.; Irvine, J.; et al. Modeling temporal and large-scale spatial variability of soil respiration from soil water availability, temperature and vegetation productivity indices. Glob. Biogeochem. Cycles 2003, 17, 1104. [Google Scholar] [CrossRef]
- Convey, P.; Peck, L.S. Antarctic environmental change and biological responses. Sci. Adv. 2019, 5, eaaZ0888. [Google Scholar] [CrossRef] [PubMed]
- Siegert, M.; Atkinson, A.; Banwell, A.; Brandon, M.; Convey, P.; Davies, B.; Downie, R.; Edwards, T.; Hubbard, B.; Marshall, G.; et al. The Antarctic Peninsula under a 1.5 °C global warming scenario. Front. Environ. Sci. 2019, 7, 102. [Google Scholar] [CrossRef]
- Shanhun, F.L.; Almond, P.C.; Clough, T.J.; Smith, C.M.S. Abiotic processes dominate CO2 fluxes in Antarctic soils. Soil Biol. Biochem. 2012, 53, 99–111. [Google Scholar] [CrossRef]
- Rosa, L.H.; da Costa Coelho, L.; Pinto, O.H.B.; Carvalho-Silva, M.; Convey, P.; Rosa, C.A.; Câmara, P.E.A.S. Ecological succession of fungal and bacterial communities in Antarctic mosses affected by a fairy ring disease. Extremophiles 2021, 25, 471–481. [Google Scholar] [CrossRef]
- Newsham, K.K.; Davey, M.L.; Hopkins, D.W.; Dennis, P.G. Regional diversity of Maritime Antarctic soil fungi and predicted responses of guilds and growth forms to climate change. Front. Microbiol. 2021, 11, 615659. [Google Scholar] [CrossRef]
- Chen, S.; Yao, H.; Han, J.; Liu, C.; Song, J.; Shi, L.; Zhu, Y.; Ma, X.; Gao, T.; Pang, X.; et al. Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species. PLoS ONE 2010, 5, e8613. [Google Scholar] [CrossRef]
- Richardson, R.T.; Lin, C.H.; Sponsler, D.B.; Quijia, J.O.; Goodell, K.; Johnson, R.M. Application of ITS2 metabarcoding to determine the provenance of pollen collected by honeybees in an agroecosystem. Appl. Plant Sci. 2015, 3, 1400066. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.J.W.T.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Academic Press: New York, NY, USA, 1990; pp. 315–322. [Google Scholar]
- Herlemann, D.; Labrenz, M.; Jürgens, K.; Bertilsson, S.; Waniek, J.J.; Andersson, A.F. Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J. 2011, 5, 1571–1579. [Google Scholar] [CrossRef]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef] [PubMed]
- Bushnell, B. BBMap: A Fast, Accurate, Splice-Aware Aligner; Lawrence Berkeley National Laboratory: Berkeley, CA, USA, 2014; Available online: https://sourceforge.net/projects/bbmap (accessed on 20 July 2024).
