Metabolomic Profiling, Volatile Fatty Acids, and Greenhouse Gas Emissions of Beef Cattle Infused with Different Essential Oil Blends
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Ethical Approval
2.2. Animals, Experimental Design, and Diet Composition
2.3. Rumen Fluid Sample Collection
2.4. Short-Chain Fatty Acid Analysis
2.5. Ammonia Nitrogen Analysis
2.6. Metabolome Analysis of Rumen Fluid
2.7. Statistical Analysis
3. Results
3.1. Volatile Fatty Acids
3.2. Greenhouse Gas Emissions, Rumen pH, and NH3-N Concentrations
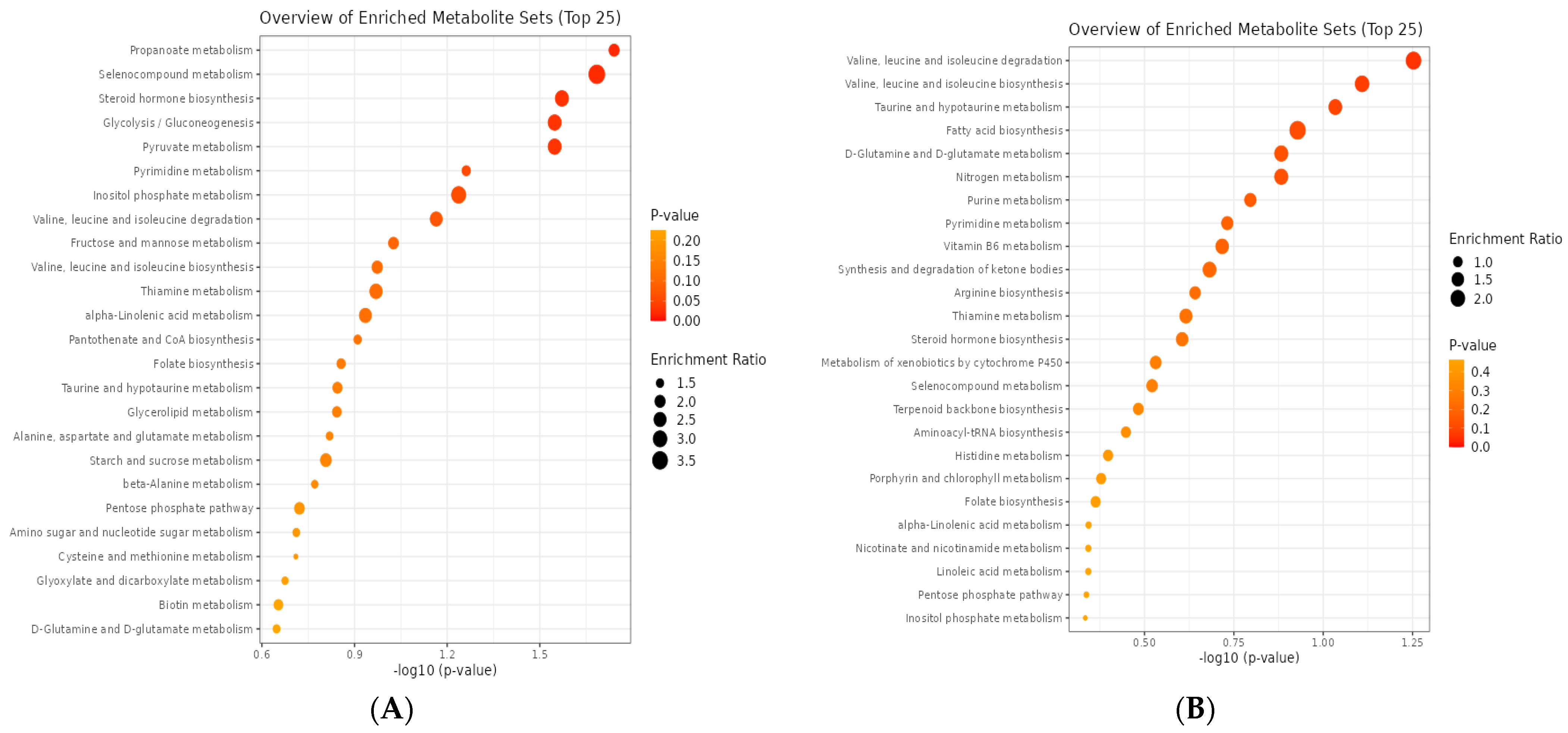
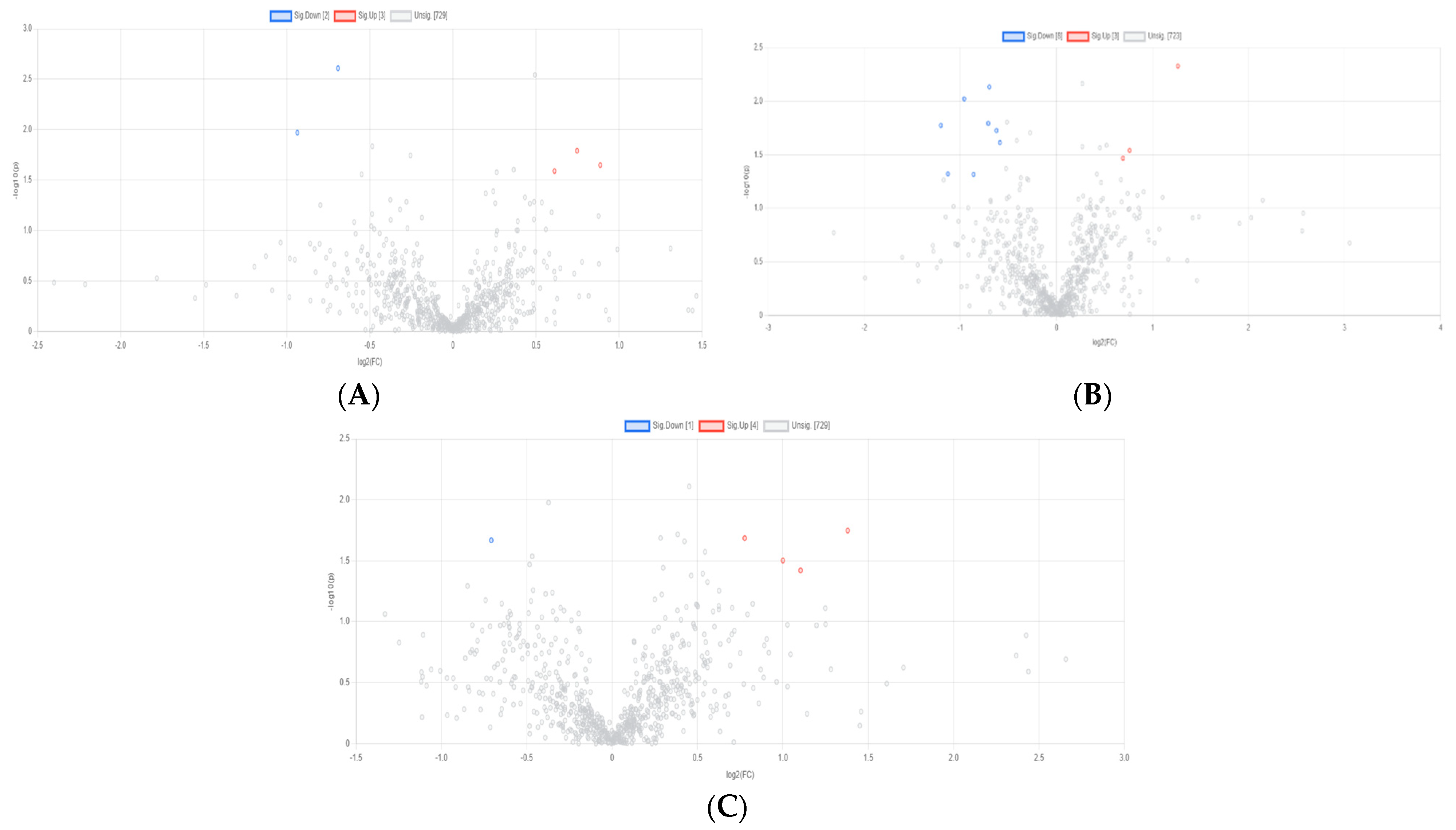
3.3. Rumen Metabolites
3.3.1. CON vs. EOB1
3.3.2. CON vs. EOB2
3.3.3. CON vs. EOB3
3.3.4. CON vs. EOB4
3.3.5. CON vs. EOB5
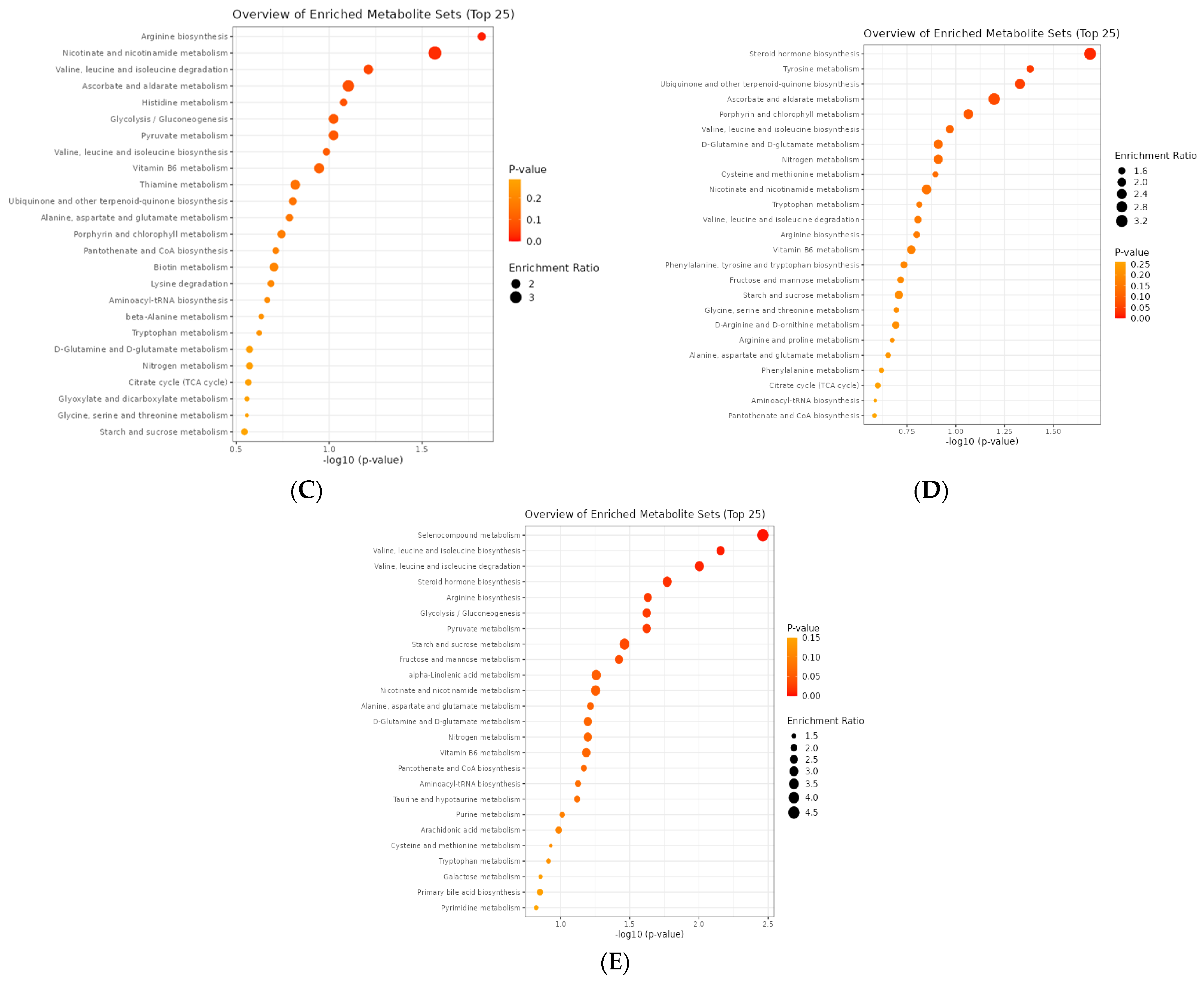
3.3.6. EOB4 vs. EOB3
3.3.7. EOB5 vs. EOB3
3.3.8. EOB5 vs. EOB4
4. Discussion
4.1. Effects of EOBs on Volatile Fatty Acids
4.2. Effects of EOBs on Rumen pH and NH3-N Concentration
4.3. Effect of EOBs on Greenhouse Gases
4.4. Effect of EOBs on Rumen Metabolites
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liu, Y.; Espinosa, C.D.; Abelilla, J.J.; Casas, G.A.; Lagos, L.V.; Lee, S.A.; Kwon, W.B.; Mathai, J.K.; Navarro, D.M.D.L.; Jaworski, N.W.; et al. Non-Antibiotic Feed Additives in Diets for Pigs: A Review. Anim. Nutr. 2018, 4, 113–125. [Google Scholar] [CrossRef] [PubMed]
- Omonijo, F.A.; Ni, L.; Gong, J.; Wang, Q.; Lahaye, L.; Yang, C. Essential Oils as Alternatives to Antibiotics in Swine Production. Anim. Nutr. 2018, 4, 126–136. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Z.; Xu, X.; Zhang, Q.; Li, P.; Zhao, P.; Li, Q.; Liu, J.; Piao, X. Effects of Essential Oil Supplementation of a Low-energy Diet on Performance, Intestinal Morphology and Microflora, Immune Properties and Antioxidant Activities in Weaned Pigs. Anim. Sci. J. 2015, 86, 279–285. [Google Scholar] [CrossRef] [PubMed]
- Tian, Q.; Piao, X. Essential Oil Blend Could Decrease Diarrhea Prevalence by Improving Antioxidative Capability for Weaned Pigs. Animals 2019, 9, 847. [Google Scholar] [CrossRef] [PubMed]
- Henderson, G.; Cox, F.; Ganesh, S.; Jonker, A.; Young, W.; Global Rumen Census Collaborators; Janssen, P.H. Rumen Microbial Community Composition Varies with Diet and Host, but a Core Microbiome Is Found across a Wide Geographical Range. Sci. Rep. 2015, 5, 14567. [Google Scholar] [CrossRef] [PubMed]
