Abstract
Peripartum dairy cows commonly experience negative energy balance (EB) and immunosuppression together with high incidences of infectious and metabolic disease. This study investigated mechanisms linking EB status with immune defense in early lactation. Data were collected from multiparous Holstein cows from six herds and leukocyte transcriptomes were analyzed using RNA sequencing. Global gene expression was related to circulating IGF-1 (as a biomarker for EB) by subdividing animals into three groups, defined as IGF-1 LOW (<35 ng/mL, n = 35), MODERATE (35–100 ng/mL, n = 92) or HIGH (>100 ng/mL, n = 43) at 14 ± 4 days in milk (DIM). Differentially expressed genes between groups were identified using CLC Genomics Workbench V21, followed by cluster and KEGG pathway analysis, focusing on the comparison between LOW and HIGH IGF-1 cows. LOW cows were older and had significantly lower dry matter intakes and EB values, whereas HIGH cows produced more milk. During the first 35 DIM, 63% of LOW cows had more than one health problem vs. 26% HIGH cows, including more with clinical mastitis and uterine infections. Gene expression analysis indicated that leukocytes in LOW cows switched energy metabolism from oxidative phosphorylation to aerobic glycolysis (PGM, LDH, and PDK4). Many antimicrobial peptides were up-regulated in LOW cows (e.g., PTX3, DMBT1, S100A8, and S100A9) together with genes associated with inflammation, platelet activation and the complement cascade. HIGH cows had greater expression of genes regulating T and B cell function and the cytoskeleton. Overall, results suggested an ongoing cycle of poor EB and higher infection rates in LOW IGF-1 cows which was reflected in altered leukocyte functionality and reduced milk production.
Keywords:
IGF-1; leukocyte; energy balance; RNA-seq; dairy cow; antimicrobial; inflammation; mastitis; PTX3; DMBT1 1. Introduction
The requirement of the mammary gland for metabolizable energy and protein in the immediate post-calving period exceeds intake by approximately 25% [1]. This results in reduced availability of amino acids and glucogenic compounds, whereas ketogenic and lipogenic compounds are generally increased [2,3,4]. During this period, dairy cows mobilize both body fat and muscle in support of lactation, with higher rates of mobilization in animals carrying more body condition at calving [5]. The associated period of negative energy balance (NEB) may last for several weeks, extending into months in severe cases [6]. During this period, the liver co-ordinates the extensive changes in metabolic pathways required via up-regulation of genes involved in fatty acid oxidation and gluconeogenesis, and down-regulation of triacylglycerol synthesis [7,8]. This may be associated with the development of hepatic steatosis, which compromises glucose production and increases inflammatory responses [9].
Dairy cows also experience changes to their somatotropic axis at this time. Pituitary-derived growth hormone (GH) usually induces hepatic IGF-1 synthesis. When cows are in NEB, however, they become insulin resistant and down-regulation of the hepatic GH receptor causes IGF-1 synthesis to fall, so reducing circulating IGF-1 [10,11]. One effect of this uncoupling is to facilitate the indirect actions of GH on lipolysis and gluconeogenesis during early lactation [12]. At the same time, local support from IGF-1 in peripheral tissues towards cell metabolism and mitosis is reduced [13,14]. The IGF-1 concentration in lactating cows reflects their nutrient intake, showing a positive correlation with both energy and protein balance [15,16,17]. Such nutritionally driven changes occur over days rather than hours, so the IGF-1 concentration in blood is relatively stable [15,18]. This makes it a more reliable metabolic marker than non-esterified fatty acids (NEFA), beta-hydroxy butyrate (BHB) and glucose, whose concentrations all alter with respect to the time of feeding [18,19].
The somatotropic axis is also known to have an important relationship with the immune system. This is modulated via peripheral resistance and down-regulation of receptors, disruption in the GH/IGF-1 signaling pathways, and dysregulation of IGF-binding protein production, which in turn alters IGF bioavailability [20]. Immune cells including peripheral blood mononuclear cells (PBMC), T- and B-lymphocytes, and NK-cells all express the IGF-1R and are therefore responsive to the effects of IGF-1 [21,22]. This can influence innate immunity, for example by modulating differentiation and proliferation of myeloid lineage cells and influencing the responsiveness of mature immune cells to antigens. For example, experiments using peritoneal exudative cells in mice found that IGF-1 could enhance phagocytosis and their ability to kill E. coli [23]. More recently, IGF-1 was shown to potentiate pro-inflammatory cytokine production induced by microbial TLR ligands in PBMC [24]. IGF-1 also affects acquired immunity via proliferative, differentiating and functional effects on mature and maturing lymphocytes in primary and secondary lymphoid organs [21,22]. Circulating IGF-1 is thus a good indicator of metabolic status in the early postpartum period [6,15,25] and is also known to prime the response of immunocompetent cells to deal with infection and respond to sepsis and tissue injury [21,22]. IGF-1 was therefore chosen as a suitable candidate biomarker to investigate the links between metabolism and infectious disease in this study.
Immune function is known to be suppressed over the peripartum period. The numbers of circulating leukocytes, including all subset T cell populations, decrease in the first two weeks after calving followed by a gradual recovery [26,27,28]. The functional capacity of blood neutrophils is reduced, in particular their ability to generate reactive oxygen species (ROS), and this is strongly associated with the liver triacylglycerol content and raised concentrations of NEFAs [29,30,31,32]. Genes important for granulocyte recruitment and interferon signaling were down-regulated in neutrophils from cows with elevated NEFA and BHB levels after calving [29,33]. There are also strong indications that cows are under increased oxidative stress at this time [34].
Mounting an effective immune defense is energetically demanding, as immune cells require an adequate supply of glucose, amino acids, fatty acids and cholesterol/oxysterols for proliferation and the production of immune molecules [35,36,37]. Neutrophils contain relatively few mitochondria, so are mainly reliant on glucose to fuel glycolytic energy production but, during an infection, pathogenic bacteria compete for a limited glucose supply [37]. Clonal expansion of T cells in response to antigen recognition requires dynamic changes in their metabolism [38,39]. As macrophages develop into a more pro-inflammatory phenotype, there is a shift towards enhanced glucose uptake and aerobic glycolysis, whereas anti-inflammatory M2 macrophages have lower glycolytic rates and higher levels of oxidative metabolism [36,40]. Both the innate and adaptive immune systems are thus affected, with reduced responsiveness of circulating T cells to mitogenic agents and less production of immunoglobulin by B cells [41]. The impairment in immune function in peripartum cows is, therefore, almost certainly linked to the reduction in nutrient availability [42].
Another contributing factor to the altered immune environment is the birth process itself. The postpartum uterus must undergo extensive remodeling to reduce in size, remove cellular debris, and restore normal architecture [43]. In 90% of cows, this early period of involution is accompanied by microbial contamination which results in uterine disease in up to half of animals [44]. The incidences of other infectious diseases including mastitis are also high in the early postpartum period [45,46,47]. During such an infection, neutrophils are rapidly recruited to the affected site(s) to kill the invading pathogens using a wide variety of mechanisms including degranulation, ROS production, phagocytosis and release of cytokines and chemokines to recruit additional leukocytes. If the local inflammatory response to an infection is insufficient to control it, then a systemic acute phase response is induced which may include fever, leukocytosis and altered expression of acute phase proteins [9,48]. This co-ordinated reaction is triggered by both pathogen-associated molecular patterns (PAMP) and host damage-associated molecular patterns (DAMP) [49,50].
The high disease incidence frequently found in early lactation adversely affects cow welfare, reduces milk production capacity and fertility and increases the likelihood of culling [1,35]. This has raised the need to improve our understanding of the links between nutrition, metabolism and the immune system during the transition period. Such adverse consequences are more common in multiparous cows [51]. In comparison with primiparous cows, these also exhibit differences in their metabolic profiles in early lactation, having lower circulating IGF-1 and higher BHB concentrations [52]. The aim of this study was thus to investigate the relationship between circulating IGF-1 (used as a biomarker of metabolic status), disease incidence and the transcriptomic profiles of leukocytes from high yielding multiparous dairy cows in early lactation.
2. Materials and Methods
2.1. Animals
Multiparous Holstein cows were recruited from six experimental farms located in the UK (Agri-Food and Biosciences Institute Hillsborough, Northern Ireland, n = 44); Denmark (Aarhus University, n = 23); Ireland (University College Dublin, n = 35); Germany (Leibniz Institute for Farm Animal Biology, FBN, n = 19); Belgium (Walloon Agricultural Research Centre, n = 18); and Italy (Consiglio per la Ricerca in Agricoltura a l’Analisi, 18 dell’Economica Agraria, CREA, n = 37). This provided a starting population of n = 176. All procedures had local ethical approval and complied with the relevant national and EU legislation under the European Union (Protection of Animals used for Scientific Purposes) Regulations 2012 (S.I. No. 543 of 2012). All cows were milked twice daily and their daily yields were recorded. Milk samples were collected twice weekly until 50 days in milk (DIM) and were analyzed for composition of protein, fat, and lactose. Body weights (BW) were recorded at least twice weekly using weigh scales. Further details of the management of each herd are provided in [53]. The average milk yield for all six herds over the 7 week study period averaged 33.3 ± 9.3 kg/day, increasing from 25.8 ± 8.1 kg/day in week 1 to 36.1 ± 9.3 kg/day in week 7 [53]. For individual herds, the milk yield in week 2 (the time of leukocyte sample collection) ranged from 28.6 ± 1.10 kg/day (CREA) to 37.3 ± 2.01 kg/day (FBN) [33].
2.2. Measurement of IGF-1 and Metabolites in Blood
Blood samples were collected by jugular or tail vein venepuncture to obtain plasma (Na heparin tubes) and serum (plain tubes), separated by centrifugation and stored at −20 °C. Concentrations of IGF-1 were determined in serum by radioimmunoassay at University College Dublin, Ireland following acid-ethanol extraction [54]. Intra-assay coefficients of variation (CV) were 12.4, 7.5 and 9.9% for low, medium and high control samples, respectively. The corresponding inter-assay CV were 7.8, 3.9 and 9.4%. The sensitivity of the assay, defined as the lowest concentration detectable, was 4 ng/mL. NEFA, BHB, glucose and fructosamine analysis was performed at the Department of Animal Science, Aarhus University, Denmark [55]. Urea was determined in plasma according to standard procedures using an auto-analyzer (ADVIA 1800® Chemistry System, Siemens Medical Solutions, Tarrytown, NY, USA). Intra- and inter-assay CV for metabolite measurements were below 3% and 4%, respectively, for both low and high control samples.
2.3. Milk Sampling and Analysis for Metabolites and Enzymes
Additional milk samples (two × 8 mL) were collected twice weekly during morning milking and stored at −20 °C for subsequent analysis at the Department of Animal Science, Aarhus University, Denmark. The results from those taken closest to 14 DIM were used in this study. Fluorometric end point analyses were used to determine milk glucose and glucose-6-phosphate, uric acid, isocitrate and BHB as described by [53]. Urea was determined by spectrophotometry [56]. The indigenous enzymes lactic dehydrogenase (LDH, EC. 1.1.1.27) and N-acetyl-β-D-glucosaminidase (NAGase, EC 3.2.1.30) were analyzed by fluorometric assays [57,58]. Intra- and inter-assay CVs were in all cases below 5% and 8%, respectively, for both low and high control samples.
2.4. Calculation of Energy Balance
Records required for energy balance (EB) calculations at approximately 14 DIM were available from four herds in this study. For these, daily dry matter intakes were recorded using electronic feeding systems. Weekly ration samples collected over the course of the sampling period were dried and shipped for NEL analysis in a single run (Cumberland Valley Agricultural Services, Franklin County, PA, USA). The energy input was calculated by multiplying the weekly NEL density of the ration with the daily dry matter intake (DMI) of the animal. The energy output was calculated according to the National Research Council guidelines [59] correcting for NEM using the last recorded body weight of the animal and the most recent milk fat, protein and lactose content together with the daily milk yield for the NEL. The energy corrected milk (ECM) yield was adjusted to 3.5% fat and 3.2% protein using the formula ECM = (0.327 × milk kg) + (12.95 × fat kg) + (7.65 × protein kg). The energy balance (in MJ/day) was determined as described previously [53].
2.5. Health Records
An illustrated scoring system covering uterine disease, mastitis and lameness was agreed in advance in order to promote consistency in diagnoses between herds. Uterine discharge and odor were assessed at approximately 14 and 35 DIM as described previously [60]. In brief, the discharge was sampled from the vagina and assessed for colour, proportion and volume of pus, and scored as: 0, clear or translucent mucus; 1, mucus containing flecks of white or off-white pus; 2, <50 mL exudate containing over 50% white or off-white mucopurulent material; or 3, >50 mL exudate containing purulent material, usually white or yellow, but occasionally sanguineous. Discharge odor was given a score 0 for normal or 1 for fetid. Lameness was scored at the same time based on a nine point scale as previously described [61]. Milk quality was assessed based on the somatic cell count (SCC) and cows were subsequently categorized into three groups as described previously [33]. Healthy cows were defined as having a SCC <100,000 cells/mL milk with no clinical symptoms. Subclinically mastitic cows were defined as having a SCC between 100,000 and 400,000 cells/mL milk with no apparent clinical symptoms. In the cows diagnosed as having clinical mastitis, their SCC was >400,000 cells/mL milk and they displayed clinical symptoms. Other diseases were classified based on the ICAR Central Health Key (https://www.icar.org/index.php/publications-technical-materials/amendments-recording-guidelines/diseases-codes-for-cows/ last accessed on 15 November 2021). Clinical diagnoses were made and recorded by farm staff and any subsequent treatments followed the normal management practices in each herd.
2.6. RNA Extraction
Blood samples at approximately 14 DIM were also taken into Tempus blood collection tubes (Thermo Fischer, Loughborough, UK), which were shaken vigorously for 15–20 s immediately upon collection, then frozen and stored at −80 °C for RNA sequence analysis (RNA-seq). All chemicals were obtained from Sigma Aldrich (Poole, Dorset, UK) unless stated otherwise. Whole-blood extraction used Tempus Spin RNA isolation Kits (Thermo Fischer, Loughborough, Leics, UK) following the manufacturer’s instructions as described previously [8]. RNA quantity and integrity were assessed using an Agilent BioAnalyzer 2000 (Agilent Technologies UK Ltd., Cheadle, UK) and Agilent RNA 6000 Nano Kit (Agilent Technologies UK Ltd., Cheadle, UK). RNA measurements were also validated using a NanoDrop 1000 (Thermo Fischer, Loughborough, Leics, UK). All selected RNA sample had a reasonable integrity (RIN number >7) and purity (260/280 between 1.8 and 2.3). The RNA was stored at −80 °C for subsequent sequencing.
2.7. RNA Sequencing
RNA-seq of the extracted whole-blood samples was performed at the University of Liege, GIGA Research Facility, Belgium. RNA-seq libraries were prepared using an epMotion liquid handling workstation (Eppendorf, Hamburg, Germany) with the Illumina TruSeq Stranded Total RNA Library Prep Ribo-Zero Gold kit (Illumina, San Diego, CA, USA). Pooled cDNA libraries were sequenced on an Illumina NextSeq 500 machine (4 lanes multiplexed) at 75 nucleotide length single-end reads to reach an average 30 million reads per sample. Raw data were stored as FASTQ files (Sanger/Illumina 1.9 [Phred + 33]) in ENA accession numbers ERP124149 and ERP122911. Sequence analysis was performed with a CLC Genomics Workbench V21 (Qiagen Digital Insights, Redwood City, CA, USA) based on the built-in workflows in its toolbox of RNA Sequencing Analysis. This included trimming the poor-quality reads, quality control and mapping the reads to a reference genome of Bos taurus assembly for both genes and transcripts (ARS-UCD1.2, Genbank). The sequencing was quantified as reads per genes and reads per kilobase of transcript per million mapped reads (RPKM) and stored in CLC Genomics Workbench as gene expression (GE) files.