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Gregory Caporaso, J. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef]
- Abarenkov, K.; Allan, Z.; Timo, P.; Raivo, P.; Filipp, I.; Nilsson, H.R.; Urmas, K. UNITE QIIME Release for Eukaryotes; UNITE Community: London, UK, 2020. [Google Scholar] [CrossRef]
- Banchi, E.; Ametrano, C.G.; Greco, S.; Stanković, D.; Muggia, L.; Pallavicini, A. PLANITS: A curated sequence reference dataset for plant ITS DNA metabarcoding. Database 2020, 2020, baz155. [Google Scholar] [CrossRef]
- Ondov, B.D.; Bergman, N.H.; Phillippy, A.M. Interactive metagenomic visualization in a web browser. BMC Bioinform. 2011, 12, 385. [Google Scholar] [CrossRef] [PubMed]
- Medinger, R.; Nolte, V.; Pandey, R.V.; Jost, S.; Ottenwälder, B.; Schlötterer, C.; Boenigk, J. Diversity in a hidden world: Potential and limitation of next-generation sequencing for surveys of molecular diversity of eukaryotic microorganisms. Mol. Ecol. 2010, 19, 32–40. [Google Scholar] [CrossRef] [PubMed]
- Weber, A.A.; Pawlowski, J. Can abundance of protists be inferred from sequence data: A case study of Foraminifera. PLoS ONE 2013, 8, e56739. [Google Scholar] [CrossRef]
- Giner, C.R.; Forn, I.; Romac, S.; Logares, R.; de Vargas, C.; Massana, R. Environmental sequencing provides reasonable estimates of the relative abundance of specific picoeukaryotes. Appl. Environ. Microbiol. 2016, 82, 4757–4766. [Google Scholar] [CrossRef]
- Deiner, K.; Bik, H.M.; Mächler, E.; Seymour, M.; Lacoursière-Roussel, A.; Altermatt, F.; Creer, S.; Bista, I.; Lodge, D.M.; de Vere, N.; et al. Environmental DNA metabarcoding: Transforming how we survey animal and plant communities. Mol. Ecol. 2017, 26, 5872–5895. [Google Scholar] [CrossRef] [PubMed]
- Hering, D.; Borja, Á.; Jones, J.I.; Pont, D.; Boets, P.; Bouchez, A.; Bruce, K.; Drakare, S.; Hänfling, B.; Kahlert, M.; et al. Implementation options for DNA-based identification into ecological status assessment under the European Water Framework Directive. Water Res. 2018, 138, 192–205. [Google Scholar] [CrossRef] [PubMed]
- Rosa, L.H.; Pinto, O.H.B.; Peter, C.; Carvalho-Silva, M.; Rosa, C.A.; Câmara, P.E.A.S. DNA metabarcoding to assess the diversity of airborne fungi present over Keller Peninsula, King George Island, Antarctica. Microb. Ecol. 2021, 82, 165–172. [Google Scholar] [CrossRef] [PubMed]
- Hammer, Ø.; Harper, D.A.T.; Ryan, P.D. PAST: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001, 4, 9. [Google Scholar] [CrossRef]
- Bardou, P.; Mariette, J.; Escudié, F.; Djemiel, C.; Klopp, C. Jvenn: An interactive Venn diagram viewer. BMC Bioinform. 2014, 15, 293. [Google Scholar] [CrossRef]
- Chapin, F.S.; Woodwell, G.M.; Randerson, J.T.; Rastetter, E.B.; Lovett, G.M.; Baldocchi, D.D.; Clark, D.A.; Harmon, M.E.; Schimel, D.S.; Valentini, R.; et al. Reconciling carbon-cycle concepts, terminology, and methods. Ecosystems 2006, 9, 1041–1050. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2024; Available online: https://www.r-project.org/ (accessed on 20 January 2024).
- Bajerski, F.; Wagner, D. Bacterial succession in Antarctic soils of two glacier forefields on Larsemann Hills, East Antarctica. FEMS Microbiol. Ecol. 2013, 85, 128–142. [Google Scholar] [CrossRef]