- Huws, S.A.; Creevey, C.J.; Oyama, L.B.; Mizrahi, I.; Denman, S.E.; Popova, M.; Muñoz-Tamayo, R.; Forano, E.; Waters, S.M.; Hess, M.; et al. Addressing Global Ruminant Agricultural Challenges Through Understanding the Rumen Microbiome: Past, Present, and Future. Front. Microbiol. 2018, 9, 2161. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, M.S.; Carvalho, M.; Bastos, M.L.; de Pinho, P.G. Metabolomics Analysis for Biomarker Discovery: Advances and Challenges. Curr. Med. Chem. 2013, 20, 257–271. [Google Scholar] [CrossRef] [PubMed]
- Foroutan, A.; Goldansaz, S.A.; Lipfert, M.; Wishart, D.S. Protocols for NMR Analysis in Livestock Metabolomics. In Metabolomics; Bhattacharya, S.K., Ed.; Methods in Molecular Biology; Springer: New York, NY, USA, 2019; Volume 1996, pp. 311–324. ISBN 978-1-4939-9487-8. [Google Scholar]
- Zhang, R.; Wu, J.; Lei, Y.; Bai, Y.; Jia, L.; Li, Z.; Liu, T.; Xu, Y.; Sun, J.; Wang, Y.; et al. Oregano Essential Oils Promote Rumen Digestive Ability by Modulating Epithelial Development and Microbiota Composition in Beef Cattle. Front. Nutr. 2021, 8, 722557. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Fu, X.; Ma, X.; Geng, S.; Jiang, X.; Huang, Q.; Hu, C.; Han, X. Intestinal Microbiome-Metabolome Responses to Essential Oils in Piglets. Front. Microbiol. 2018, 9, 1988. [Google Scholar] [CrossRef]
- Scollan, N.D.; Greenwood, P.L.; Newbold, C.J.; Ruiz, D.R.Y.; Shingfield, K.J.; Wallace, R.J.; Hocquette, J.F. Future Research Priorities for Animal Production in a Changing World. Anim. Prod. Sci. 2011, 51, 1–5. [Google Scholar] [CrossRef]
- Knapp, J.R.; Laur, G.L.; Vadas, P.A.; Weiss, W.P.; Tricarico, J.M. Invited Review: Enteric Methane in Dairy Cattle Production: Quantifying the Opportunities and Impact of Reducing Emissions. J. Dairy Sci. 2014, 97, 3231–3261. [Google Scholar] [CrossRef]
- Gerber, P.J.; Hristov, A.N.; Henderson, B.; Makkar, H.; Oh, J.; Lee, C.; Meinen, R.; Montes, F.; Ott, T.; Firkins, J.; et al. Technical Options for the Mitigation of Direct Methane and Nitrous Oxide Emissions from Livestock: A Review. Animal 2013, 7, 220–234. [Google Scholar] [CrossRef]