2.8. Relationship between Gene Expression with Circulating IGF-1
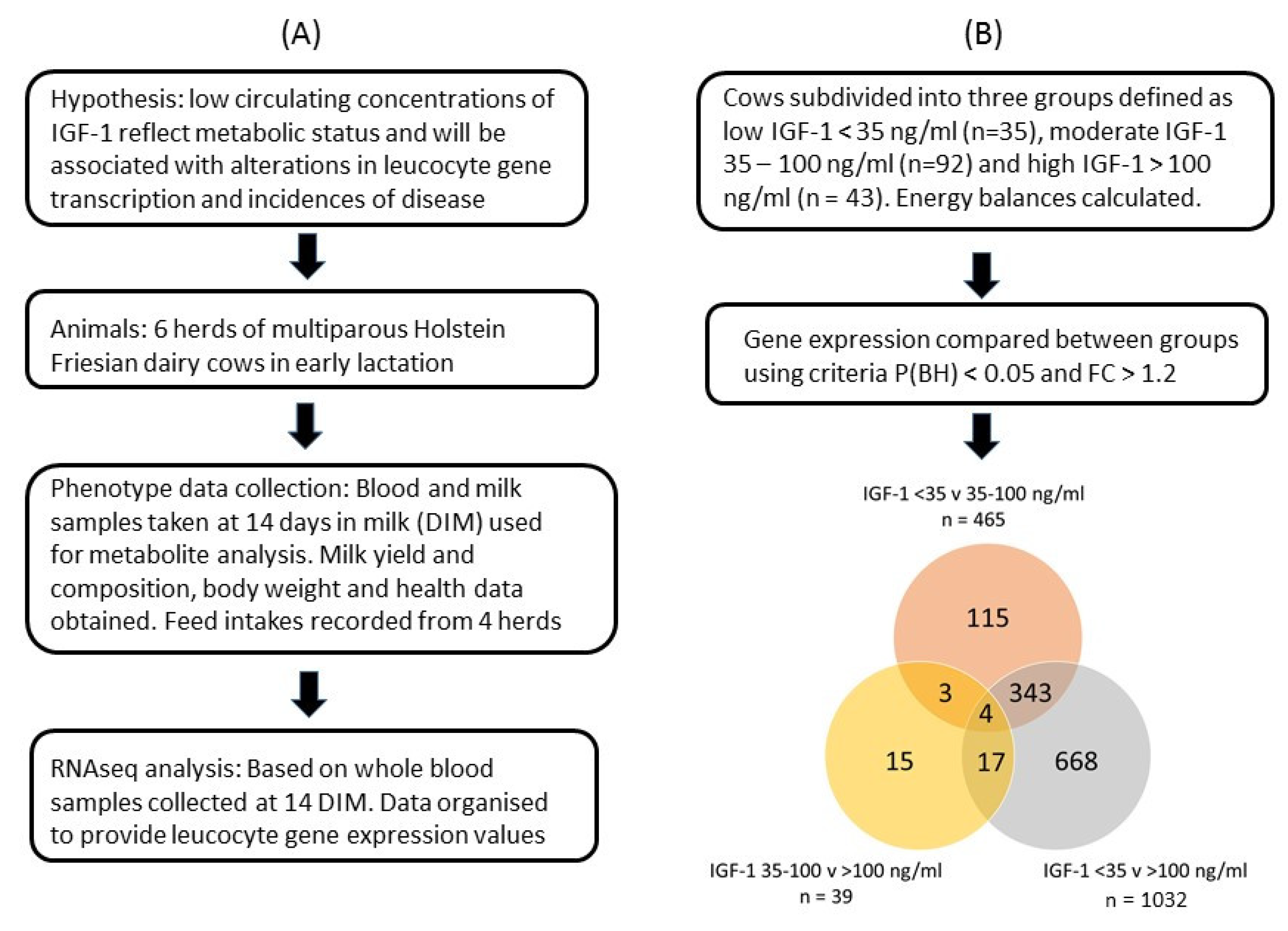
The GE files derived from each sample were used for analysis of differential gene expression in leucocytes collected approximately 14 DIM using CLC Genomics Workbench V21 (Qiagen Digital Insights, Redwood City, CA, USA). Global gene expression was related to the circulating IGF-1 concentration measured in blood samples taken at the same time. For this, cows were subdivided into three groups defined as low IGF-1 < 35 ng/mL (LOW, n = 35), moderate IGF-1 35–100 ng/mL (MOD, n = 92) and high IGF-1 > 100 ng/mL (HIGH, n = 43). These thresholds were selected based on our previous work showing associations between the IGF-1 concentrations in early lactation with the likelihood of developing endometritis [62] and subsequent fertility [63]. Four cows did not have an IGF-1 value, so they were removed from this study. Principal component analysis with normalized RNA-seq reads expressed as RPKM showed that the overall expression values of two cows were markedly different from the others. These were considered as outliers and excluded in the subsequent analysis. This left 170 cows in the analysis. The expression values were normalized with trimmed mean and Z-score normalizations across all samples. Gene expression levels were then compared between the IGF-1 groups using negative binomial generalized linear modeling. IGF-1 groups were set as the test variable and herd as the controlling variable to minimize the differences of gene expression arising from the differences of herds. False discovery rates (FDR) for multiple tests were controlled with the Benjamini–Hochberg (BH) adjustment and significance was considered at p(BH) < 0.05. Our previous studies of leucocyte gene expression [8] showed that a sample size of 15 cows per analytical group achieved 85% power at p < 0.05 with FDR correction to detect the true differential expression with fold changes > 1.2 between group pairs. Differentially expressed genes (DEG) with p(BH) < 0.05 and absolute fold change > 1.2 in pairwise comparisons between the three IGF-1 groups were selected for subsequent analysis. The DEG were taken forward for pathway analysis using DAVID bioinformatics resources version 6.8 (https://david.ncifcrf.gov/ accessed on 16 Aug 2021) [64,65] with Bos taurus as background. Fisher’s exact test with BH adjustment was used and statistical significance was considered at p(BH) < 0.05. An overview of the study design and data analysis is provided in Figure 1.
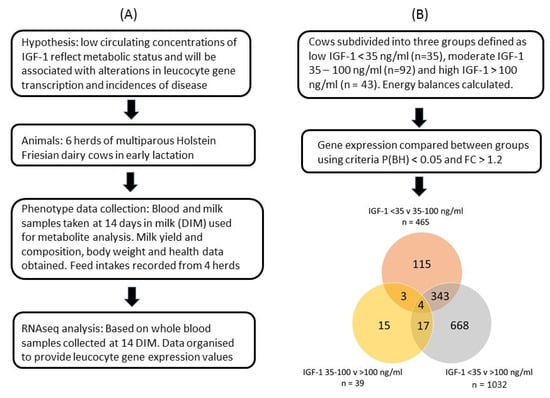
Figure 1.
Overview of (A) the study design and (B) the data analysis. Venn diagram indicates numbers of differentially expressed genes (DEG) identified in each group and their overlap between groups.
2.9. Statistical Analysis
The values for milk yield and composition, blood and milk metabolites and EB at 14 DIM were expressed as the mean ± standard error of mean (SE). Differences between the IGF-1 groups were compared with one-way analysis of variance (ANOVA) using SPSS V27 software package (Chicago, IL, USA). The homogeneity of variance for each variable was assessed with Levene’s test. Where homogeneity was not achieved, logarithmic transform was performed before ANOVA. Where ANOVA showed significance, multiple comparisons with Tukey’s HSD test were performed to identify the source of differences. Health data were tabulated and analyzed using Pearson’s chi-squared test. In both cases significance was considered where p < 0.05.
3. Results
This paper relates to the results obtained from blood samples collected from each cow at 14 ± 4 days DIM and health records relating to calving and the first 5 weeks postpartum. The blood samples were used for: (i) measurement of circulating IGF-1 and metabolite concentrations and (ii) RNA-seq analysis of whole-blood leucocyte populations.
3.1. Cow Data
Cows were subdivided into three groups based on their circulating IGF-1 concentration. Those in the LOW group had significantly higher parity, averaging 3.9 ± 0.27 lactations in comparison with 2.6 ± 0.17 for the HIGH cows (mean ± SEM, p < 0.001, Table 1). Cows in the HIGH group had significantly higher milk yields by the second week after calving. Their milk also contained more protein, had a slightly lower fat:protein ratio and a higher lactose content compared to LOW group cows (Table 1). The energy corrected milk 7 day averages for week 2 did not, however, differ between IGF-1 groups (LOW, 38.0 ± 1.29 kg; MOD, 39.3 ± 0.95 kg; HIGH, 41.9 ± 1.34 kg; p = 0.119).

Table 1.
Lactation number, milk yield and milk quality data from weeks one and two of lactation according to the IGF-1 group as measured in plasma at 14 days in milk.
Blood metabolite measurements made at the same time as the IGF-1, at approximately 14 DIM, showed that the HIGH cows also had higher circulating concentrations of glucose, fructosamine and urea than the other two groups (Table 2). NEFA concentrations were, however, greatest in the LOW group, with a trend towards raised concentrations of BHB. These findings were supported by milk metabolite measurements, which showed higher concentrations of free glucose, urea and uric acid in the HIGH group, whereas milk BHB and isocitrate were significantly raised in the LOW IGF-1 cows (Table 2). Cows from four of the six herds used in this study (n = 117) also had their feed intakes recorded. These together with cow weights and milk yield and content were then used to calculate energy balances. These data showed that the cows in all three IGF-1 groups had similar average body weights, but the LOW group cows consumed less feed in early lactation, with a DMI on average 3.4 kg/d lower than that of the HIGH group cows. This was associated with a significantly lower EB value in the LOW IGF-1 cows (Table 2).

Table 2.
Blood and milk metabolites, feed intakes and calculated energy balance at 14 days in milk according to the IGF-1 group.
3.2. Health
Among the LOW IGF-1 cows, only 3/35 (9%) had no recorded health issues during the first 5 weeks of lactation, 10/35 (29%) had one recorded health problem and 22/35 (63%) had more than one problem, with up to four recorded in total. This compared with 6/43 (14%) of the HIGH IGF-1 cows with no problems, 26/43 (60%) with one problem and 11/43 (26%) with two problems. Breaking this down further, there were no differences in the proportions of cows in each IGF-1 group according to various calving parameters (assisted calving, stillbirths, twins or retained placenta) (Table 3).

Table 3.
Health data in the first 35 days in milk (DIM) according to the circulating IGF-1 concentration at 14 days in milk †.
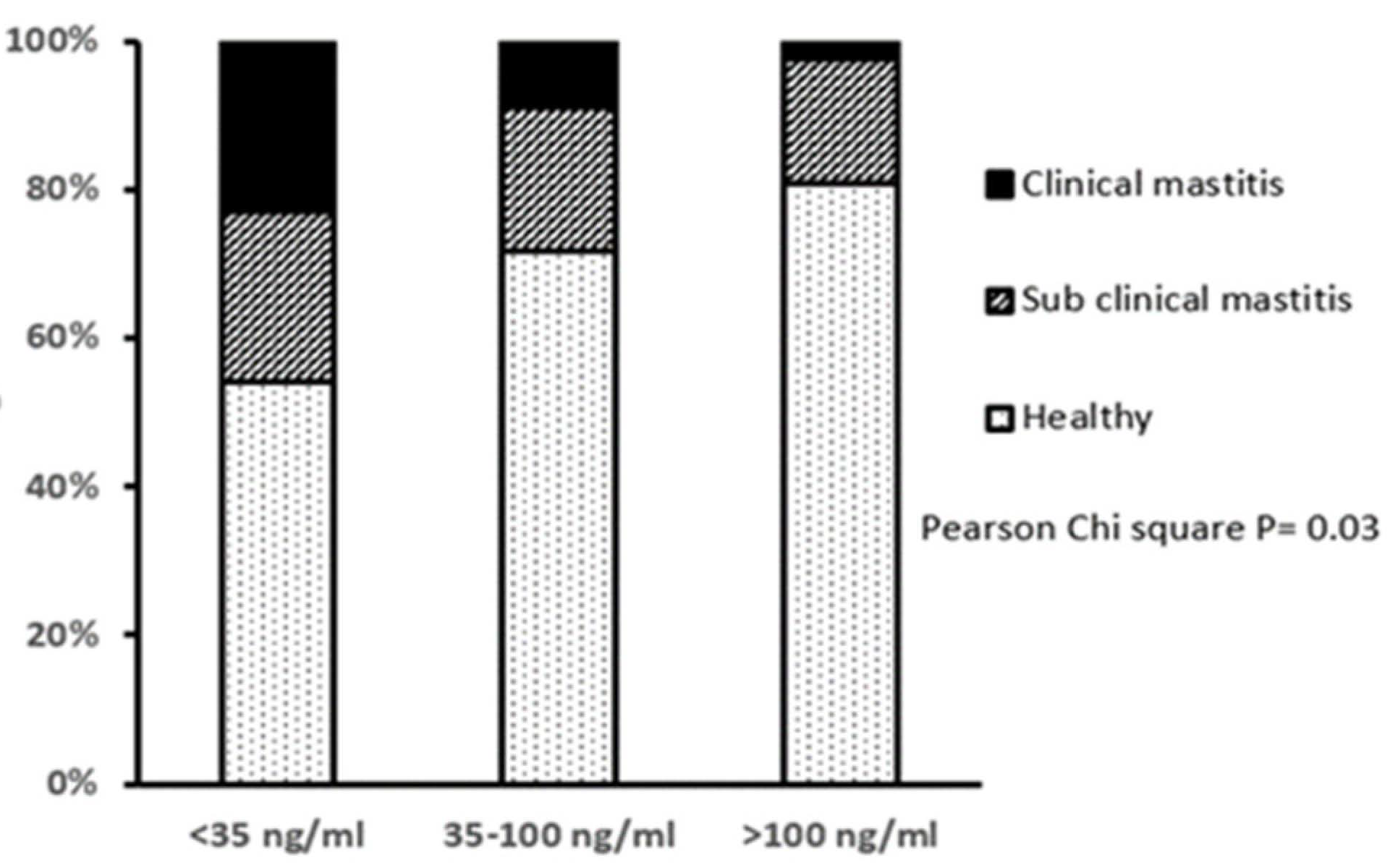
Approximately 29% of all cows had a lameness score >3 at day 14, assessed on a scale of 0–9, implying some degree of lameness, but there were no differences in the proportions of lame animals between the three IGF-1 groups. However, LOW IGF-1 cows had a higher proportion of cases of displaced abomasum and the only two cases of milk fever reported in this study. Based on the herd records and measurements of SCC at 14 DIM, the LOW IGF-1 cows also had a higher proportion with clinical mastitis in early lactation and correspondingly fewer classified as having healthy milk (Figure 2).
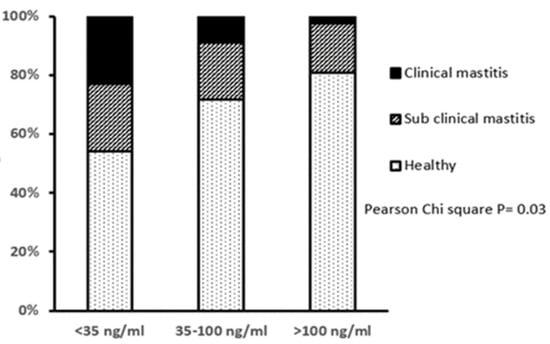
Figure 2.
Relationship between circulating IGF-1 concentration at 14 days in milk and the incidence of mastitis in early lactation. Cows were grouped by their circulating IGF-1 concentration as LOW (<35 ng/mL, n = 35), MODERATE (35–100 ng/mL, n = 92) or HIGH (>100 ng/mL, n = 43). The mastitis classifications were: Healthy (SCC < 100,000 cells/mL milk, no clinical symptoms); subclinical mastitis (SCC 100,000 to 400,000 cells/mL milk, no clinical symptoms) or clinical mastitis (SCC > 400,000 cells/mL milk, with clinical symptoms). There was a significant difference in the proportions of cows with different mastitis classification between IGF-1 groups, with fewer healthy and more mastitic cows in the LOW IGF-1 group.