- Chong, C.W.; Pearce, D.A.; Convey, P.; Yew, W.C.; Tan, I.K.P. Patterns in the distribution of soil bacterial 16S rRNA gene sequences from different regions of Antarctica. Geoderma 2012, 181–182, 45–55. [Google Scholar] [CrossRef]
- Doytchinov, V.V.; Dimov, S.G. Microbial community composition of the Antarctic ecosystems: Review of the Bacteria, Fungi, and Archaea identified through an NGS-based metagenomics approach. Life 2022, 12, 916. [Google Scholar] [CrossRef]
- Varliero, G.; Lebre, P.H.; Adams, B.; Chown, S.L.; Convey, P.; Dennis, P.G.; Fan, D.; Ferrari, B.; Frey, B.; Hogg, I.D.; et al. Biogeographic survey of soil bacterial communities across Antarctica. Microbiome 2024, 12, 9. [Google Scholar] [CrossRef]
- Krauze, P.; Wagner, D.; Yang, S.; Spinola, D.; Kühn, P. Influence of prokaryotic microorganisms on initial soil formation along a glacier forefield on King George Island, Maritime Antarctica. Sci. Rep. 2021, 11, 13135. [Google Scholar] [CrossRef]
- Cannone, N.; Wagner, D.; Hubberten, H.W.; Guglielmin, M. Biotic and abiotic factors influencing soil properties across a latitudinal gradient in Victoria Land, Antarctica. Geoderma 2008, 144, 50–65. [Google Scholar] [CrossRef]
- Almeida, I.C.; Schaefer, C.E.G.; Fernandes, R.B.; Pereira, T.T.; Nieuwendam, A.; Pereira, A.B. Active layer thermal regime at different vegetation covers at Lions Rump, King George Island, Maritime Antarctica. Geomorphology 2014, 225, 36–46. [Google Scholar] [CrossRef]
- Dedysh, S.N.; Damsté, J.S.S. Acidobacteria. In Bergey’s Manual of Systematics of Archaea and Bacteria; ELS: Boralesgamuwa, Sri Lanka, 2018. [Google Scholar] [CrossRef]
- Pascual, J.; Huber, K.J.; Overmann, J. Pyrinomonadaceae. In Bergey’s Manual of Systematics of Archaea and Bacteria 2018; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2018. [Google Scholar] [CrossRef]
- Gran-Scheuch, A.; Ramos-Zuñiga, J.; Fuentes, E.; Bravo, D.; Pérez-Donoso, J.M. Effect of co-contamination by PAHs and heavy metals on bacterial communities of diesel contaminated soils of South Shetland Islands, Antarctica. Microorganisms 2020, 8, 1749. [Google Scholar] [CrossRef]
- Bendia, A.G.; Moreira, J.C.F.; Ferreira, J.C.N.; Romano, R.G.; Ferreira, I.G.C.; Franco, D.C.; Evangelista, H.; Montone, R.C.; Pellizari, V.H. Insights into Antarctic microbiomes: Diversity patterns for terrestrial and marine habitats. An. Acad. Bras. Cienc. 2023, 95 (Suppl. S3), e20211442. [Google Scholar] [CrossRef]
- Han, J.; Jung, J.; Hyun, S.; Park, H.; Park, W. Effects of nutritional input and diesel contamination on soil enzyme activities and microbial communities in Antarctic soils. J. Microbiol. 2012, 50, 916–924. [Google Scholar] [CrossRef] [PubMed]
- Naloka, K.; Kuntaveesuk, A.; Muangchinda, C.; Kuntaveesuk, A.; Muangchinda, C.; Chavanich, S.; Viyakarn, V.; Chen, B.; Pinyakong, O. Pseudomonas and Pseudarthrobacter are the key players in synergistic phenanthrene biodegradation at low temperatures. Sci. Rep. 2024, 14, 11976. [Google Scholar] [CrossRef]
- Bridge, P.D.; Spooner, B.M.; Roberts, P.J. List of Non-Lichenised Fungi from the Antarctic Region; British Antarctic Survey: Cambridgeshire, UK, 2008; Available online: https://legacy.bas.ac.uk/bas_research/data/access/fungi/ (accessed on 17 September 2024).