- Hristov, A.N.; Oh, J.; Firkins, J.L.; Dijkstra, J.; Kebreab, E.; Waghorn, G.; Makkar, H.P.S.; Adesogan, A.T.; Yang, W.; Lee, C.; et al. SPECIAL TOPICS—Mitigation of Methane and Nitrous Oxide Emissions from Animal Operations: I. A Review of Enteric Methane Mitigation Options1. J. Anim. Sci. 2013, 91, 5045–5069. [Google Scholar] [CrossRef]
- Kholif, A.E.; Kassab, A.Y.; Azzaz, H.H.; Matloup, O.H.; Hamdon, H.A.; Olafadehan, O.A.; Morsy, T.A. Essential Oils Blend with a Newly Developed Enzyme Cocktail Works Synergistically to Enhance Feed Utilization and Milk Production of Farafra Ewes in the Subtropics. Small Rumin. Res. 2018, 161, 43–50. [Google Scholar] [CrossRef]
- Cardozo, P.W.; Calsamiglia, S.; Ferret, A.; Kamel, C. Screening for the Effects of Natural Plant Extracts at Different pH on in Vitro Rumen Microbial Fermentation of a High-Concentrate Diet for Beef Cattle1. J. Anim. Sci. 2005, 83, 2572–2579. [Google Scholar] [CrossRef] [PubMed]
- AOAC. Official Methods of Analysis, 17th ed.; The Association of Official Analytical Chemists: Gaithersburg, MD, USA, 2000. [Google Scholar]
- Alabi, J.O.; Okedoyin, D.O.; Anotaenwere, C.C.; Wuaku, M.; Gray, D.; Adelusi, O.O.; Ike, K.A.; Olagunju, L.K.; Dele, P.A.; Anele, U.Y. Essential Oil Blends with or without Fumaric Acid Influenced In Vitro Rumen Fermentation, Greenhouse Gas Emission, and Volatile Fatty Acids Production of a Total Mixed Ration. Ruminants 2023, 3, 373–384. [Google Scholar] [CrossRef]
- Siqueira, M.; Chagas, J.; Monnerat, J.P.; Monteiro, C.; Mora-Luna, R.; Dubeux, J.; DiLorenzo, N.; Ruiz-Moreno, M.; Ferreira, M. Nutritive Value, In Vitro Fermentation, and Methane Production of Cactus Cladodes, Sugarcane Bagasse, and Urea. Animals 2021, 11, 1266. [Google Scholar] [CrossRef]
- Olagunju, L.K.; Isikhuemhen, O.S.; Dele, P.A.; Anike, F.N.; Ike, K.A.; Shaw, Y.; Brice, R.M.; Orimaye, O.E.; Wuaku, M.; Essick, B.G.; et al. Effects of the Incubation Period of Pleurotus Ostreatus on the Chemical Composition and Nutrient Availability of Solid-State-Fermented Corn Stover. Animals 2023, 13, 2587. [Google Scholar] [CrossRef]
- Lynch, J.M.; Barbano, D.M.; Fleming, J.R.; Barbano Laboratory; California Department of Food and Agriculture; Dairy One; Dairy Quality Control Institute, Inc.; Land O’Lakes; State of Wisconsin Department of Agriculture. Determination of the Total Nitrogen Content of Hard, Semihard, and Processed Cheese by the Kjeldahl Method: Collaborative Study. J. AOAC Int. 2002, 85, 445–455. [Google Scholar] [CrossRef]