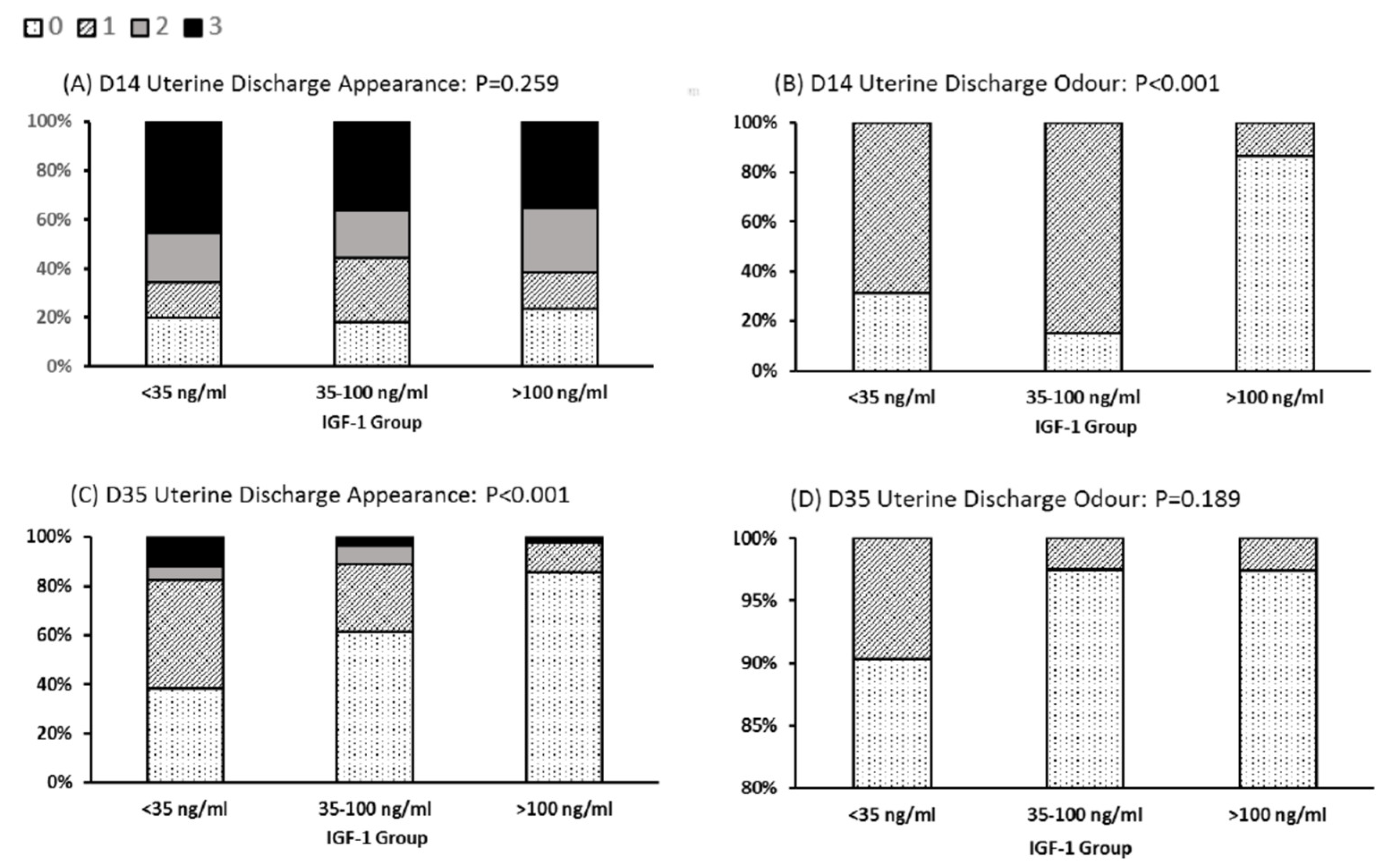
There were also differences noted in the character of the discharge from the reproductive tract (Figure 3). The LOW IGF-1 cows contained a significantly higher proportion with a fetid odor in their uterine discharge at day 14 (69% in IGF-1 < 35 ng/mL vs. 13.5% in cows with IGF-1 > 100 ng/mL). For 86% of cows in the HIGH IGF-1 category, the uterine discharge at day 35 was clear. This compared with only 38% of LOW IGF-1 cows having clear mucus at this time, with 62% classified as having mucus containing mucopurulent material (Figure 3).
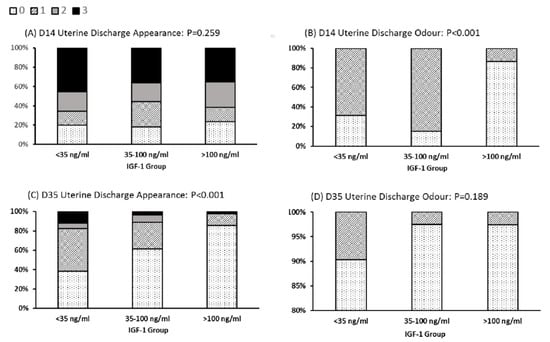
Figure 3.
Relationships between IGF-1 category and uterine discharge appearance and odor. Cows were grouped by their circulating IGF-1 concentration as LOW (<35 ng/mL, n = 35), MODERATE (35–100 ng/mL, n = 92) or HIGH (>100 ng/mL, n = 43). Uterine discharge was sampled from the vagina at 14 and 35 days in milk (D14, D35) and scored as: 0, clear or translucent mucus; 1, containing flecks of white or off-white pus; 2, <50 mL exudate containing over 50% white or off-white mucopurulent material; or 3, >50 mL exudate containing purulent material, usually white or yellow, but occasionally sanguineous (A,C). Discharge odor was scored 0 for normal or 1 for fetid (B,D). The graphs are plotted to show the percentage of animals in each IGF-1 category with each score. Note the difference in y-axis scale used for (D). There was a significant difference in classification (Pearson’s chi-squared test) among IGF-1 groups with respect to discharge odor at Day 14 and discharge appearance at D35.
3.3. Gene Expression Data in Leucocytes
Gene expression data in circulating leukocytes were compared between the three IGF-1 groups. A comparison of gene expression levels between the LOW and HIGH IGF-1 groups generated a list of 1032 DEG. There were 465 DEG between the LOW and MOD groups but only 39 DEG between the MOD and HIGH groups (Figure 1). These results indicated that most of the differences in gene expression lay between the cows with LOW IGF-1 and the remainder. Subsequent analysis as reported here therefore focused on the comparison between cows with LOW and HIGH IGF-1. This included 817 genes which were more highly expressed in the LOW cows (i.e., up-regulated with low IGF-1) and 215 which were more highly expressed in the HIGH cows (i.e., down-regulated in cows with low IGF-1). Only four genes were common between all three comparisons. Three of these were most highly expressed in the LOW IGF-1 cows (CFAP69, cilia and flagella-associated protein 69; DMBT1, deleted in malignant brain tumors 1 and MERTK, MER proto-oncogene, tyrosine kinase) and one was most highly expressed in the HIGH IGF-1 cows (SEMA6B, semaphorin 6B).
3.4. Genes Up-Regulated in LOW IGF-1 Cows
The full list of 817 genes is provided in Supplementary Table S1, with the top genes summarized in Table 4. PTX3 and DMBT1, which both have antimicrobial activity, were the two most highly up-regulated genes and were also the most highly significant. Other up-regulated genes encode proteins with a role in inflammation and coagulation (FGA, LOC112445470, LOC511106, LOC107131803) and TF encodes transferrin, an iron transport protein. Two genes with important immunological functions were highly significant: DCSTAMP is involved in osteoclastogenesis and myeloid differentiation, while PROK2 acts on prokineticin receptors on monocytes, macrophages and dendritic cells to induce chemotaxis and release of inflammatory cytokines. The three up-regulated genes with the highest expression levels all encode S100 calcium-binding proteins (S100A8, S100A9 and S100A12) and other highly expressed genes were HP, RNAse6 and CTSB. Haptoglobin plays a key role in iron metabolism and all three of these genes encode proteins with antimicrobial activity. Two other highly significant genes, GATM and PDK4, have important roles in metabolism. Of the highly expressed genes, PFM encodes a pyruvate kinase important in glycolysis and TALDO1 is involved in the pentose phosphate pathway.

Table 4.
Topmost significantly up-regulated genes in leucocytes of cows with LOW (<35 ng/mL, n = 35) in comparison with HIGH (>100 ng/mL, n = 43) circulating IGF-1 at 14 days in milk.
3.5. Genes Down-Regulated in LOW IGF-1 Cows
Comparison of gene expression levels between the LOW and HIGH IGF-1 groups generated a list of 215 DEG which were down-regulated in cows with low IGF-1. The full list is provided in Supplementary Table S2 and the top genes are summarized in Table 5. A number of genes with roles in immune signaling (ST6GAL1, CXXC5, BLA-DQB, CD52, MS4A1, KIAA0040, SPIB, BANK1, PTPRCAP, CD79B, SEMA6B) and autoimmunity (MYH6, LBH) were less highly expressed in the cows with LOW IGF-1. Two further genes encoded proteins identified as being multidrug resistance-associated protein 4-like (LOC112449072, LOC100847574). Of these, LOC112449072 featured in both the greatest fold change and the most significant lists. The purinergic receptor P2RY10 is involved in ATP release from cells and inflammation. LTB and LBH, encoding lyphotoxin beta and limb bud and heart development, respectively, also have potential roles in inflammation. Several other less highly expressed genes in LOW IGF-1 cows are involved in the cytoskeleton and cell motility (DMTN, REM1, MYLIP, ARPC5L). HBZ had the highest fold change. This gene encodes zeta-globin, an alpha-like hemoglobin normally expressed during embryonic development while ALAS2 encodes an erythroid-specific mitochondrially located enzyme which catalyzes the first step in the heme biosynthetic pathway. Highly down-regulated genes encoding two other mitochondrial enzymes were COQ2 and BCAT2.

Table 5.
Top most significantly down-regulated genes in leucocytes of cows with LOW (<35 ng/mL, n = 35) in comparison with HIGH (>100 ng/mL, n = 43) circulating IGF-1 at 14 days in milk.
3.6. Cluster Analysis
Cluster analysis was performed in DAVID using a gene list which combined all the up-regulated and down-regulated DEG between LOW and HIGH IGF-1 cows. This analysis identified five significant annotation clusters with enrichment scores between 7.69 and 2.77 (Table 6). The genes in each cluster are listed in Supplementary Tables S3 and S4. Clusters 1 and 2 were both large clusters with considerable overlap, with 82 genes (19%) common to both. These focused around disulfide bonds, glycoproteins, glycosylation sites and transmembrane proteins. Out of the total of 425 genes in these two clusters, 340 (80%) were up-regulated in the LOW IGF-1 cows and 85 (20%) were down-regulated. Based on the GO term cellular components, most were associated with either a membrane (plasma, Golgi, endoplasmic reticulum, lysosome) or the extracellular space, but they had a very wide variety of molecular functions. As the majority of proteins encoded by immune-related genes are located in or at the plasma membrane, this is expected from an immune cell population.

Table 6.
Functional cluster analysis of all genes which showed significant differential expression in leukocytes collected from dairy cows with LOW (<35 ng/mL) or HIGH (>100 ng/mL) circulating IGF-1 concentrations, sampled at approximately 14 days in milk.
Cluster 3 identified genes associated with scavenger receptor activity. Of these CD163, MARCO and SCARF1 were all up-regulated in LOW IGF-1 cows. CD163 encodes a protein which is important in the clearance of hemoglobin/haptoglobin complexes by macrophages, MARCO encodes a receptor which may bind both Gram-negative and Gram-positive bacteria, while SCARF1 regulates the uptake of chemically modified low density lipoproteins. The gene list also included four members of the WC1 co-receptor family (LOC786796, WC1, WC1.3, WC-7). These are group B scavenger receptors which are found exclusively on gamma delta T cells, all of which were less highly expressed in the LOW IGF-1 cows.
Cluster 4 identified 20 genes involved in defense responses to bacteria, of which all but one were up-regulated in LOW IGF-1 cows. These mainly encoded antimicrobial proteins (CATHL4, DEFB1, DEFB13, DEFB4A, DEFB7, DEFB10, HP, LAP, S100A8, and S100A9). The cluster also included genes encoding two components of the IgE receptor (FCGR1A and FCER1G), the TNFα receptor (TNFRSF1A) and BCL3 and NOD2, which are both involved in the NFκB pathway. ELANE was the only down-regulated gene. This encodes an elastase which hydrolyzes proteins in azurophil granules, specialized lysosomes found in neutrophils.
Cluster 5 contained 28 genes relating to “Inflammatory response” and “Innate immunity”, of which all but four were up-regulated in LOW IGF-1 cows and six were common to Cluster 4. In addition to the antimicrobial proteins already mentioned, this cluster included additional antimicrobial genes (PTX3, S100A12), genes encoding key cytokine and chemokine signaling molecules (TLR4, CD14, CCL8, CXCL8, IL2RA), two members of the complement system (C2, CFB) and the scavenger receptor CD163. Other genes involved in inflammation included two components of the coagulation factor fibrinogen (FGA, FGB). The down-regulated genes were BOLA-DQB, BLK, LGR4, and TLR10.
3.7. KEGG Pathway Analysis
KEGG pathway analysis using all identified DEG between LOW and HIGH IGF-1 cows initially revealed four pathways significant at p(BH) < 0.05. These are summarized in Table 7 and the full gene lists are provided in Supplementary Tables S5 and S6.

Table 7.
KEGG pathway analysis based on all genes which showed significant differential expression in leukocytes collected from dairy cows with LOW (<35 ng/mL) vs. HIGH (>100 ng/mL) circulating IGF-1 concentrations, sampled at approximately 14 days in milk.
The pathway “bta04640:Hematopoietic cell lineage” is again to be expected in a leukocyte cell population. Three other highly significant pathways were “bta04611:Platelet activation”, “bta04610:Complement and coagulation cascades” and “bta04060:Cytokine–cytokine receptor interaction”. A further three pathways were identified at p(BH) = 0.064. These were “bta04015:Rap1 signaling pathway”, “bta04978:Mineral absorption”, and “bta04512:ECM-receptor interaction”.
No significant metabolic pathways were identified in the initial analysis. As this was unexpected, a manual search of the entire list of 1,032 DEG was therefore made, which identified 149 genes (14.4%) which had some known metabolic function (Supplementary Table S7). These were then input into DAVID as a new gene list. Of these, 44 genes were present in KEGG pathway “bta01100:Metabolic pathways” of which 36 were up-regulated and 8 down-regulated in LOW IGF-1 compared to HIGH cows (Supplementary Tables S8 and S9). A further five significant metabolic pathways were identified, as summarized in Table 8: “bta01230:Biosynthesis of amino acids”, “bta01200:Carbon metabolism”, “bta00410:beta-Alanine metabolism”, “bta00620:Pyruvate metabolism” and “bta00770:Pantothenate and CoA biosynthesis”. All of these pathways participate in energy generation. In addition the pathway “bta02010:ABC transporters” was significant. ABC transporters are integral membrane proteins that allow ATP-dependent movement of various molecules across the plasma membrane.

Table 8.
KEGG pathway analysis of DEG # which showed significant differential expression in leukocytes collected from dairy cows with LOW (<35 ng/mL) vs. HIGH (>100 ng/mL) circulating IGF-1 concentrations, sampled at approximately 14 days in milk.
4. Discussion
4.1. Gene Expression in Peripheral Blood
Whole peripheral blood has been widely used in gene expression studies due to its initial ease of collection and because it can be processed directly without the requirement to separate out specific cell types. The whole-blood leucocyte population includes T and B lymphocytes, natural killer cells, platelets, monocytes and granulocytes (neutrophils, eosinophils and, basophils). In dairy cows the average leucocyte population is between 5–12 cells per ml of blood, predominantly consisting of lymphocytes and neutrophils [66]. The transcriptional changes measured between different cows in this study therefore represent changes in gene expression within particular cell types together with changes in their relative proportions. This proportion will change as cells of different lineages may proliferate, undergo apoptosis or migrate to or from tissues into blood in response to different signals [66,67]. It was not possible to collect additional blood samples for analysis of leukocyte populations within this study. Despite this limitation, many previous studies in both human and livestock have now reported that transcriptional signatures of whole blood can reliably differentiate individuals with a variety of diseases [68,69].
4.2. Phenotypic Changes
Milk production capacity should increase progressively until approximately the fifth lactation [70]. However, the older LOW IGF-1 cows, with a mean lactation number of 3.9, produced less milk than the HIGH cows by the second week of lactation and their milk contained less protein. This was associated with reduced feed intakes, with DMIs being on average approximately 3 kg/d lower in the LOW cows. Voluntary DMIs during late pregnancy fall to a nadir around calving then increase again in early lactation, during which time a variety of metabolic and humoral signals and environmental factors affect appetite [3,5]. Blood and milk metabolite measurements showed that the LOW IGF-1 cows had lower glucose, fructosamine and urea concentrations together with raised BHB and NEFA. These indices show that the LOW IGF-1 cows were experiencing a shortage of both protein and energy, which would cause them to mobilize more body tissue. The higher NEFA concentration may itself be one factor reducing feed intake [3]. In addition, endotoxemia and sepsis cause acute phase inflammatory responses, which drive a shift towards catabolism [48]. During this process a number of cytokines can cause hypophagia, in particular tumor necrosis factor-α (TNF-α), interleukin-1β (IL-1β), and interleukin-6 (IL-6) [71,72]. Treatment of cattle steers with lipopolysaccharide (LPS, an endotoxin produced by Gram negative bacteria) to induce an acute phase response reduced their feed intake by 60% within 24 h and this was accompanied by a reduction of 24% in circulating IGF-1 [73].
In terms of health, the LOW IGF cows experienced higher incidences of displaced abomasum and uterine infections, confirming our earlier studies [28,74]. The LOW IGF-1 cows were also more likely to be affected by either clinical or subclinical mastitis in early lactation. Based on the SCC data, it is possible that some cows may have had a mammary infection during the dry period which was carried over into the next lactation. This has been shown to influence inflammatory responses during the transition period [75], but was not monitored in our study. These results therefore support earlier work cited in the Introduction establishing a link between health and metabolism. It should be noted, however, that our experimental design could not distinguish cause from effect. A two-way interaction seems likely with nutrient deficiency compromising the immune system while infections and inflammation reduce appetite, so worsening the situation.