- Rosa, L.H.; da Silva, T.H.; Ogaki, M.B.; Pinto, O.H.B.; Stech, M.; Convey, P.; Carvalho-Silva, M.; Rosa, C.A.; Câmara, P. DNA metabarcoding uncovers fungal diversity in soils of protected and non-protected areas on Deception Island, Antarctica. Sci. Rep. 2020, 10, 21986. [Google Scholar] [CrossRef]
- Câmara, P.E.A.S.; Carvalho-Silva, M. 180 years of botanical investigations in Antarctica and the role of Brazil. Acta Bot. Bras. 2020, 34, 430–436. [Google Scholar] [CrossRef]
- Câmara, P.E.A.S.; Carvalho-Silva, M.; Pinto, O.H.; Amorim, E.T.; Henriques, D.K.; da Silva, T.H.; Pellizzari, F.; Convey, P.; Rosa, L.H. Diversity and ecology of Chlorophyta (Viridiplantae) assemblages in protected and non-protected sites in Deception Island (Antarctica, South Shetland Islands) assessed using an NGS approach. Microbial Ecol. 2021, 81, 323–334. [Google Scholar] [CrossRef]
- Andreeva, V.M.; Kurbatova, L.E. Terrestrial and aerophilic nonmotile green microalgae (Chlorophyta) from regions of investigation of Russian Antarctic expedition. Nov. Sist. Nizsh. Rast. 2014, 46, 12–26. [Google Scholar] [CrossRef]
- Ye, M.; Cao, S.Q.; Jiang, S.T.; Pan, L.J.; Luo, S.Z.; Li, X.J. Species diversity of Lachnum (Helotiales, Hyaloscyphaceae) from temperate China. J. Zhejiang Univ. Sci. B 2006, 7, 20–27. [Google Scholar] [CrossRef]
- Bruyant, P.; Moënne-Loccoz, Y.; Almario, J. Root-associated Helotiales fungi: Overlooked players in plant nutrition. Soil Biol. Biochem. 2024, 191, 109363. [Google Scholar] [CrossRef]
- Kirk, P.M.; Cannon, P.F.; Minter, D.W.; Stalpers, J.A. Dictionary of the Fungi, 10th ed.; CAB International: Wallingford, UK, 2011. [Google Scholar]
- Rosa, L.H.; de Sousa, J.R.P.; de Menezes, G.C.A.; da Costa Coelho, L.; Carvalho-Silva, M.; Convey, P.; Câmara, P.E.A.S. Opportunistic fungi found in fairy rings are present on different moss species in the Antarctic Peninsula. Polar Biol. 2020, 43, 587–596. [Google Scholar] [CrossRef]
- Borden, K.A.; Mafa-Attoye, T.G.; Dunfield, K.E.; Thevathasan, N.V.; Gordon, A.M.; Isaac, M.E. Root Functional Trait and Soil Microbial Coordination: Implications for Soil Respiration in Riparian Agroecosystems. Front. Plant Sci. 2021, 12, 681113. [Google Scholar] [CrossRef]
- Darcy, J.L.; Schmidt, S.K. Nutrient limitation of microbial phototrophs on a debris-covered glacier. Soil Biol. Biochem. 2016, 95, 156–163. [Google Scholar] [CrossRef]
- Sun, L.; Ataka, M.; Han, M.; Han, Y.; Gan, D.; Xu, T.; Guo, Y.; Zhu, B. Root exudation as a major competitive fine-root functional trait of 18 coexisting species in a subtropical forest. New Phytol. 2020, 229, 259–271. [Google Scholar] [CrossRef]
- Yergeau, É.; Newsham, K.K.; Pearce, D.A.; Kowalchuk, G.A. Patterns of bacterial diversity across a range of Antarctic terrestrial habitats. Environ. Microbiol. 2007, 9, 2670–2682. [Google Scholar] [CrossRef] [PubMed]
- Wild, B.; Alves, R.E.; Bárta, J.; Čapek, P.; Gentsch, N.; Guggenberger, G.; Hugelius, G.; Knoltsch, A.; Kuhry, P.; Lashchinskiy, N.; et al. Amino acid production exceeds plant nitrogen demand in Siberian tundra. Environ. Res. Lett. 2018, 13, 034002. [Google Scholar] [CrossRef]