- Chen, D.; Su, X.; Wang, N.; Li, Y.; Yin, H.; Li, L.; Li, L. Chemical Isotope Labeling LC-MS for Monitoring Disease Progression and Treatment in Animal Models: Plasma Metabolomics Study of Osteoarthritis Rat Model. Sci. Rep. 2017, 7, 40543. [Google Scholar] [CrossRef]
- Idowu, M.; Taiwo, G.; Sidney, T.; Morenikeji, O.B.; Pech Cervantes, A.; Estrada-Reyes, Z.M.; Wilson, M.; Ogunade, I.M. The Differential Plasma and Ruminal Metabolic Pathways and Rumen Bacterial Taxa Associated with Divergent Residual Body Weight Gain Phenotype in Crossbred Beef Steers. Transl. Anim. Sci. 2023, 7, txad054. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Li, R.; Zhou, J.; Zuniga, A.; Stanislaus, A.E.; Wu, Y.; Huan, T.; Zheng, J.; Shi, Y.; Wishart, D.S.; et al. MyCompoundID: Using an Evidence-Based Metabolome Library for Metabolite Identification. Anal. Chem. 2013, 85, 3401–3408. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Wang, X.; Li, X.; Zhao, C.; Hu, P.; Chen, H.; Liu, Z.; Liu, G.; Wang, Z. Correlation between Composition of the Bacterial Community and Concentration of Volatile Fatty Acids in the Rumen during the Transition Period and Ketosis in Dairy Cows. Appl. Environ. Microbiol. 2012, 78, 2386–2392. [Google Scholar] [CrossRef]
- Storm, A.C.; Kristensen, N.B.; Hanigan, M.D. A Model of Ruminal Volatile Fatty Acid Absorption Kinetics and Rumen Epithelial Blood Flow in Lactating Holstein Cows. J. Dairy Sci. 2012, 95, 2919–2934. [Google Scholar] [CrossRef]
- Kholif, A.E.; Olafadehan, O.A. Essential Oils and Phytogenic Feed Additives in Ruminant Diet: Chemistry, Ruminal Microbiota and Fermentation, Feed Utilization and Productive Performance. Phytochem. Rev. 2021, 20, 1087–1108. [Google Scholar] [CrossRef]
- Castillejos, L.; Calsamiglia, S.; Ferret, A.; Losa, R. Effects of Dose and Adaptation Time of a Specific Blend of Essential Oil Compounds on Rumen Fermentation. Anim. Feed. Sci. Technol. 2007, 132, 186–201. [Google Scholar] [CrossRef]
- Jiang, F.; Gao, Y.; Peng, Z.; Ma, X.; You, Y.; Hu, Z.; He, A.; Liao, Y. Isoacids Supplementation Improves Growth Performance and Feed Fiber Digestibility Associated with Ruminal Bacterial Community in Yaks. Front. Microbiol. 2023, 14, 1175880. [Google Scholar] [CrossRef]