4.3. Markers of Energy Metabolism
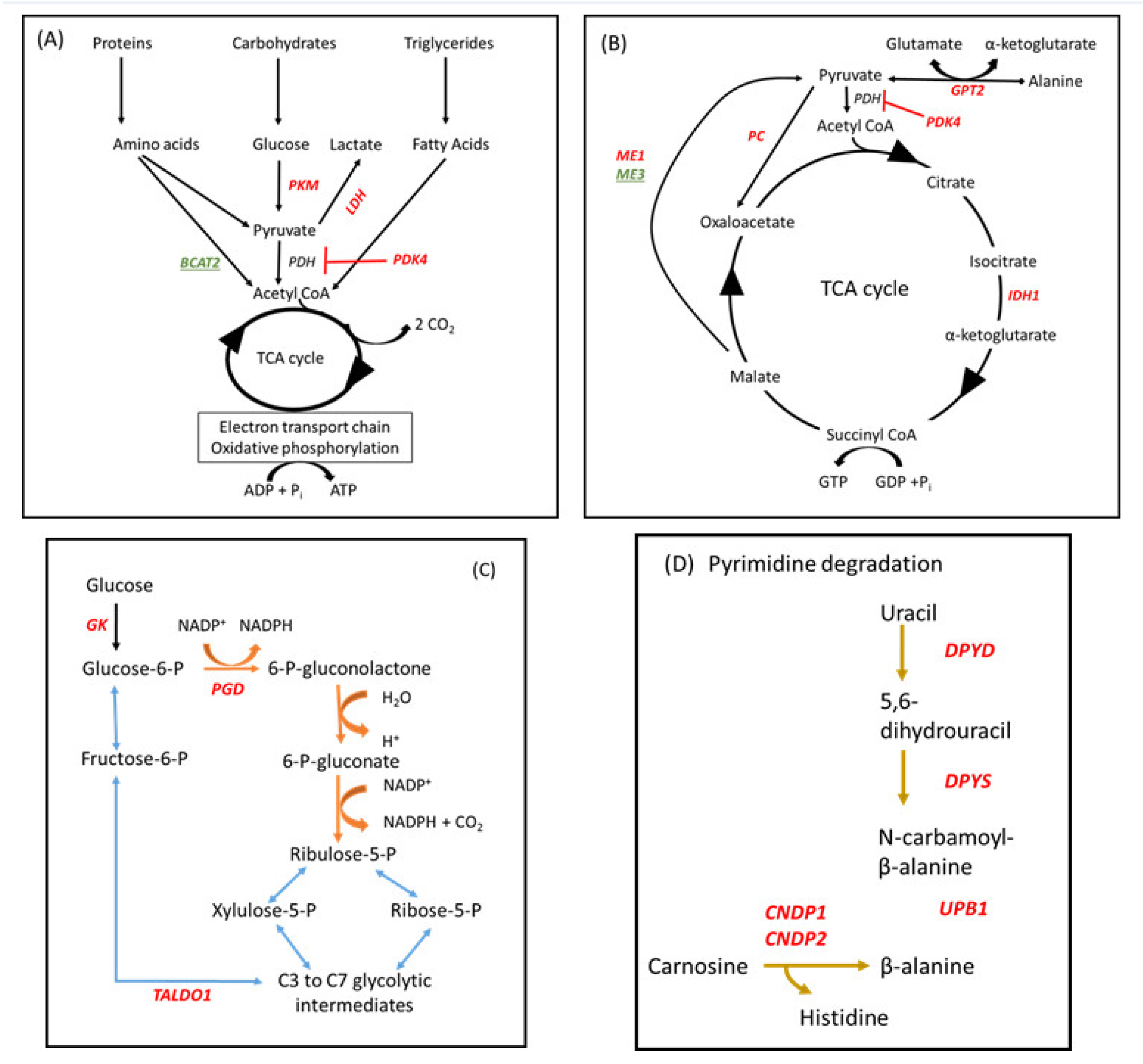
Immunometabolism encompasses both the immediate response to an infection and the longer term inflammatory responses triggered by cell injury or infection [76]. As mitochondria are the main organelles of energy metabolism, the overall ability of a cell to respond appropriately becomes compromised if any sort of stressor causes mitochondrial dysfunction. Important genes and pathways which were differentially affected between LOW and HIGH IGF-1 cows in our study are illustrated in Figure 4.
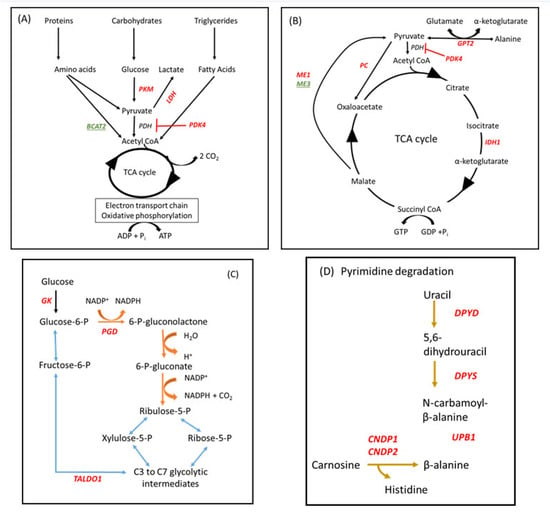
Figure 4.
Metabolic pathways which were differentially influenced in leucocytes between cows with LOW (<35 ng/mL) and HIGH (>100 ng/mL) IGF-1 in early lactation #. # Genes encoding enzymes which were up- or down-regulated in LOW IGF-1 cows are shown in red or green font, respectively. (A,B) Energy sources used for adenosine triphosphate (ATP) and guanine triphosphate (GTP) synthesis via the tricarboxylic acid (TCA) cycle. In LOW IGF-1 cows PKM2, PDK4, LDH, GPT2, PC, ME1, IDH1 were all up-regulated whereas BCAT2 and ME3 were down-regulated. (C) The pentose phosphate shunt (PPS) provides an alternative pathway for oxidation of glucose. It has two phases, an oxidative phase (shown in brown), in which NADPH oxidase and ribulose-5-phosphate (a pentose sugar) are generated, and a non-oxidative phase (shown in blue), in which unused ribulose-5-phosphate is converted to other sugar intermediates and returned to the glycolytic pathway. GK, PGD, and TALDO1 were all up-regulated in LOW IGF-1 cows. (D) Part of pathway bta00410:beta-alanine metabolism. DPYD, DPYS, UPB1, CNDP1 and CNDP2 were all up-regulated in LOW IGF-1 cows. See text for further information.
Three genes which were highly up-regulated in leucocytes of LOW IGF-1 cows encode enzymes which are absolutely central to the reprogramming of metabolism associated with mounting an immune response, namely PKM2, PDK4, and GATM. In resting macrophages, most pyruvate is directed to the tricarboxylic acid (TCA) cycle via acetyl-CoA to generate ATP via oxidative phosphorylation (OXPHOS), although some is converted to lactate. In activated immune cells, such as M1 macrophages and dendritic cells stimulated by LPS, their energy metabolism switches from OXPHOS to aerobic glycolysis, similar to the “Warburg effect”, causing conversion of the majority of the pyruvate generated into lactate, with glycolytic intermediates providing the source of adenosine triphosphate (ATP) [77,78]. This is associated with up-regulation of pyruvate kinase M2 (PKM2) the rate-limiting enzyme in the glycolytic pathway [79]. At the same time, the mitochondrial enzyme pyruvate dehydrogenase kinase isozyme 4, encoded by PDK4, is up-regulated in response to an increased lipid supply in the form of free fatty acids, inactivating the pyruvate dehydrogenase complex and helping to conserve glucose by limiting the conversion of pyruvate to acetyl-CoA [80] (Figure 4A,B). Under anaerobic conditions, lactate dehydrogenase promotes the NADH-dependent reduction of pyruvate to lactate. Another mitochondrial enzyme GATM encodes glycine amidinotransferase (AGAT) which is central to creatine biosynthesis [81]. Creatine then combines with a phosphoryl group to generate phosphocreatine, which can donate its phosphate group to convert adenosine diphosphate (ADP) into ATP, so acting as a rapidly available source of energy.
Key genes contributing to the pentose phosphate shunt (PPS) were also up-regulated in LOW IGF-1 cows (Figure 4C). This provides an alternative pathway for oxidation of glucose and generates both nicotinamide adenine dinucleotide phosphate (NADPH) oxidase and ribulose-5-phosphate. Glucokinase (GK) converts glucose to glucose-6-phosphate, the starting point for the PPS, while phosphogluconate dehydrogenase (PGD) is the rate-limiting enzyme in this pathway. Some ribose-5-phosphate is used for nucleotide synthesis but unused glycolytic intermediaries can be recycled by transaldolase 1 (TALDO1). The relative production of NADPH varies with metabolic demand and it subsequently acts as a reducing agent in many biosynthetic pathways including fatty acid synthesis, detoxification and ROS generation. For example, the PPS is enhanced in macrophages stimulated with LPS and this is crucial in providing anti-oxidant cellular defense [76,82].
Two of the KEGG pathways identified in this study centered on amino acid metabolism. A number of identified DEG (GPT2, BCAT2, PC, IDH1, and PKM) are involved in regulating the balance of usage between proteins and carbohydrates to provide energy for cell metabolism via the TCA cycle (Figure 4A,B). Glutamic-pyruvic transaminase 2 (GPT2) catalyzes the reversible transamination between the glucogenic amino acid alanine and 2-oxoglutarate to generate pyruvate and glutamate. Pyruvate carboxylase (PC) converts pyruvate to oxaloacetate and is activated by acetyl CoA during fasting. Isocitrate dehydrogenase (IDH1) is one of three regulated enzymes contributing to the TCA cycle, catalyzing an oxidative decarboxylation reaction which generates NADH. Malic enzyme catalyzes the oxidative decarboxylation of malate to pyruvate. ME1 encodes a cytosolic, NADP(+)-dependent isoform which was up-regulated whereas ME3 encodes a mitochondrial NADP(+)-dependent isoform which was down-regulated. This reaction links the glycolytic and citric acid cycles and generates NADPH. BCAT2 encodes a branched-chain aminotransferase which catalyzes the first step in the breakdown of branched-chain amino acids (valine, leucine and isoleucine, BCAAs). Intermediary branched-chain α-keto acids are subsequently degraded further to generate propionyl CoA (valine and isoleucine) or acetyl CoA (leucine and isoleucine), which can then enter the TCA cycle. As BCAT2 was down-regulated in LOW IGF-1 cows, this would be expected to increase circulating BCAA. Multiple studies have found a positive correlation between circulating concentrations of BCAA, obesity and insulin resistance in human populations [83,84]. Other DEG included ASS1 and CPS1, which both encode enzymes involved in the urea cycle. DPYD, DPYS and UPB1 encode three consecutive enzymes required for the degradation of uracil to β-alanine, while CNDP1 and CNDP2 both encode carnosine dipeptidases, which convert carnosine to β-alanine (Figure 4D). In LOW IGF-1 cows this would reduce availability of carnosine, a dipeptide which has beneficial antioxidant effects including acting as a ROS scavenger [85]. β-alanine itself may, however, be involved in synaptic transmission [86].
COQ2 was a highly down-regulated gene in LOW IGF-1 cows. This encodes coenzyme Q2, a polyprenyltransferase that forms part of the coenzyme Q10 pathway. CoQ10 is an obligate component of the mitochondrial electron transport chain and CoQ depletion in MCF-7 cells was shown to decreases the ATP level and increase oxidative stress. It also induced a glycolytic phenotype causing an increased glucose uptake rate, increased lactate production, and led to mitochondrial dysfunction [87,88]. Overall, these results provide strong evidence that the energy and protein deficiency experienced by LOW IGF-1 cows leads to significant alterations in their leucocyte gene expression, indicative of a switch from OXPHOS to aerobic glycolysis and an increased reliance on glucogenic carbon. This is likely to be associated with increased oxidative damage in comparison with leucocytes from HIGH IGF-1 cows.
4.4. ABC Transporters
Two genes (LOC112449072 and LOC100847574) more highly expressed in HIGH IGF-1 cows encode multidrug resistance-associated protein 4-like proteins which belong to the superfamily of ATP-binding cassette (ABC) transporters. These are integral membrane proteins which allow ATP-dependent movement of various molecules across the plasma membrane. ATP, UTP, ADP, and NAD+ are all released into the extracellular space by damaged and dying cells, but also by regulated mechanisms which include ABC transporters. It is therefore possible that these transporter proteins are involved in secretion of ATP into the extracellular space [89] where it acts as an inflammatory mediator [90]. Our study also identified the ABC transporter pathway, which included six up-regulated (ABCA3, ABCA5, ABCA6, ABCA9, and ABCA10) and one down-regulated (ABCG1) gene in LOW IGF-1 cows. Some of the encoded proteins have been implicated in cholesterol efflux to high density lipoprotein, which reduces accumulation of cholesteryl esters in macrophages. Combined deletion of ABCA1 and ABCG1 in mice leads to excessive lipid accumulation in tissue macrophage foam cells causing inflammation [91,92]. The precise roles of many of these individual ABC transporters do, however, need further investigation.
4.5. Antimicrobial Activity
There was strong evidence for up-regulation of many genes which encode proteins with antimicrobial activity in the LOW IGF-1 cows. Pentraxin 3, encoded by PTX3, is a member of the long pentraxin family, which plays an essential role in innate immunity and is an established biomarker for several inflammatory conditions [93]. Expression in dairy cows increased in mammary tissue following experimental infection with Staphylococcus aureus [94] and also in endometrium of postpartum cows with metritis [95]. DMBT1 encodes a multifunctional glycoprotein known as deleted in malignant brain tumors 1 or salivary scavenger and agglutinin (SALSA). This protein is able to bind both bacteria and viruses directly and can also interact with various endogenous ligands, including IgA, lactoferrin, fibrin and components of the complement system [96,97]. It plays an important role in both immune defense and inflammation while deposition in the vascular epithelium assists with vascular repair [98]. S100A8, S100A9 and S100A12 are all calcium-binding proteins belonging to the S100 protein family. They act as antimicrobials and in wound healing [99,100,101] and can amplify endotoxin-induced up-regulation of cytokines, chemokines and other inflammatory mediators [102,103]. Their respective genes are amongst the most highly expressed in endometrium of early postpartum cows in severe NEB [28] or with endometrial inflammation [104]. S100A8 and S100A9 proteins were localized to epithelial and stromal cells in addition to leucocytes [105] and gene expression in bovine endometrium in vitro was up-regulated in response to LPS [106].
Additional antimicrobial genes were up-regulated in LOW IGF-1 cows, contributing to Cluster 4 “defense response to bacteria” (CATHL4, DEFB1, DEFB4A, DEFB7, DEFB10, DEFB13 and LAP). These encode cathelicidin 4 and several β-defensins, which belong to a large group of cationic peptides with amphipathic properties, enabling them to permeate pathogen membranes and cause cell death [107,108]. They also act as DAMPs to activate host responses to cell damage [109]. All of the genes identified here apart from DEFB13 have previously been associated with bovine mastitis [33,110,111] and/or endometrial infection and inflammation in postpartum cows [28,104]. Neutrophil expression of DEFB1, -4 and -10 also increased on the day of calving [112].
RNAse4 and CTSB were highly expressed in leucocytes of LOW IGF-1 cows. RNAse4 encodes a protein with broad-spectrum antimicrobial activity which is also secreted into milk, so contributing to host defense in the mammary gland [113]. CTSB encodes cathepsin B, an endogenous cysteine protease with both endopeptidase and exopeptidase activity, providing the ability to kill pathogens [114]. The protein is abundant in milk from cows with subclinical mastitis associated with Staphylococcus aureus or Streptococcus uberis infection [115].
Two other highly up-regulated genes were TF and HP, which encode transferrin (also known as lactoferrin) and haptoglobin, respectively, which are principally involved in iron homeostasis. The pathway “Mineral absorption” included CYBRD1, STEAP1 and STEAP2, which all encode proteins that participate in iron uptake, while HMOX1 encodes heme oxygenase, an essential enzyme in heme catabolism. Sufficient iron is essential for many cellular processes but iron in excess is harmful as it catalyzes free radical formation and also increases availability for microbial proliferation [116]. Macrophages play a key role in maintaining iron homeostasis by phagocytosing senescent erthyrocytes and recycling the iron [117]. Transferrin is able to sequester free iron from the environment to inhibit bacterial growth [118] and it also has direct bactericidal actions and can prevent viral entry into cells [119]. Haptoglobin binds free plasma hemoglobin, which is then captured by CD163, a ligand-specific transmembrane scavenger receptor found on monocytes [48], which was also up-regulated in LOW IGF-1 cows. This may help to protect tissues from free hemoglobin-mediated oxidative damage.
Although hemoglobin is mainly associated with erythrocytes, it can also be found in PBMC in which it localizes to the mitochondria with the dual function of increasing mitochondrial respiratory rate and protecting from H2O2-induced cytotoxicity and mitochondrial DNA damage [120]. HBZ expression is normally found in embryonic erythroid cells but the gene can be partially re-activated postnatally by acetylation and inhibition of histone de-acetylases [121]. ALAS2 encodes a mitochondrial enzyme 5’-aminolevulinate synthase 2 that regulates the initial step of heme biosynthesis and the gene is up-regulated in blood leukocytes of human patients with sepsis [122]. In contrast, both HBZ and ALAS2 in the present study were more highly up-regulated in cows with HIGH IGF-1, possibly providing a greater level of cytoprotection.