- Namsaraev, Z.; Bobrik, A.; Kozlova, A.; Krylova, A.; Rudenko, A.; Mitina, A.; Saburov, A.; Patrushev, M.; Karnachuk, O.; Toshchakov, S. Carbon emission and biodiversity of Arctic soil microbial communities of the Novaya Zemlya and Franz Josef Land archipelagos. Microorganisms 2023, 11, 482. [Google Scholar] [CrossRef]

| Non-Vegetated Soils | Vegetated Soils | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diversity Indices | Site 1 | Site 2 | Site 3 | Site 4 | Site 5 | Site 6 | Site 7 | |||||||
| Prok | Euk | Prok | Euk | Prok | Euk | Prok | Euk | Prok | Euk | Prok | Euk | Prok | Euk | |
| Number of DNA reads | 244,718 | 370,446 | 195,018 | 408,136 | 216,630 | 427,360 | 173,407 | 306,007 | 62,644 | 162,660 | 263,922 | 690,288 | 233,629 | 700,190 |
| Number of ASVs | 239 | 214 | 198 | 211 | 178 | 175 | 176 | 201 | 170 | 124 | 198 | 134 | 214 | 171 |
| Fisher’s-α (diversity) | 26.14 | 21.99 | 21.76 | 21.41 | 19.06 | 17.3 | 19.34 | 20.96 | 21.28 | 13.16 | 20.97 | 12.25 | 23.22 | 16 |
| Margalef (richness) | 19.18 | 16.61 | 16.17 | 16.25 | 14.41 | 13.42 | 14.51 | 15.83 | 15.3 | 10.25 | 15.78 | 9.9 | 17.23 | 12.63 |
| Simpson’s (dominance) | 0.97 | 0.91 | 0.97 | 0.84 | 0.97 | 0.83 | 0.96 | 0.79 | 0.97 | 0.82 | 0.97 | 0.72 | 0.97 | 0.75 |
| Soil Samples | ||||
|---|---|---|---|---|
| Non-Vegetated Soil | Vegetated Soil | |||
| Diversity Indices | Prok | Euk | Prok | Euk |
| Number of DNA reads | 656,366 | 1,205,942 | 733,602 | 1,859,145 |
| Number of ASVs | 307 | 316 | 279 | 279 |
| Fisher’s-α (diversity) | 30.8 | 29.79 | 27.36 | 24.86 |
| Margalef (richness) | 22.85 | 22.5 | 20.58 | 19.26 |
| Simpson’s (dominance) | 0.97 | 0.88 | 0.97 | 0.76 |
| µmol CO2 m−2 s−1 | ||||
| NEE | 0.13 ± 0.043 | 0.19 ± 0.297 | ||
| ER | 0.206 ± 0.060 | 0.567 ± 0.346 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rosa, L.H.; Gonçalves, V.N.; Barreto, D.L.C.; Francelino, M.R.; Baldi, C.G.O.; Mello, D.C.; Santos, K.C.R.; Lopes, F.A.C.; Carvalho-Silva, M.; Convey, P.; et al. Carbon Dioxide Fluxes Associated with Prokaryotic and Eukaryotic Communities in Ice-Free Areas on King George Island, Maritime Antarctica. DNA 2025, 5, 15. https://doi.org/10.3390/dna5010015
Rosa LH, Gonçalves VN, Barreto DLC, Francelino MR, Baldi CGO, Mello DC, Santos KCR, Lopes FAC, Carvalho-Silva M, Convey P, et al. Carbon Dioxide Fluxes Associated with Prokaryotic and Eukaryotic Communities in Ice-Free Areas on King George Island, Maritime Antarctica. DNA. 2025; 5(1):15. https://doi.org/10.3390/dna5010015
Chicago/Turabian StyleRosa, Luiz H., Vívian N. Gonçalves, Débora Luiza Costa Barreto, Marcio Rocha Francelino, Clara Glória Oliveira Baldi, Danilo Cesar Mello, Kárita C. R. Santos, Fabyano A. C. Lopes, Micheline Carvalho-Silva, Peter Convey, and et al. 2025. "Carbon Dioxide Fluxes Associated with Prokaryotic and Eukaryotic Communities in Ice-Free Areas on King George Island, Maritime Antarctica" DNA 5, no. 1: 15. https://doi.org/10.3390/dna5010015
APA StyleRosa, L. H., Gonçalves, V. N., Barreto, D. L. C., Francelino, M. R., Baldi, C. G. O., Mello, D. C., Santos, K. C. R., Lopes, F. A. C., Carvalho-Silva, M., Convey, P., & Câmara, P. E. A. S. (2025). Carbon Dioxide Fluxes Associated with Prokaryotic and Eukaryotic Communities in Ice-Free Areas on King George Island, Maritime Antarctica. DNA, 5(1), 15. https://doi.org/10.3390/dna5010015