- Gleason, C.B.; Beckett, L.M.; White, R.R. Rumen Fermentation and Epithelial Gene Expression Responses to Diet Ingredients Designed to Differ in Ruminally Degradable Protein and Fiber Supplies. Sci. Rep. 2022, 12, 2933. [Google Scholar] [CrossRef]
- Roy, D.; Tomar, S.K.; Kumar, V. Rumen Modulatory Effect of Thyme, Clove and Peppermint Oils in Vitro Using Buffalo Ruminal Liquor. Vet. World 2015, 8, 203–207. [Google Scholar] [CrossRef]
- Membrive, C.M.B. Anatomy and Physiology of the Rumen. In Rumenology; Millen, D.D., De Beni Arrigoni, M., Lauritano Pacheco, R.D., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 1–38. ISBN 978-3-319-30531-8. [Google Scholar]
- Tager, L.R.; Krause, K.M. Effects of Essential Oils on Rumen Fermentation, Milk Production, and Feeding Behavior in Lactating Dairy Cows. J. Dairy Sci. 2011, 94, 2455–2464. [Google Scholar] [CrossRef] [PubMed]
- Dijkstra, J.; Van Gastelen, S.; Dieho, K.; Nichols, K.; Bannink, A. Review: Rumen Sensors: Data and Interpretation for Key Rumen Metabolic Processes. Animal 2020, 14, s176–s186. [Google Scholar] [CrossRef] [PubMed]
- Salter, D.N.; Daneshvar, K.; Smith, R.H. The Origin of Nitrogen Incorporated into Compounds in the Rumen Bacteria of Steers given Protein- and Urea-Containing Diets. Br. J. Nutr. 1979, 41, 197–209. [Google Scholar] [CrossRef] [PubMed]
- Temmar, R.; Rodríguez-Prado, M.; Forgeard, G.; Rougier, C.; Calsamiglia, S. Interactions among Natural Active Ingredients to Improve the Efficiency of Rumen Fermentation In Vitro. Animals 2021, 11, 1205. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, Z.; Qin, W.; Gao, X.; Wu, J.; Zhao, S.; Jiao, T. Effects of Oregano Essential Oil, Cobalt and Synergistic of Both of Them on Rumen Degradation Rate and Fermentation Characteristics for Corn Silage. Ital. J. Anim. Sci. 2022, 21, 1476–1488. [Google Scholar] [CrossRef]
- Cardozo, P.W.; Calsamiglia, S.; Ferret, A.; Kamel, C. Effects of Natural Plant Extracts on Ruminal Protein Degradation and Fermentation Profiles in Continuous Culture1. J. Anim. Sci. 2004, 82, 3230–3236. [Google Scholar] [CrossRef]