4.6. Vascular Damage and Inflammation
Two of the most significant KEGG pathways identified were “Platelet activation” and “Complement and coagulation cascades”, which have overlapping components. Following vascular injury, platelets initially adhere to the extracellular matrix (ECM), with the glycoprotein von Willebrand factor (VWF) forming a bridge between exposed collagen and the platelet glycoprotein receptor complex on the platelet membrane [123]. This process can be triggered through the activation of multiple pathways by the binding of several agonists to their receptors, such as thromboxane A2, adenosine diphosphate (ADP), and thrombin. Platelets express many adhesion molecules to facilitate their interactions between each other, leukocytes and the endothelium, causing aggregation. Almost all the genes identified which play a role in vasodilation and platelet aggregation were up-regulated in the LOW IGF-1 cows. They included VWF itself, F5, FGA FGB, ITGA2, ITGB3, P2RY1, PLA2G4A, PRKG1, PTGIR, PTGS1, SELP, TBXAS1 and TFPI.
The complement system proteins are involved in opsonization of pathogens, lysis of infectious organisms, phagocytosis of microbes and damaged cells and induction of an inflammatory response. DEG directly involved in the complement cascade in the present study included CFI, C1R, C2, C3AR1, C8G, CD55, CFB, CFI, and CR2. All except CR2 were up-regulated in LOW IGF-1 cows. This is supported by the finding that blood complement was increased in periparturient cows with a high liver activity index, shown to be associated with inflammatory and immune parameters [124]. A proteomic analysis identified nine complement proteins in PBMC from postpartum dairy cows [125].
Three highly up-regulated genes in LOW IGF-1 cows were LOC107131803, LOC112445470 and LOC511106, which were identified as Serpin B3-like, Serpin B4-like and Serpin peptidase inhibitor, clade B like, respectively. SERPINB3 and SERPINB4 are serine/cysteine protease inhibitors and in humans their expression is up-regulated in association with a number of inflammatory conditions [126]. Other serpin family members play key functions as serine protease inhibitors in thrombotic and fibrinolytic cascades in response to infection and vascular injury [127].
The Rap1 signaling pathway was also identified in the analysis. Rap1 is a small GTPase that couples extracellular signals to intracellular responses to control a large array of cellular-signaling pathways. Based on the DEG identified in our study, it is likely to be playing an important role in the inflammatory processes described above including cell-matrix interactions and angiogenesis. A variety of growth factors, adenosine, dopamine and lysophosphatidic acid (LPA) all contribute to the control of Rap1 cycling between an inactive GDP-bound and an active GTP-bound conformation (Supplementary Figure S1). Activated Rap1 then influences adhesion and migration by regulating the function of integrins and other adhesion molecules and it also affects cell proliferation and survival via altered MAP kinase (MAPK) activity [128]. Rap1 has also been implicated in regulating glucose homeostasis and lipid metabolism during heat stress in Holstein cows [129].
4.7. Cytokine and Chemokine Signaling Pathways
Kegg pathway analysis identified “Cytokine–cytokine receptor interaction” as significant. The gene list included transforming growth factor-beta (TGF-beta) superfamily receptors (ACVR1, ACVR1B, ACVR2B, and BMPR2); chemokines and their receptors (CCL8, CXCL8, CCR1, and CCR4); cytokines and their receptors (CSF1R, IL12B, TSLP, IL1RAP, IL2RA, IL3RA, IL15RA, and IL18R1) and members of the TNF-receptor superfamily (LTB, LTBR, RELT, TNFRSF1A, TNFRSF1B, and TNFRSF9). Feeding cows high energy diets during the dry period is known to increase circulating concentrations of pro-inflammatory mediators such as IL1B, TNF and CXCL2 [130]. TNFα is a key inflammatory cytokine which has a direct effect on the bovine liver to promote triacylglycerol accumulation and decrease glucose production [131], while TNFSF13 and TNFSF13B encode proteins termed APRIL and BAFF which are important for B cell activation. The influence of TNFα on appetite was discussed above. Colony stimulating factor 1 is another important cytokine which controls the production, differentiation, and function of macrophages via its receptor CSF1R [132].
4.8. Adaptive Immunity
Several prominent genes with lower expression in LOW IGF-1 cows (and conversely up-regulated in HIGH IGF-1 cows) are known to be involved in T and B cell activation. The major histocompatibility complex (MHC, known as BoLA in cattle) is a set of highly polymorphic genes that bind pathogen-derived peptides and present them for recognition by specialized T cells, resulting in T and B cell activation and expansion. BOLA-DQB plays a role in antigen presentation [133]. CD52 encodes a glycoprotein widely expressed on the cell surface, which can modulate T cell activation [134]. MS4A1 encodes membrane spanning 4-domains A1, also known as CD20, which plays a role in the development and differentiation of B cells into plasma cells. SPIB encodes a transcriptional activator which acts as a lymphoid-specific enhancer and regulates the expression of many components of the B cell receptor signaling pathway [135]. BANK1 (B cell scaffold protein with ankyrin repeats 1) is an adaptor protein which is primarily expressed in B cells where it is also involved in many signaling pathways. These include TLR signaling, B cell receptor-induced calcium mobilization and tyrosine phosphorylation of inositol 1,4,5-trisphosphate receptors. BANK1 also negatively regulates CD40-mediated B cell activation, so preventing hyperactive B cell responses [136]. Other genes encoding proteins playing key roles in T and B cell activation and function included PTPRCAP [137], CD79B [138] and ST6GAL1 [139]. ST6Gal knock out mice are severely immunosuppressed [140]. These results corroborate the idea that HIGH IGF-1 cows have a greater capacity to mount an effective adaptive immune response.
4.9. Cytoskeleton and Cell Motility
Cell migration is crucial for leucocyte functionality and two of the four genes which discriminated between all three IGF-1 groups (CFAP69 up-regulated and SEMA6B down-regulated in LOW IGF-1 cows, respectively) have roles related to the cytoskeleton and cell motility. A number of the other genes most down-regulated in LOW IGF-1 cows (DMTN, REM1, MYLIP, ARPC5L) also came into this functional category. CFAP69 encodes cilia and flagella-associated protein 69, which forms part of the cytoskeletal scaffold, with knockout resulting in loss of motility [141]. Semaphorin signaling results in changes to the cytoskeletal and adhesive machinery that modulate cellular adhesion and influence cell shape, morphology and motility. Some semaphorins can influence vascular permeability and are involved in establishing interactions between platelets that allow thrombus formation during hemostasis [142]. Within immune cell populations, they have roles in initiation of T cell mediated antibody responses, B cell function and macrophage activation by antigen-specific T cells at sites of inflammation [143]. Semaphorin 6B encoded by SEMA6B is a transmembrane protein which is a key target gene for vitamin D in PBMC [144]. It was one of the most highly down-regulated genes in LOW IGF-1 cows (−6.4 fold) but there is currently little information available regarding its specific functions. Several other SEMA genes (SEMA4F, SEMA4D, and SEMA3C) were up-regulated in the LOW IGF-1 cows. Of these, SEMA4D was very highly expressed.
The cytoskeleton is composed of actin and tubulin networks and intermediate filaments. These are involved in the development of adhesive contacts, establishment of polarity and cell motility. Cytoskeleton remodeling involves cycles of actin polymerization and depolymerization, which are regulated by a variety of proteins and requires tight coordination [145]. The Arp2/3 complex is involved in generating branched actin filament networks [146]. The small GTPases Rac and Rho antagonize each other at multiple levels and control cell polarization and different migration modes: Rac activity dominates at the leading edge during mesenchymal migration, while amoeboid movement is characterized by high levels of active Rho [147]. Actomyosin interactions are also closely involved in the regulated assembly and disassembly of the actin network [148]. The genes more highly expressed in cows with HIGH IGF-1 included ARPC5L (encoding actin related protein 2/3 complex subunit 5 like), REM1 (encoding a member of the RAS-like GTP-binding protein family) and DMTN (encoding dematin actin-binding protein). All of these are known to be involved in regulating the actin cytoskeleton, while myosin regulatory light chain interacting protein (encoded by MYLIP) is a cytoskeletal effector protein that links actin to membrane-bound proteins at the cell surface. The differential expression of these various genes between leukocytes derived from HIGH and LOW IGF-1 cows therefore suggests that there are likely to be differences in adhesive properties and migratory capacity between their respective cell populations.
4.10. Potential Links to Health and Fertility
Our own previous work has indicated that the IGF-1 concentration in the immediate post-calving period is strongly linked to subsequent fertility, a relationship which was much more pronounced in multiparous than primiparous cows [63]. Key genes of the somatotropic axis regulating the production and bioavailability of IGF-1 showed differential hepatic expression in cows with high or low genetic merit for fertility traits [149] and circulating concentrations of IGF-1 were greater throughout the gestation-lactation cycle in the more fertile cows [150]. This relationship could in part be caused by a direct effect on the reproductive tract. The IGF system is important in promoting the repair of the uterine endometrium after calving [28,151]. Circulating IGF-1 is also considered critical for follicular maturation as it amplifies the gonadotropin stimulus [152,153]. Cows with low IGF-1 after calving therefore remain anovulatory for longer [154,155,156]. Overall, cows with a low nadir in IGF-1 in the first 2 weeks after calving subsequently take longer to conceive in both high yielding [63] and pasture based [156] systems.
There is also a more indirect relationship between the somatotropic axis and fertility via compromised immunity, as evidenced here. LOW IGF-1 cows are clearly more likely to succumb to infections after calving, particularly affecting the uterus and mammary gland. The association between low IGF-1 and clinical endometritis supports an earlier study which showed that multiparous cows in which the plasma IGF-I concentration fell below 40 ng/mL in the week after calving were nearly three times more likely to suffer from a subsequent uterine infection [62]. The data presented here show an increased likelihood that they may also develop clinical or subclinical mastitis. In addition, previous studies have shown associations between low IGF-1 pre-calving with clinical ketosis [157] and between low IGF-1 postpartum with both displaced abomasum and an increased risk of culling [74].
These diseases may well contribute to persistent inflammatory changes, which have an adverse effect on the cow’s ability to conceive some months later. In this respect, it is interesting to compare our results with a study of fertility in beef heifers which measured transcriptional profiles in white blood cells collected at the time of insemination [158]. Their analysis revealed 18 DEG between heifers which did or did not conceive. ALAS2, BOLA-DQB and DMBT1 were common to the lists of highly differentially expressed genes identified in our study. Of these DMBT1 was one of the most highly up-regulated genes in both studies and therefore warrants further investigation as a biomarker for fertility. FCER1A and P2RY12 were also on their list and we found genes for two other components of the IgE receptor, FCGR1A and FCER1G were up-regulated in LOW IGF-1 cows while three other purinergic receptors were identified, with P2RY1 and P2RY2 up-regulated and P2RY10 down-regulated.
5. Conclusions
A growing number of studies have measured selected inflammatory mediators in peripartum cows but few to date have used a similar global approach or based it on such a sizeable population as that used here. Our study has linked transcriptomic analysis of circulating leukocytes to the IGF-1 concentration, used as a biomarker of metabolic function. This has confirmed changes in metabolic pathways in LOW compared to HIGH IGF-1 cows, with immune cells switching their energy metabolism from oxidative phosphorylation to aerobic glycolysis, and making greater use of the pentose phosphate shunt. Our analysis has also extended the range of candidate genes and pathways regulating immunity which were differentially affected. Many antimicrobial peptides were up-regulated in LOW vs. HIGH IGF-1 cows. On the other hand, the HIGH IGF-1 cows had higher expression of more genes regulating T and B cell function and the cytoskeleton. A large number of genes associated with inflammation were also differentially expressed, particularly those associated with platelet activation and the complement cascade. Overall, the results suggest that the LOW IGF-1 cows are engaged in an energetically demanding battle against ongoing infection(s) involving the innate immune system. This reduces their appetite and milk yield and is likely to have an adverse effect on their subsequent fertility. On the other hand, the HIGH IGF-1 cows have higher nutrient availability and can use this to promote use of their adaptive immune system to better resist infection. Our results therefore revealed several mechanisms which help to explain why and how innate and adaptive immunity can become compromised in postpartum dairy cows with low circulating IGF-1 levels in early lactation. The data presented here thus support other available evidence in suggesting that the metabolic status of individual cows is linked directly to the high incidence of health disorders during this critical period [1,3,51,159]. Furthermore, 35/170 (20%) of all cows in this study fell into the LOW IGF-1 category, indicating the need for improved peripartum management to reduce the incidence of such animals.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/ruminants1020012/s1, Table S1: List of DEG with FC ≥ 1.2 and FDR p < 0.05 with higher expression in cows with LOW IGF-1 (<35 ng/mL, n = 35) in comparison with HIGH IGF-1 (>100 ng/mL, n = 43) in leucocytes of cows at 14 days in milk, based on the global transcriptome; Table S2.: List of DEG with FC ≥ 1.2 and FDR p < 0.05 with lower expression in cows with LOW IGF-1 (<35 ng/mL, n = 35) in comparison with HIGH IGF-1 (>100 ng/mL, n = 43) in leucocytes of cows at 14 days in milk, based on the global transcriptome; Table S3: Cluster analysis using DAVID based on list of 1,032 DEG with FC ≥ 1.2 and FDR p < 0.05 between cows with LOW IGF-1 (<35 ng/mL, n = 35) and HIGH IGF-1 (>100 ng/mL, n = 43) in leukocytes of cows at 14 days in milk, based on the global transcriptome; Table S4: Lists of genes in clusters 4–6 from analysis using DAVID based on list of 1,032 DEG with FC ≥ 1.2 and FDR p < 0.05 between cows with LOW IGF-1 (<35 ng/mL, n = 35) and HIGH IGF-1 (>100 ng/mL, n = 43) in leukocytes of cows at 14 days in milk, based on the global transcriptome; Table S5: Kegg pathway analysis using DAVID based on list of 1,032 DEG with FC ≥ 1.2 and FDR p < 0.05 between cows with LOW IGF-1 (<35 ng/mL, n = 35) and HIGH IGF-1 (>100 ng/mL, n = 43) in leukocytes of cows at 14 days in milk, based on the global transcriptome; Table S6: Lists of genes in significant KEGG pathways from analysis using DAVID based on list of 1032 DEG with FC ≥ 1.2 and FDR p < 0.05 between cows with LOW IGF-1 (<35 ng/mL, n = 35) and HIGH IGF-1 (>100 ng/mL, n = 43) in leukocytes of cows at 14 days in milk, based on the global transcriptome; Table S7: Sub list of metabolic-related DEG with FC ≥ 1.2 and FDR p < 0.05 in comparison between cows with LOW (<35 ng/mL, n = 35) and HIGH IGF-1 (>100 ng/mL, n = 43) in leucocytes collected at 14 days in milk, based on the global transcriptome; Table S8: Kegg pathway analysis using DAVID based on sublist of 149 metabolic-related DEG with FC ≥ 1.2 and FDR p > 0.05 between cows with LOW IGF-1 (<35 ng/mL, n = 35) and HIGH IGF-1 (>100 ng/mL, n = 43) in leukocytes of cows at 14 days in milk, based on the global transcriptome; Table S9: Lists of genes in significant KEGG pathways from analysis using DAVID based on list of 149 metabolic-related DEG with FC ≥ 1.2 and FDR p > 0.05 between cows with LOW IGF-1 (<35 ng/mL, n = 35) and HIGH IGF-1 (>100 ng/mL, n = 43) in leukocytes of cows at 14 days in milk, based on the global transcriptome; Figure S1: Modified representation of Kegg pathway bta04015: Rap1 signaling pathway, highlighting DEG identified in this study.
Author Contributions
Conceptualization, D.C.W., K.L.I. and M.A.C.; methodology, L.B., L.F., M.S., H.T., L.T. and Z.C.; formal analysis, D.C.W., L.B., M.S., H.T., L.T. and Z.C.; investigation, F.B., M.A.C., C.F., C.G., K.L.I., C.M., M.S., F.S., H.T., L.T. and Z.C.; data curation, L.B., L.F., M.H., M.S., H.T. and Z.C.; writing—original draft preparation, D.C.W. and Z.C.; writing—review and editing, D.C.W., C.F., L.F., C.G., M.H., K.L.I., C.M. and Z.C.; project administration, D.C.W., F.B., M.A.C., C.F., K.L.I. and C.M.; funding acquisition, M.A.C., D.C.W. and K.L.I. All authors have read and agreed to the published version of the manuscript.
Funding
This project received funding from the European Union’s Seventh Framework Programme (Brussels, Belgium) for research, technological development, and demonstration under grant agreement no. 613689. The views expressed in this publication are the sole responsibility of the authors and do not necessarily reflect the views of the European Commission.
Institutional Review Board Statement
The data in this study were collected as a part of the Genotype plus Environment (GplusE) FP7-Project (http://www.gpluse.eu (accessed on 20 Nov 2021)). All procedures had local ethical approval and complied with the relevant national and EU legislation under the European Union (Protection of Animals used for Scientific Purposes) Regulations 2012 (S.I. No. 543 of 2012). University College Dublin (UCD), Schools of Veterinary Medicine; and Agriculture and Food Science), Belfield. Dublin 4, Ireland. The University farm is a licenced premise for the conduct of animal based research under the Cruelty to Animals Act 1876 as amended by European Communities regulations 2002 and 2005 (as required under the European Union Directive 2010/63/EU). Animal experiments were approved by the University College Dublin animal research ethics committee. The licence to perform this experiment was ref. B100/2177. The Agri-Food and Biosciences Institute (AFBI), Hillsborough, Co. Down, UK, BT26 6DR. Animal experimentation was performed under the Animals (Scientific Procedures) Act 1986 and covered by Home Office Project Licence number PPL2754 and a Certificate of Designation for the Establishment. The work was also approved by the AFBI Ethical Review Committee. Aarhus University (AU), Department of Animal Science, Denmark. The work was approved by the Danish Animal Experiments Inspectorate, number 2014-15-0201-00282 and complied with the Danish and EU Laws concerning animal experimentation and care of experimental animals. Consiglio per la Ricerca e la Sperimentazione in Agricoltura, Italy (CREA). The cows involved in this project were supervised in compliance with the Italian law and regulations regarding experimental work on animals. The CREA-PCM is authorized to use farm animals for experimental design (as stated in DM 26/96-4 of Italian Welfare Ministry). Walloon Agricultural Research Centre, Department for Quality of Agricultural Products (CRA-W). The experiment was carried out in accordance with the EU Directive 2010/63/EU for animal experiments and the protocol (14-1617) was approved by the ethical commission of Liège University. Leibniz Institute for Farm Animal Biology—FBN Dummerstorf. All procedures were approved by the research ethical committee of FBN based on the German and EU legislation (European Union Directive 2010/63/EU) concerning the use of research animals in relation to the welfare and care of experimental animals. The reference number was LALLF—7221.3-1-053/13.
Data Availability Statement
The RNA-seq fastq data can be obtained from the European Nucleotide Archive at https://www.ebi.ac.uk/ena/browser/home (accessed on 9/10/2021) by searching with the accession numbers ERP124149 and ERP122911.
Acknowledgments
Many thanks go to the farm staff who contributed and to all members of the GplusE Consortium. (http://www.gpluse.eu/index.php/contributors/ (accessed on 16 November 2021): Mark Crowe, Niamh McLoughlin, Alan Fahey, Fiona Carter, Elizabeth Matthews, Andreia Santoro, Colin Byrne, Pauline Rudd, Roisin O’Flaherty, Sinead Hallinan, D Claire Wathes, Laura Buggiotti, Mazdak Salavati, Zhangrui Cheng, Ali Fouladi, Geoff Pollott, Dirk Werling, Beatriz Sanz Bernardo, Conrad Ferris, Alistair Wylie, Matt Bell, Mieke Vaneetvelde, Kristof Hermans, Miel Hostens, Geert Opsomer, Sander Moerman, Jenne De Koster, Hannes Bogaert, Jan Vandepitte, Leila Vandevelde, Bonny Vanranst, Klaus Ingvartsen, Martin Tang Sorensen, Johanna Hoglund, Susanne Dahl, Soren Ostergaard, Janne Rothmann, Mogens Krogh, Else Meyer, Leslie Foldager, Charlotte Gaillard, Jehan Ettema, Tine Rousing, Torben Larsen, Victor H. Silva de Oliveira, Cinzia Marchitelli, Federica Signorelli, Francesco Napolitano, Bianca Moioli, Alessandra Crisà, Luca Buttazzoni, Jennifer McClure, Daragh Matthews, Francis Kearney, Andrew Cromie, Matt McClure, Shujun Zhang, Xing Chen, Huanchun Chen, Junlong Zhao, Liguo Yang, Guohua Hua, Chen Tan, Guiqiang Wang, Michel Bonneau, Marlène Sciarretta, Armin Pearn, Arnold Evertson, Linda Kosten, Anders Fogh, Thomas Andersen, Matthew Lucy, Chris Elsik, Gavin Conant, Jerry Taylor, Deborah Triant, Nicolas Gengler, Michel Georges, Frederic Colinet, Marilou Ramos Pamplona, Hedi Hammami, Catherine Bastin, Haruko Takeda, Aurelie Laine, Anne-Sophie Van Laere, Rodrigo Mota, Saied Naderi Darbagshahi, Frederic Dehareng, Clement Grelet, Amelie Vanlierde, Eric Froidmont, Frank Becker, Martin Schulze, Sergio Palma Vera, and Andrea Pompozzi.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Drackley, J.K. Biology of Dairy Cows During the Transition Period: The Final Frontier? J. Dairy Sci. 1999, 82, 2259–2273. [Google Scholar] [CrossRef]
- Loor, J.J.; Dann, H.M.; Guretzky, N.A.J.; Everts, R.E.; Oliveira, R.; Green, C.A.; Litherland, N.B.; Rodriguez-Zas, S.L.; Lewin, H.A.; Drackley, J.K. Plane of nutrition prepartum alters hepatic gene expression and function in dairy cows as assessed by longitudinal transcript and metabolic profiling. Physiol. Genom. 2006, 27, 29–41. [Google Scholar] [CrossRef] [PubMed]
- Ingvartsen, K.L. Feeding- and management-related diseases in the transition cow: Physiological adaptations around calving and strategies to reduce feeding-related diseases. Anim. Feed Sci. Technol. 2006, 126, 175–213. [Google Scholar] [CrossRef]
- Habel, J.; Sundrum, A. Mismatch of Glucose Allocation between Different Life Functions in the Transition Period of Dairy Cows. Animals 2020, 10, 1028. [Google Scholar] [CrossRef]
- Ingvartsen, K.L.; Andersen, J.B. Integration of metabolism and intake regulation: A review focusing on periparturient animals. J. Dairy Sci. 2000, 83, 1573–1597. [Google Scholar] [CrossRef]
- Taylor, V.J.; Beever, D.E.; Wathes, D.C. Physiological Adaptations to Milk Production that Affect the Fertility of High Yielding Dairy Cows. In Dairying, Using Science to Meet Consumer Needs, Kebreab, E., Mills, J., Beever, D., Eds.; Cambridge University Press: Cambridge, UK. Br. Soc. Anim. Sci. Occas. Publ. 2004, 29, 37–71. [Google Scholar] [CrossRef]
- Loor, J.J.; Dann, H.M.; Everts, R.E.; Oliveira, R.; Green, C.A.; Guretzky, N.A.J.; Rodriguez-Zas, S.L.; Lewin, H.A.; Drackley, J.K. Temporal gene expression profiling of liver from periparturient dairy cows reveals complex adaptive mechanisms in hepatic function. Physiol. Genom. 2005, 23, 217–226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wathes, D.C.; Cheng, Z.; Salavati, M.; Buggiotti, L.; Takeda, H.; Tang, L.; Becker, F.; Ingvartsen, K.I.; Ferris, C.; Hostens, M.; et al. Relationships between metabolic profiles and gene expression in liver and leukocytes of dairy cows in early lactation. J. Dairy Sci. 2021, 104, 3596–3616. [Google Scholar] [CrossRef]
- Trevisi, E.; Amadori, M.; Cogrossi, S.; Razzuoli, E.; Bertoni, G. Metabolic stress and inflammatory response in high-yielding, periparturient dairy cows. Res. Vet. Sci. 2012, 93, 695–704. [Google Scholar] [CrossRef]
- Kobayashi, Y.; Boyd, C.K.; Bracken, C.J.; Lamberson, W.R.; Keisler, D.H.; Lucy, M.C. Reduced Growth Hormone Receptor (GHR) Messenger Ribonucleic Acid in Liver of Periparturient Cattle Is Caused by a Specific Down-Regulation of GHR 1A That Is Associated with Decreased Insulin-Like Growth Factor I. Endocrinology 1999, 140, 3947–3954. [Google Scholar] [CrossRef]
- Fenwick, M.A.; Fitzpatrick, R.; Kenny, D.A.; Diskin, M.G.; Patton, J.; Murphy, J.J.; Wathes, D.C. Interrelationships between negative energy balance (NEB) and IGF regulation in liver of lactating dairy cows. Domest. Anim. Endocrinol. 2008, 34, 31–44. [Google Scholar] [CrossRef]
- Bell, A.W. Regulation of organic nutrient metabolism during transition from late pregnancy to early lactation. J. Anim. Sci. 1995, 73, 2804–2819. [Google Scholar] [CrossRef]
- Thissen, J.-P.; Ketelslegers, J.-M.; Underwood, L.E. Nutritional Regulation of the Insulin-Like Growth Factors. Endocr. Rev. 1994, 15, 80–101. [Google Scholar] [CrossRef] [PubMed]
- Etherton, T.D.; Bauman, D.E. Biology of Somatotropin in Growth and Lactation of Domestic Animals. Physiol. Rev. 1998, 78, 745–761. [Google Scholar] [CrossRef] [PubMed]
- Ronge, H.; Blum, J.; Clement, C.; Jans, F.; Leuenberger, H.; Binder, H. Somatomedin C in dairy cows related to energy and protein supply and to milk production. Anim. Sci. 1988, 47, 165–183. [Google Scholar] [CrossRef] [Green Version]
- Meier, S.; Kolver, E.S.; Verkerk, G.A.; Roche, J.R. Effects of divergent Holstein-Friesian strain and diet on diurnal patterns of plasma metabolites and hormones. J. Dairy Res. 2010, 77, 432–437. [Google Scholar] [CrossRef]
- Zbinden, R.S.; Falk, M.; Münger, A.; Dohme-Meier, F.; Van Dorland, H.A.; Bruckmaier, R.M.; Gross, J.J. Metabolic load in dairy cows kept in herbage-based feeding systems and suitability of potential markers for compromised well-being. J. Anim. Physiol. Anim. Nutr. 2017, 101, 767–778. [Google Scholar] [CrossRef] [PubMed]
- Taylor, V.J.; Beever, D.E.; Bryant, M.J.; Wathes, D.C. Pre-pubertal measurements of the somatotrophic axis as predictors of milk production in Holstein–Friesian dairy cows. Domest. Anim. Endocrinol. 2006, 31, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Sheahan, A.J.; Boston, R.C.; Roche, J.R. Diurnal patterns of grazing behavior and humoral factors in supplemented dairy cows. J. Dairy Sci. 2013, 96, 3201–3210. [Google Scholar] [CrossRef]
- Witkowska-Sędek, E.; Pyrżak, B. Chronic inflammation and the growth hormone/insulin-like growth factor-1 axis. Cent. Eur. J. Immunol. 2020, 45, 469–475. [Google Scholar] [CrossRef]
- Clark, R. The Somatogenic Hormones and Insulin-Like Growth Factor-1: Stimulators of Lymphopoiesis and Immune Function. Endocr. Rev. 1997, 18, 157–179. [Google Scholar] [CrossRef] [PubMed]
- Heemskerk, V.H.; Daemen, M.A.R.C.; Buurman, W.A. Insulin-like growth factor-1 (IGF-1) and growth hormone (GH) in immunity and inflammation. Cytokine Growth Factor Rev. 1999, 10, 5–14. [Google Scholar] [CrossRef]
- Inoue, T.; Saito, H.; Hashiguchi, Y.; Fukatsu, K.; Inaba, T.; Lin, M.-T.; Han, L.; Furukawa, S.; Muto, T. Growth hormone and insulin-like growth factor I augment Escherichia coli-killing activity of murine peritoneal exudative cells. Shock 1996, 6, 345–350. [Google Scholar] [CrossRef]
- Wolters, T.L.C.; Netea, M.G.; Hermus, A.R.M.M.; Smit, J.W.A.; Netea-Maier, R.T. IGF1 potentiates the pro-inflammatory response in human peripheral blood mononuclear cells via MAPK. J. Mol. Endocrinol. 2017, 59, 129–139. [Google Scholar] [CrossRef]
- De Koster, J.; Salavati, M.; Grelet, C.; Crowe, M.A.; Matthews, E.; O’Flaherty, R.; Opsomer, G.; Foldager, L.; Gplus, E.; Hostens, M. Prediction of metabolic clusters in early-lactation dairy cows using models based on milk biomarkers. J. Dairy Sci. 2019, 102, 2631–2644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, T.Q.; Weston, P.G.; Lund, L.A.; Brodie, B.; McKenna, D.J.; Wagner, W.C. Association between neutrophil functions and periparturient disorders in cows. Am. J. Vet. Res. 1994, 55, 934–943. [Google Scholar] [PubMed]
- Mallard, B.A.; Dekkers, J.C.; Ireland, M.J.; Leslie, K.E.; Sharif, S.; VanKampen, C.L.; Wagter, L.; Wilkie, B.N. Alteration in immune responsiveness during the peripartum period and its ramification on dairy cow and calf health. J. Dairy Sci. 1998, 81, 585–595. [Google Scholar] [CrossRef]
- Wathes, D.C.; Cheng, Z.; Chowdhury, W.; Fenwick, M.A.; Fitzpatrick, R.; Morris, D.G.; Patton, J.; Murphy, J.J. Negative energy balance alters global gene expression and immune responses in the uterus of postpartum dairy cows. Physiol. Genom. 2009, 39, 1–13. [Google Scholar] [CrossRef]
- Crookenden, M.A.; Moyes, K.M.; Kuhn-Sherlock, B.; Lehnert, K.; Walker, C.G.; Loor, J.J.; Mitchell, M.D.; Murray, A.; Dukkipati, V.S.R.; Vailati-Riboni, M.; et al. Transcriptomic analysis of circulating neutrophils in metabolically stressed peripartal grazing dairy cows. J. Dairy Sci. 2019, 102, 7408–7420. [Google Scholar] [CrossRef]
- Zerbe, H.; Schneider, N.; Leibold, W.; Wensing, T.; Kruip, T.A.M.; Schuberth, H.J. Altered functional and immunophenotypical properties of neutrophilic granulocytes in postpartum cows associated with fatty liver. Theriogenology 2000, 54, 771–786. [Google Scholar] [CrossRef]
- Hammon, D.S.; Evjen, I.M.; Dhiman, T.R.; Goff, J.P.; Walters, J.L. Neutrophil function and energy status in Holstein cows with uterine health disorders. Vet. Immunol. Immunopathol. 2006, 113, 21–29. [Google Scholar] [CrossRef]
- McCarthy, M.M.; Yasui, T.; Felippe, M.J.B.; Overton, T.R. Associations between the degree of early lactation inflammation and performance, metabolism, and immune function in dairy cows. J. Dairy Sci. 2016, 99, 680–700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, Z.; Wylie, A.; Ferris, C.; Ingvartsen, K.L.; Wathes, D.C.; GplusE Consortium. Effect of diet and nonesterified fatty acid levels on global transcriptomic profiles in circulating peripheral blood mononuclear cells in early lactation dairy cows. J. Dairy Sci. 2021, 104, 10059–10075. [Google Scholar] [CrossRef]
- Bernabucci, U.; Ronchi, B.; Lacetera, N.; Nardone, A. Influence of Body Condition Score on Relationships Between Metabolic Status and Oxidative Stress in Periparturient Dairy Cows. J. Dairy Sci. 2005, 88, 2017–2026. [Google Scholar] [CrossRef] [Green Version]
- Ingvartsen, K.L.; Moyes, K. Nutrition, immune function and health of dairy cattle. Animal 2013, 7, 112–122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loftus, R.M.; Finlay, D.K. Immunometabolism: Cellular Metabolism Turns Immune Regulator. J. Biol. Chem. 2016, 291, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Walls, J.; Sinclair, L.; Finlay, D. Nutrient sensing, signal transduction and immune responses. Semin. Immunol. 2016, 28, 396–407. [Google Scholar] [CrossRef] [Green Version]
- Dimeloe, S.; Burgener, A.-V.; Grählert, J.; Hess, C. T-cell metabolism governing activation, proliferation and differentiation; a modular view. Immunology 2017, 150, 35–44. [Google Scholar] [CrossRef] [Green Version]
- Desdín-Micó, G.; Soto-Heredero, G.; Mittelbrunn, M. Mitochondrial activity in T cells. Mitochondrion 2018, 41, 51–57. [Google Scholar] [CrossRef]