- Benchaar, C.; Hassanat, F.; Petit, H.V. Dose–Response to Eugenol Supplementation to Dairy Cow Diets: Methane Production, N Excretion, Ruminal Fermentation, Nutrient Digestibility, Milk Production, and Milk Fatty Acid Profile. Anim. Feed. Sci. Technol. 2015, 209, 51–59. [Google Scholar] [CrossRef]
- Hart, K.J.; Jones, H.G.; Waddams, K.E.; Worgan, H.J.; Zweifel, B.; Newbold, C.J. An Essential Oil Blend Decreases Methane Emissions and Increases Milk Yield in Dairy Cows. Open J. Anim. Sci. 2019, 9, 259–267. [Google Scholar] [CrossRef]
- Rossi, C.A.S.; Grossi, S.; Dell’Anno, M.; Compiani, R.; Rossi, L. Effect of a Blend of Essential Oils, Bioflavonoids and Tannins on In Vitro Methane Production and In Vivo Production Efficiency in Dairy Cows. Animals 2022, 12, 728. [Google Scholar] [CrossRef]
- Brice, R.M.; Dele, P.A.; Ike, K.A.; Shaw, Y.A.; Olagunju, L.K.; Orimaye, O.E.; Subedi, K.; Anele, U.Y. Effects of Essential Oil Blends on In Vitro Apparent and Truly Degradable Dry Matter, Efficiency of Microbial Production, Total Short-Chain Fatty Acids and Greenhouse Gas Emissions of Two Dairy Cow Diets. Animals 2022, 12, 2185. [Google Scholar] [CrossRef]
- Castro-Montoya, J.; Peiren, N.; Cone, J.W.; Zweifel, B.; Fievez, V.; De Campeneere, S. In Vivo and in Vitro Effects of a Blend of Essential Oils on Rumen Methane Mitigation. Livest. Sci. 2015, 180, 134–142. [Google Scholar] [CrossRef]
- Staerfl, S.; Kreuzer, M.; Soliva, C. In Vitro Screening of Unconventional Feeds and Various Natural Supplements for Their Ruminal Methane Mitigation Potential When Includedin a Maize-Silage Based Diet. J. Anim. Feed. Sci. 2010, 19, 651–664. [Google Scholar] [CrossRef]
- Wang, B.; Jia, M.; Fang, L.; Jiang, L.; Li, Y. Effects of Eucalyptus Oil and Anise Oil Supplementation on Rumen Fermentation Characteristics, Methane Emission, and Digestibility in Sheep1. J. Anim. Sci. 2018, 96, 3460–3470. [Google Scholar] [CrossRef] [PubMed]
- Shah, A.M.; Ma, J.; Wang, Z.; Hu, R.; Wang, X.; Peng, Q.; Amevor, F.K.; Goswami, N. Production of Hydrogen Sulfide by Fermentation in Rumen and Its Impact on Health and Production of Animals. Processes 2020, 8, 1169. [Google Scholar] [CrossRef]
- Berg, J.M.; Tymoczko, J.L.; Stryer, L.; Stryer, L. Biochemistry, 5th ed.; W.H. Freeman: New York, NY, USA, 2002; ISBN 978-0-7167-3051-4. [Google Scholar]