- O’Neill, L.A.J.; Hardie, D.G. Metabolism of inflammation limited by AMPK and pseudo-starvation. Nature 2013, 493, 346–355. [Google Scholar] [CrossRef]
- Nonnecke, B.J.; Kimura, K.; Goff, J.P.; Kehrli, M.E., Jr. Effects of the Mammary Gland on Functional Capacities of Blood Mononuclear Leukocyte Populations from Periparturient Cows. J. Dairy Sci. 2003, 86, 2359–2368. [Google Scholar] [CrossRef]
- Ingvartsen, K.L.; Moyes, K.M. Factors contributing to immunosuppression in the dairy cow during the periparturient period. Jpn. J. Vet. Res. 2015, 63 (Suppl. S1), S15–S24. [Google Scholar]
- Gier, H.T.; Marion, G.B. Uterus of the cow after parturition: Involutional changes. Am. J. Vet. Res. 1968, 29, 83–96. [Google Scholar] [PubMed]
- Sheldon, I.M.; Lewis, G.S.; LeBlanc, S.; Gilbert, R.O. Defining postpartum uterine disease in cattle. Theriogenology 2006, 65, 1516–1530. [Google Scholar] [CrossRef] [PubMed]
- Vangroenweghe, F.; Lamote, I.; Burvenich, C. Physiology of the periparturient period and its relation to severity of clinical mastitis. Domest. Anim. Endocrinol. 2005, 29, 283–293. [Google Scholar] [CrossRef]
- Leblanc, S. Monitoring Metabolic Health of Dairy Cattle in the Transition Period. J. Reprod. Dev. 2010, 56, S29–S35. [Google Scholar] [CrossRef] [Green Version]
- Smith, G.L.; Friggens, N.C.; Ashworth, C.J.; Chagunda, M.G.G. Association between body energy content in the dry period and post-calving production disease status in dairy cattle. Animal 2017, 11, 1590–1598. [Google Scholar] [CrossRef] [Green Version]
- Ceciliani, F.; Ceron, J.J.; Eckersall, P.D.; Sauerwein, H. Acute phase proteins in ruminants. J. Proteom. 2012, 75, 4207–4231. [Google Scholar] [CrossRef]
- Brodsky, I.E.; Medzhitov, R. Targeting of immune signalling networks by bacterial pathogens. Nat. Cell Biol. 2009, 11, 521–526. [Google Scholar] [CrossRef]
- Krysko, D.V.; Agostinis, P.; Krysko, O.; Garg, A.D.; Bachert, C.; Lambrecht, B.N.; Vandenabeele, P. Emerging role of damage-associated molecular patterns derived from mitochondria in inflammation. Trends Immunol. 2011, 32, 157–164. [Google Scholar] [CrossRef]
- Ingvartsen, K.L.; Dewhurst, R.J.; Friggens, N.C. On the relationship between lactational performance and health: Is it yield or metabolic imbalance that cause production diseases in dairy cattle? A position paper. Livest. Prod. Sci. 2003, 83, 277–308. [Google Scholar] [CrossRef]
- Wathes, D.C.; Cheng, Z.; Bourne, N.; Taylor, V.J.; Coffey, M.P.; Brotherstone, S. Differences between primiparous and multiparous dairy cows in the inter-relationships between metabolic traits, milk yield and body condition score in the periparturient period. Domest. Anim. Endocrinol. 2007, 33, 203–225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krogh, M.A.; Hostens, M.; Salavati, M.; Grelet, C.; Sorensen, M.T.; Wathes, D.C.; Ferris, C.P.; Marchitelli, C.; Signorelli, F.; Napolitano, F.; et al. Between- and within-herd variation in blood and milk biomarkers in Holstein cows in early lactation. Animal 2020, 14, 1067–1075. [Google Scholar] [CrossRef]
- Beltman, M.E.; Forde, N.; Furney, P.; Carter, F.; Roche, J.F.; Lonergan, P.; Crowe, M.A. Characterisation of endometrial gene expression and metabolic parameters in beef heifers yielding viable or non-viable embryos on Day 7 after insemination. Reprod. Fertil. Dev. 2010, 22, 987–999. [Google Scholar] [CrossRef] [Green Version]
- Bjerre-Harpøth, V.; Storm, A.C.; Vestergaard, M.; Larsen, M.; Larsen, T. Effect of postpartum propylene glycol allocation to over-conditioned Holstein cows on concentrations of milk metabolites. J. Dairy Res. 2016, 83, 156–164. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, N.I.; Larsen, T.; Bjerring, M.; Ingvartsen, K.L. Quarter Health, Milking Interval, and Sampling Time During Milking Affect the Concentration of Milk Constituents. J. Dairy Sci. 2005, 88, 3186–3200. [Google Scholar] [CrossRef] [Green Version]
- Larsen, T. Determination of lactate dehydrogenase (LDH) activity in milk by a fluorometric assay. J. Dairy Res. 2005, 72, 209–216. [Google Scholar] [CrossRef] [PubMed]
- Larsen, T.; Moyes, K.M. Fluorometric determination of uric acid in bovine milk. J. Dairy Res. 2010, 77, 438–444. [Google Scholar] [CrossRef]
- NRC. Nutrient Requirements of Dairy Cattle, 7th ed.; National Academies Press: Washington, DC, USA, 2001; Volume 1. [Google Scholar]
- Williams, E.J.; Fischer, D.P.; Pfeiffer, D.U.; England, G.C.W.; Noakes, D.E.; Dobson, H.; Sheldon, I.M. Clinical evaluation of postpartum vaginal mucus reflects uterine bacterial infection and the immune response in cattle. Theriogenology 2005, 63, 102–117. [Google Scholar] [CrossRef]
- Manson, F.J.; Leaver, J.D. The influence of concentrate amount on locomotion and clinical lameness in dairy cattle. Anim. Sci. 1988, 47, 185–190. [Google Scholar] [CrossRef]
- Beltman, M.E.; McNally, J.C.; Kelly, E.; Crowe, M.A. Relationship between plasma concentrations of IGF-I and clinical endometritis, and response to progesterone synchrony in dairy cows during early lactation. J. Dairy Sci. 2020, 103, 9493–9501. [Google Scholar] [CrossRef]
- Taylor, V.J.; Cheng, Z.; Pushpakumara, P.G.A.; Wathes, D.C.; Beever, D.E. Relationships between the plasma concentrations of insulin-like growth factor-I in dairy cows and their fertility and milk yield. Vet. Rec. 2004, 155, 583–588. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Roland, L.; Drillich, M.; Iwersen, M. Hematology as a diagnostic tool in bovine medicine. J. Vet. Diagn. Investig. 2014, 26, 592–598. [Google Scholar] [CrossRef] [Green Version]
- Berry, M.P.R.; Graham, C.; McNab, F.W.; Xu, Z.; Bloch, S.A.A.; Oni, T.; Wilkinson, K.; Banchereau, R.; Skinner, J.; Wilkinson, R.; et al. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature 2010, 466, 973–977. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blankley, S.; Berry, M.P.R.; Graham, C.M.; Bloom, C.I.; Lipman, M.; O’Garra, A. The application of transcriptional blood signatures to enhance our understanding of the host response to infection: The example of tuberculosis. Philos. Trans. R. Soc. B Biol. Sci. 2014, 369, 20130427. [Google Scholar] [CrossRef]
- Park, H.-E.; Park, H.-T.; Jung, Y.H.; Yoo, H.S. Gene expression profiles of immune-regulatory genes in whole blood of cattle with a subclinical infection of Mycobacterium avium subsp. paratuberculosis. PLoS ONE 2018, 13, e0196502. [Google Scholar] [CrossRef] [Green Version]
- Ray, D.E.; Halbach, T.J.; Armstrong, D.V. Season and Lactation Number Effects on Milk Production and Reproduction of Dairy Cattle in Arizona. J. Dairy Sci. 1992, 75, 2976–2983. [Google Scholar] [CrossRef]
- Langhans, W. Bacterial products and the control of ingestive behavior: Clinical implications. Nutrition 1996, 12, 303–315. [Google Scholar] [CrossRef]
- Johnson, R.W. Immune and endocrine regulation of food intake in sick animals. Domest. Anim. Endocrinol. 1998, 15, 309–319. [Google Scholar] [CrossRef]
- Elsasser, T.H.; Caperna, T.J.; Rumsey, T.S. Endotoxin administration decreases plasma insulin-like growth factor (IGF)-I and IGF-binding protein-2 in Angus × Hereford steers independent of changes in nutritional intake. J. Endocrinol. 1995, 144, 109–117. [Google Scholar] [CrossRef] [PubMed]
- Lyons, N.A.; Cooke, J.S.; Wilson, S.; van Winden, S.C.; Gordon, P.J.; Wathes, D.C. Relationships between metabolite and IGF1 concentrations with fertility and production outcomes following left abomasal displacement. Vet. Rec. 2014, 174, 657. [Google Scholar] [CrossRef]
- Cattaneo, L.; Lopreiato, V.; Piccioli-Cappelli, F.; Trevisi, E.; Minuti, A. Plasma albumin-to-globulin ratio before dry-off as a possible index of inflammatory status and performance in the subsequent lactation in dairy cows. J. Dairy Sci. 2021, 104, 8228–8242. [Google Scholar] [CrossRef]
- Enagy, C.; Ehaschemi, A. Time and Demand are Two Critical Dimensions of Immunometabolism: The Process of Macrophage Activation and the Pentose Phosphate Pathway. Front. Immunol. 2015, 6, 164. [Google Scholar] [CrossRef] [Green Version]
- Kelly, B.; O’Neill, L.A.J. Metabolic reprogramming in macrophages and dendritic cells in innate immunity. Cell Res. 2015, 25, 771–784. [Google Scholar] [CrossRef] [Green Version]
- Yi, Z.; Wu, Y.; Zhang, W.; Wang, T.; Gong, J.; Cheng, Y.; Miao, C. Activator-Mediated Pyruvate Kinase M2 Activation Contributes to Endotoxin Tolerance by Promoting Mitochondrial Biogenesis. Front. Immunol. 2021, 11, 595316. [Google Scholar] [CrossRef] [PubMed]
- Xie, M.; Yu, Y.; Kang, R.; Zhu, S.; Yang, L.; Zeng, L.; Sun, X.; Yang, M.; Billiar, T.R.; Wang, H.; et al. PKM2-dependent glycolysis promotes NLRP3 and AIM2 inflammasome activation. Nat. Commun. 2016, 7, 13280. [Google Scholar] [CrossRef]
- Holness, M.J.; Sugden, M.C. Regulation of pyruvate dehydrogenase complex activity by reversible phosphorylation. Biochem. Soc. Trans. 2003, 31, 1143–1151. [Google Scholar] [CrossRef] [PubMed]
- Longo, N.; Ardon, O.; Vanzo, R.; Schwartz, E.; Pasquali, M. Disorders of creatine transport and metabolism. Am. J. Med. Genet. Part C Semin. Med. Genet. 2011, 157, 72–78. [Google Scholar] [CrossRef]
- Baardman, J.; Verberk, S.G.S.; Prange, K.H.M.; van Weeghel, M.; van der Velden, S.; Ryan, D.G.; Wust, R.C.I.; Neele, A.E.; Speijer, D.; Denis, S.W.; et al. A Defective Pentose Phosphate Pathway Reduces Inflammatory Macrophage Responses during Hypercholesterolemia. Cell Rep. 2018, 25, 2044–2052.e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, X.; Han, Q.; Liu, Y.; Sun, C.; Gang, X.; Wang, G. The Relationship between Branched-Chain Amino Acid Related Metabolomic Signature and Insulin Resistance: A Systematic Review. J. Diabetes Res. 2016, 2016, 2794591. [Google Scholar] [CrossRef] [PubMed]
- HoleČek, M. Branched-chain amino acids in health and disease: Metabolism, alterations in blood plasma, and as supplements. Nutr. Metab. 2018, 15, 33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jukić, I.; Kolobarić, N.; Stupin, A.; Matić, A.; Kozina, N.; Mihaljević, Z.; Mihalj, M.; Šušnjara, P.; Stupin, M.; Ćurić, Ž.B.; et al. Carnosine, Small but Mighty—Prospect of Use as Functional Ingredient for Functional Food Formulation. Antioxidants 2021, 10, 1037. [Google Scholar] [CrossRef]
- Schnackerz, K.D.; Dobritzsch, D. Amidohydrolases of the reductive pyrimidine catabolic pathway: Purification, characteriza-tion, structure, reaction mechanisms and enzyme deficiency. Biochim. Biophys. Acta (BBA) Proteins Proteom. 2008, 1784, 431–444. [Google Scholar] [CrossRef]
- Liparulo, I.; Bergamini, C.; Bortolus, M.; Calonghi, N.; Gasparre, G.; Kurelac, I.; Masin, L.; Rizzardi, N.; Rugolo, M.; Wang, W.; et al. Coenzyme Q biosynthesis inhibition induces HIF-1α stabilization and metabolic switch toward glycolysis. FEBS J. 2021, 288, 1956–1974. [Google Scholar] [CrossRef]
- Wang, W.; Liparulo, I.; Rizzardi, N.; Bolignano, P.; Calonghi, N.; Bergamini, C.; Fato, R. Coenzyme Q Depletion Reshapes MCF-7 Cells Metabolism. Int. J. Mol. Sci. 2020, 22, 198. [Google Scholar] [CrossRef]
- Abraham, E.H.; Prat, A.G.; Gerweck, L.; Seneveratne, T.; Arceci, R.J.; Kramer, R.; Guidotti, G.; Cantiello, H.F. The multidrug resistance (mdr1) gene product functions as an ATP channel. Proc. Natl. Acad. Sci. USA 1993, 90, 312–316. [Google Scholar] [CrossRef] [Green Version]
- Giuliani, A.L.; Sarti, A.C.; Di Virgilio, F. Ectonucleotidases in Acute and Chronic Inflammation. Front. Pharmacol. 2021, 11, 619458. [Google Scholar] [CrossRef]
- Out, R.; Hoekstra, M.; Habets, K.; Meurs, I.; de Waard, V.; Hildebrand, R.B.; Wang, Y.; Chimini, G.; Kuiper, J.; Van Berkel, T.J.; et al. Combined Deletion of Macrophage ABCA1 and ABCG1 Leads to Massive Lipid Accumulation in Tissue Macrophages and Distinct Atherosclerosis at Relatively Low Plasma Cholesterol Levels. Arter. Thromb. Vasc. Biol. 2008, 28, 258–264. [Google Scholar] [CrossRef] [Green Version]
- Groenen, A.G.; Halmos, B.; Tall, A.R.; Westerterp, M. Cholesterol efflux pathways, inflammation, and atherosclerosis. Crit. Rev. Biochem. Mol. Biol. 2021, 56, 426–439. [Google Scholar] [CrossRef] [PubMed]
- Bottazzi, B.; Inforzato, A.; Messa, M.; Barbagallo, M.; Magrini, E.; Garlanda, C.; Mantovani, A. The pentraxins PTX3 and SAP in innate immunity, regulation of inflammation and tissue remodelling. J. Hepatol. 2016, 64, 1416–1427. [Google Scholar] [CrossRef] [Green Version]
- Lutzow, Y.C.S.; Donaldson, L.; Gray, C.P.; Vuocolo, T.; Pearson, R.D.; Reverter, A.; Byrne, K.A.; Sheehy, P.A.; Windon, R.; Tellam, R.L. Identification of immune genes and proteins involved in the response of bovine mammary tissue to Staphylococcus aureus infection. BMC Vet. Res. 2008, 4, 18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Genís, S.; Arís, A.; Kaur, M.; Cerri, R.L.A. Effect of metritis on endometrium tissue transcriptome during puerperium in Holstein lactating cows. Theriogenology 2018, 122, 116–123. [Google Scholar] [CrossRef] [Green Version]
- Leito, J.T.; Ligtenberg, A.J.; Nazmi, K.; De Blieck-Hogervorst, J.M.; Veerman, E.C.; Amerongen, A.V.N. A common binding motif for various bacteria of the bacteria-binding peptide SRCRP2 of DMBT1/gp-340/salivary agglutinin. Biol. Chem. 2008, 389, 1193–1200. [Google Scholar] [CrossRef]
- Reichhardt, M.P.; Holmskov, U.; Meri, S. SALSA—A dance on a slippery floor with changing partners. Mol. Immunol. 2017, 89, 100–110. [Google Scholar] [CrossRef] [Green Version]
- Müller, H.; Hu, J.; Popp, R.; Schmidt, M.H.H.; Müller-Decker, K.; Mollenhauer, J.; Fisslthaler, B.; Eble, J.A.; Fleming, I. Deleted in Malignant Brain Tumors 1 is Present in the Vascular Extracellular Matrix and Promotes Angiogenesis. Arter. Thromb. Vasc. Biol. 2012, 32, 442–448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kligman, D.; Hilt, D.C. The S100 protein family. Trends Biochem. Sci. 1988, 13, 437–443. [Google Scholar] [CrossRef]
- Sohnle, P.G.; Hunter, M.J.; Hahn, B.; Chazin, W.J. Zinc-Reversible Antimicrobial Activity of Recombinant Calprotectin (Migration Inhibitory Factor–Related Proteins 8 and 14). J. Infect. Dis. 2000, 182, 1272–1275. [Google Scholar] [CrossRef]
- Thorey, I.S.; Roth, J.; Regenbogen, J.; Halle, J.-P.; Bittner, M.; Vogl, T.; Kaesler, S.; Bugnon, P.; Reitmaier, B.; Durka, S.; et al. The Ca2+-binding Proteins S100A8 and S100A9 Are Encoded by Novel Injury-regulated Genes. J. Biol. Chem. 2001, 276, 35818–35825. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Foell, D.; Wittkowski, H.; Vogl, T.; Roth, J. S100 proteins expressed in phagocytes: A novel group of damage-associated molecular pattern molecules. J. Leukoc. Biol. 2007, 81, 28–37. [Google Scholar] [CrossRef]
- Ehrchen, J.M.; Sunderkötter, C.; Foell, D.; Vogl, T.; Roth, J. The endogenous Toll-like receptor 4 agonist S100A8/S100A9 (calprotectin) as innate amplifier of infection, autoimmunity, and cancer. J. Leukoc. Biol. 2009, 86, 557–566. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, M.; Peter, S.; Gärtner, M.A.; Michel, G.; Jung, M.; Einspanier, R.; Gabler, C. Increased mRNA expression of selected antimicrobial peptides around ovulation and during inflammatory processes in the bovine endometrium postpartum. Theriogenology 2016, 86, 2040–2053. [Google Scholar] [CrossRef] [PubMed]
- Swangchan-Uthai, T.; Lavender, C.R.; Cheng, Z.; Fouladi-Nashta, A.A.; Wathes, D.C. Time Course of Defense Mechanisms in Bovine Endometrium in Response to Lipopolysaccharide. Biol. Reprod. 2012, 87, 135. [Google Scholar] [CrossRef] [PubMed]
- Swangchan-Uthai, T.; Chen, Q.; Kirton, S.E.; Fenwick, M.A.; Cheng, Z.; Patton, J.; Fouladi-Nashta, A.A.; Wathes, D.C. Influence of energy balance on the antimicrobial peptides S100A8 and S100A9 in the endometrium of the post-partum dairy cow. Reproduction 2013, 145, 527–539. [Google Scholar] [CrossRef] [PubMed]
- Selsted, M.E.; Tang, Y.Q.; Morris, W.L.; McGuire, P.A.; Novotny, M.J.; Smith, W.; Henschen, A.H.; Cullor, J.S. Purification, primary structures, and antibacterial activities of beta-defensins, a new family of antimicrobial peptides from bovine neu-trophils. J. Biol. Chem. 1993, 268, 6641–6648. [Google Scholar] [CrossRef]
- Hilchie, A.L.; Wuerth, K.; Hancock, R.E. Immune modulation by multifaceted cationic host defense (antimicrobial) peptides. Nat. Chem. Biol. 2013, 9, 761–768. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Oppenheim, J.J. Alarmins and antimicrobial immunity. Med. Mycol. 2009, 47, S146–S153. [Google Scholar] [CrossRef] [Green Version]
- Gurao, A.; Kashyap, S.K.; Singh, R. β-defensins: An innate defense for bovine mastitis. Vet. World 2017, 10, 990–998. [Google Scholar] [CrossRef] [Green Version]
- Isobe, N. Control mechanisms for producing antimicrobial factors in ruminant mammary gland. Anim. Sci. J. 2017, 88, 937–943. [Google Scholar] [CrossRef] [Green Version]
- Crookenden, M.A.; Walker, C.G.; Heiser, A.; Murray, A.; Dukkipati, V.S.R.; Kay, J.K.; Meier, S.; Moyes, K.M.; Mitchell, M.D.; Loor, J.J.; et al. Effects of precalving body condition and prepartum feeding level on gene expression in circulating neutrophils. J. Dairy Sci. 2017, 100, 2310–2322. [Google Scholar] [CrossRef] [Green Version]
- Gupta, S.K.; Haigh, B.J.; Seyfert, H.-M.; Griffin, F.J.; Wheeler, T.T. Bovine milk RNases modulate pro-inflammatory responses induced by nucleic acids in cultured immune and epithelial cells. Dev. Comp. Immunol. 2017, 68, 87–97. [Google Scholar] [CrossRef]
- Alnakip, M.; Quintela-Baluja, M.; Böhme, K.; Fernandezno, I.C.; Caamaño-Antelo, S.; Calo-Mata, P.; Barros-Velázquez, J. The Immunology of Mammary Gland of Dairy Ruminants between Healthy and Inflammatory Conditions. J. Veter-Med. 2014, 2014, 659801. [Google Scholar] [CrossRef] [Green Version]
- Abdelmegid, S.; Kelton, D.; Caswell, J.; Kirby, G. Proteomic 2D-DIGE Analysis of Milk Whey from Dairy Cows with Staphylococcus aureus Mastitis Reveals Overexpression of Host Defense Proteins. Microorganisms 2020, 8, 1883. [Google Scholar] [CrossRef]
- Weinberg, E.D. The hazards of iron loading. Metallomics 2010, 2, 732–740. [Google Scholar] [CrossRef] [Green Version]
- Montalbetti, N.; Simonin, A.; Kovacs, G.; Hediger, M.A. Mammalian iron transporters: Families SLC11 and SLC40. Mol. Aspects Med. 2013, 34, 270–287. [Google Scholar] [CrossRef] [PubMed]
- Weinberg, E.D. Iron and infection. Microbiol. Rev. 1978, 42, 45–66. [Google Scholar] [CrossRef]
- Zarzosa-Moreno, D.; Avalos-Gómez, C.; Ramírez-Texcalco, L.S.; Torres-López, E.; Ramírez-Mondragón, R.; Hernández-Ramírez, J.O.; Serrano-Luna, J.; De La Garza, M. Lactoferrin and Its Derived Peptides: An Alternative for Combating Virulence Mechanisms Developed by Pathogens. Molecules 2020, 25, 5763. [Google Scholar] [CrossRef]
- Brunyanszki, A.; Erdelyi, K.; Szczesny, B.; Olah, G.; Salomao, R.; Herndon, D.N.; Szabo, C. Upregulation and Mitochondrial Sequestration of Hemoglobin Occur in Circulating Leukocytes during Critical Illness, Conferring a Cytoprotective Phenotype. Mol. Med. 2015, 21, 666–675. [Google Scholar] [CrossRef] [PubMed]
- King, A.J.; Songdej, D.; Downes, D.J.; A Beagrie, R.; Liu, S.; Buckley, M.; Hua, P.; Suciu, M.C.; Oudelaar, A.M.; Hanssen, L.L.P.; et al. Reactivation of a developmentally silenced embryonic globin gene. Nat. Commun. 2021, 12, 4439. [Google Scholar] [CrossRef] [PubMed]
- Leite, G.G.F.; Scicluna, B.P.; Van Der Poll, T.; Salomão, R. Genetic signature related to heme-hemoglobin metabolism pathway in sepsis secondary to pneumonia. NPJ Syst. Biol. Appl. 2019, 5, 26. [Google Scholar] [CrossRef] [Green Version]
- Yun, S.-H.; Sim, E.-H.; Goh, R.-Y.; Park, J.-I.; Han, J.-Y. Platelet Activation: The Mechanisms and Potential Biomarkers. BioMed Res. Int. 2016, 2016, 9060143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trevisi, E.; Zecconi, A.; Bertoni, G.; Piccinini, R. Blood and milk immune and inflammatory profiles in periparturient dairy cows showing a different liver activity index. J. Dairy Res. 2010, 77, 310–317. [Google Scholar] [CrossRef] [PubMed]
- Kra, G.; Nemes-Navon, N.; Daddam, J.R.; Livshits, L.; Jacoby, S.; Levin, Y.; Zachut, M.; Moallem, U. Proteomic analysis of peripheral blood mononuclear cells and inflammatory status in postpartum dairy cows supplemented with different sources of omega-3 fatty acids. J. Proteom. 2021, 246, 104313. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Sheshadri, N.; Zong, W.-X. SERPINB3 and B4: From biochemistry to biology. Semin. Cell Dev. Biol. 2017, 62, 170–177. [Google Scholar] [CrossRef] [Green Version]
- Yaron, J.R.; Zhang, L.; Guo, Q.; Haydel, S.E.; Lucas, A.R. Fibrinolytic Serine Proteases, Therapeutic Serpins and Inflammation: Fire Dancers and Firestorms. Front. Cardiovasc. Med. 2021, 8, 648947. [Google Scholar] [CrossRef]
- Quilliam, L.A.; Rebhun, J.F.; Castro, A.F. A growing family of guanine nucleotide exchange factors is responsible for activation of ras-family GTPases. Prog. Nucleic Acid Res. Mol. Biol. 2002, 71, 391–444. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Lee, S.; Son, J.; Lim, H.; Kim, E.; Kim, D.; Ha, S.; Hur, T.; Lee, S.; Choi, I. Analysis of circulating-microRNA expression in lactating Holstein cows under summer heat stress. PLoS ONE 2020, 15, e0231125. [Google Scholar] [CrossRef]
- Zenobi, M.G.; Gardinal, R.; Zuniga, J.E.; Mamedova, L.K.; Driver, J.P.; Barton, B.A.; Santos, J.E.P.; Staples, C.R.; Nelson, C.D. Effect of prepartum energy intake and supplementation with ruminally protected choline on innate and adaptive immunity of multiparous Holstein cows. J. Dairy Sci. 2020, 103, 2200–2216. [Google Scholar] [CrossRef]
- Bradford, B.J.; Mamedova, L.K.; Minton, J.E.; Drouillard, J.S.; Johnson, B.J. Daily Injection of Tumor Necrosis Factor-α Increases Hepatic Triglycerides and Alters Transcript Abundance of Metabolic Genes in Lactating Dairy Cattle. J. Nutr. 2009, 139, 1451–1456. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muñoz-Garcia, J.; Cochonneau, D.; Télétchéa, S.; Moranton, E.; Lanoe, D.; Brion, R.; Lézot, F.; Heymann, M.-F.; Heymann, D. The twin cytokines interleukin-34 and CSF-1: Masterful conductors of macrophage homeostasis. Theranostics 2021, 11, 1568–1593. [Google Scholar] [CrossRef] [PubMed]
- Ellis, S.A.; Ballingall, K.T. Cattle MHC: Evolution in action? Immunol. Rev. 1999, 167, 159–168. [Google Scholar] [CrossRef] [PubMed]
- Furukawa, A.; Wisel, S.A.; Tang, Q. Impact of Immune-Modulatory Drugs on Regulatory T Cell. Transplantation 2016, 100, 2288–2300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Willis, S.N.; Tellier, J.; Liao, Y.; Trezise, S.; Light, A.; O’Donnell, K.; Garrett-Sinha, L.A.; Shi, W.; Tarlinton, D.M.; Nutt, S.L. Environmental sensing by mature B cells is controlled by the transcription factors PU.1 and SpiB. Nat. Commun. 2017, 8, 1426. [Google Scholar] [CrossRef] [PubMed]
- Aiba, Y.; Yamazaki, T.; Okada, T.; Gotoh, K.; Sanjo, H.; Ogata, M.; Kurosaki, T. BANK Negatively Regulates Akt Activation and Subsequent B Cell Responses. Immunity 2006, 24, 259–268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Altin, J.G.; Sloan, E.K. The role of CD45 and CD45-associated molecules in T cell activation. Immunol. Cell Biol. 1997, 75, 430–445. [Google Scholar] [CrossRef]
- Maity, P.C.; Blount, A.; Jumaa, H.; Ronneberger, O.; Lillemeier, B.F.; Reth, M. B cell antigen receptors of the IgM and IgD classes are clustered in different protein islands that are altered during B cell activation. Sci. Signal. 2015, 8, ra93. [Google Scholar] [CrossRef]
- Holdbrooks, A.T.; Ankenbauer, K.E.; Hwang, J.; Bellis, S.L. Regulation of inflammatory signaling by the ST6Gal-I sialyltransferase. PLoS ONE 2020, 15, e0241850. [Google Scholar] [CrossRef]
- Hennet, T.; Chui, D.; Paulson, J.C.; Marth, J.D. Immune regulation by the ST6Gal sialyltransferase. Proc. Natl. Acad. Sci. USA 1998, 95, 4504–4509. [Google Scholar] [CrossRef] [Green Version]
- Joachimiak, E.; Osinka, A.; Farahat, H.; Świderska, B.; Sitkiewicz, E.; Poprzeczko, M.; Fabczak, H.; Wloga, D. Composition and function of the C1b/C1f region in the ciliary central apparatus. Sci. Rep. 2021, 11, 11760. [Google Scholar] [CrossRef]
- Wannemacher, K.M.; Wang, L.; Zhu, L.; Brass, L.F. The role of semaphorins and their receptors in platelets: Lessons learned from neuronal and immune synapses. Platelets 2011, 22, 461–465. [Google Scholar] [CrossRef] [PubMed]
- Alto, L.T.; Terman, J.R. Semaphorins and their signaling mechanisms. In Semaphorin Signaling; Terman, J.R., Ed.; Methods in Molecular Biology; Humana Press: New York, NY, USA, 2017; Volume 1493, pp. 1–25. ISBN 978-1-4939-6448-2. [Google Scholar] [CrossRef] [Green Version]
- Koivisto, O.; Hanel, A.; Carlberg, C. Key Vitamin D Target Genes with Functions in the Immune System. Nutrients 2020, 12, 1140. [Google Scholar] [CrossRef] [Green Version]
- Rivière, T.; Bader, A.; Pogoda, K.; Walzog, B.; Maier-Begandt, D. Structure and Emerging Functions of LRCH Proteins in Leukocyte Biology. Front. Cell Dev. Biol. 2020, 8, 584134. [Google Scholar] [CrossRef] [PubMed]
- Tur-Gracia, S.; Martinez-Quiles, N. Emerging functions of cytoskeletal proteins in immune diseases. J. Cell Sci. 2021, 134, jcs253534. [Google Scholar] [CrossRef]
- Schaks, M.; Giannone, G.; Rottner, K. Actin dynamics in cell migration. Essays Biochem. 2019, 63, 483–495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reymann, A.-C.; Boujemaa-Paterski, R.; Martiel, J.-L.; Guérin, C.; Cao, W.; Chin, H.F.; De La Cruz, E.M.; Théry, M.; Blanchoin, L. Actin Network Architecture Can Determine Myosin Motor Activity. Science 2012, 336, 1310–1314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cummins, S.B.; Waters, S.M.; Evans, A.C.O.; Lonergan, P.; Butler, S.T. Genetic merit for fertility traits in Holstein cows: III. Hepatic expression of somatotropic axis genes during pregnancy and lactation. J. Dairy Sci. 2012, 95, 3711–3721. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cummins, S.B.; Lonergan, P.; Evans, A.C.O.; Berry, D.P.; Evans, R.D.; Butler, S.T. Genetic merit for fertility traits in Holstein cows: I. Production characteristics and reproductive efficiency in a pasture-based system. J. Dairy Sci. 2012, 95, 1310–1322. [Google Scholar] [CrossRef] [PubMed]
- Wathes, D.C.; Cheng, Z.; Fenwick, M.A.; Fitzpatrick, R.; Patton, J. Influence of energy balance on the somatotrophic axis and matrix metalloproteinase expression in the endometrium of the postpartum dairy cow. Reproduction 2011, 141, 269–281. [Google Scholar] [CrossRef] [Green Version]
- Fortune, J.E.; Rivera, G.M.; Evans, A.C.O.; Turzillo, A.M. Differentiation of Dominant Versus Subordinate Follicles in Cattle. Biol. Reprod. 2001, 65, 648–654. [Google Scholar] [CrossRef]
- Wathes, D.C.; Fenwick, M.; Cheng, Z.; Bourne, N.; Llewellyn, S.; Morris, D.G.; Kenny, D.; Murphy, J.; Fitzpatrick, R. Influence of negative energy balance on cyclicity and fertility in the high producing dairy cow. Theriogenology 2007, 68, S232–S241. [Google Scholar] [CrossRef] [PubMed]
- Beam, S.W.; Butler, W.R. Energy Balance and Ovarian Follicle Development Prior to the First Ovulation Postpartum in Dairy Cows Receiving Three Levels of Dietary Fat. Biol. Reprod. 1997, 56, 133–142. [Google Scholar] [CrossRef] [Green Version]
- Pushpakumara, P.G.A.; Gardner, N.H.; Reynolds, C.K.; Beever, D.E.; Wathes, D.C. Relationships between transition period diet, metabolic parameters and fertility in lactating dairy cows. Theriogenology 2003, 60, 1165–1185. [Google Scholar] [CrossRef]
- Patton, J.; Kenny, D.A.; McNamara, S.; Mee, J.F.; O’Mara, F.P.; Diskin, M.G.; Murphy, J.J. Relationships Among Milk Production, Energy Balance, Plasma Analytes, and Reproduction in Holstein-Friesian Cows. J. Dairy Sci. 2007, 90, 649–658. [Google Scholar] [CrossRef] [Green Version]
- Piechotta, M.; Mysegades, W.; Ligges, U.; Lilienthal, J.; Hoeflich, A.; Miyamoto, A.; Bollwein, H. Antepartal insulin-like growth factor 1 and insulin-like growth factor binding protein 2 concentrations are indicative of ketosis in dairy cows. J. Dairy Sci. 2015, 98, 3100–3109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dickinson, S.E.; Griffin, B.A.; Elmore, M.F.; Kriese-Anderson, L.; Elmore, J.B.; Dyce, P.W.; Rodning, S.P.; Biase, F.H. Transcriptome profiles in peripheral white blood cells at the time of artificial insemination discriminate beef heifers with different fertility potential. BMC Genom. 2018, 19, 129. [Google Scholar] [CrossRef] [Green Version]
- LeBlanc, S.J.; Lissemore, K.D.; Kelton, D.F.; Duffield, T.F.; Leslie, K.E. Major advances in disease prevention in dairy cattle. J. Dairy Sci. 2006, 89, 1267–1279. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).