- Blanco, A.; Blanco, G. Carbohydrate Metabolism. In Medical Biochemistry; Elsevier: Amsterdam, The Netherlands, 2017; pp. 283–323. ISBN 978-0-12-803550-4. [Google Scholar]
- Hobson, R.M.; Saunders, B.; Ball, G.; Harris, R.C.; Sale, C. Effects of β-Alanine Supplementation on Exercise Performance: A Meta-Analysis. Amino Acids 2012, 43, 25–37. [Google Scholar] [CrossRef]
- Boldyrev, A.A.; Aldini, G.; Derave, W. Physiology and Pathophysiology of Carnosine. Physiol. Rev. 2013, 93, 1803–1845. [Google Scholar] [CrossRef]
- Malinovsky, A.V. Why Threonine Is an Essential Amino Acid in Mammals and Birds: Studies at the Enzyme Level. Biochemistry 2018, 83, 795–799. [Google Scholar] [CrossRef]
- Li, D.; Mao, X.; Zeng, X.; Qiao, S.; Wu, G. Specific Roles of Threonine in Intestinal Mucosal Integrity and Barrier Function. Front. Biosci. 2011, 3, 1192–1200. [Google Scholar] [CrossRef]
- Hu, S.; He, W.; Wu, G. Hydroxyproline in Animal Metabolism, Nutrition, and Cell Signaling. Amino Acids 2022, 54, 513–528. [Google Scholar] [CrossRef]
- Wu, G. Important Roles of Dietary Taurine, Creatine, Carnosine, Anserine and 4-Hydroxyproline in Human Nutrition and Health. Amino acids 2020, 52, 329–360. [Google Scholar] [CrossRef]
- Perry, R.J.; Borders, C.B.; Cline, G.W.; Zhang, X.-M.; Alves, T.C.; Petersen, K.F.; Rothman, D.L.; Kibbey, R.G.; Shulman, G.I. Propionate Increases Hepatic Pyruvate Cycling and Anaplerosis and Alters Mitochondrial Metabolism. J. Biol. Chem. 2016, 291, 12161–12170. [Google Scholar] [CrossRef] [PubMed]
- Gray, L.R.; Tompkins, S.C.; Taylor, E.B. Regulation of Pyruvate Metabolism and Human Disease. Cell. Mol. Life Sci. 2014, 71, 2577–2604. [Google Scholar] [CrossRef] [PubMed]
- Wozniak, G.G.; Strahl, B.D. Hitting the ‘Mark’: Interpreting Lysine Methylation in the Context of Active Transcription. Biochim. Et Biophys. Acta BBA Gene Regul. Mech. 2014, 1839, 1353–1361. [Google Scholar] [CrossRef] [PubMed]
- Sidney, T.; Taiwo, G.; Idowu, M.; Amusan, I.; Pech Cervantes, A.; Ogunade, I. Rumen Fluid Amine/Phenol-Metabolome of Beef Steers with Divergent Residual Feed Intake Phenotype. Ruminants 2023, 3, 1–8. [Google Scholar] [CrossRef]
- Tomé, D.; Bos, C. Lysine Requirement through the Human Life Cycle. J. Nutr. 2007, 137, 1642S–1645S. [Google Scholar] [CrossRef] [PubMed]
- Brenes, A.; Roura, E. Essential Oils in Poultry Nutrition: Main Effects and Modes of Action. Anim. Feed. Sci. Technol. 2010, 158, 1–14. [Google Scholar] [CrossRef]
- Ochoa-Velasco, C.E.; Avila-Sosa, R.; Navarro-Cruz, A.R.; López-Malo, A.; Palou, E. Biotic and Abiotic Factors to Increase Bioactive Compounds in Fruits and Vegetables. In Food Bioconversion; Elsevier: Amsterdam, The Netherlands, 2017; pp. 317–349. [Google Scholar]
- Wink, M. Modes of Action of Herbal Medicines and Plant Secondary Metabolites. Medicines 2015, 2, 251–286. [Google Scholar] [CrossRef] [PubMed]
- Ferrazzano, G.F.; Amato, I.; Ingenito, A.; Zarrelli, A.; Pinto, G.; Pollio, A. Plant Polyphenols and Their Anti-Cariogenic Properties: A Review. Molecules 2011, 16, 1486–1507. [Google Scholar] [CrossRef] [PubMed]
- Manach, C.; Scalbert, A.; Morand, C.; Rémésy, C.; Jiménez, L. Polyphenols: Food Sources and Bioavailability. Am. J. Clin. Nutr. 2004, 79, 727–747. [Google Scholar]
- Dorantes-Iturbide, G.; Orzuna-Orzuna, J.F.; Lara-Bueno, A.; Mendoza-Martínez, G.D.; Miranda-Romero, L.A.; Lee-Rangel, H.A. Essential Oils as a Dietary Additive for Small Ruminants: A Meta-Analysis on Performance, Rumen Parameters, Serum Metabolites, and Product Quality. Vet. Sci. 2022, 9, 475. [Google Scholar] [CrossRef]
- Nehme, R.; Andrés, S.; Pereira, R.B.; Ben Jemaa, M.; Bouhallab, S.; Ceciliani, F.; López, S.; Rahali, F.Z.; Ksouri, R.; Pereira, D.M.; et al. Essential Oils in Livestock: From Health to Food Quality. Antioxidants 2021, 10, 330. [Google Scholar] [CrossRef] [PubMed]
- Kriseldi, R.; Silva, M.; Lee, J.; Adhikari, R.; Williams, C.; Corzo, A. Understanding the Interactive Effects of Dietary Leucine with Isoleucine and Valine in the Modern Commercial Broiler. Poult. Sci. 2022, 101, 102140. [Google Scholar] [CrossRef]
- Nie, C.; He, T.; Zhang, W.; Zhang, G.; Ma, X. Branched Chain Amino Acids: Beyond Nutrition Metabolism. Int. J. Mol. Sci. 2018, 19, 954. [Google Scholar] [CrossRef]
- Bettio, L.E.B.; Gil-Mohapel, J.; Rodrigues, A.L.S. Guanosine and Its Role in Neuropathologies. Purinergic Signal. 2016, 12, 411–426. [Google Scholar] [CrossRef] [PubMed]
- Massari, C.M.; Zuccarini, M.; Di Iorio, P.; Tasca, C.I. Guanosine Mechanisms of Action: Toward Molecular Targets. Front. Pharmacol. 2021, 12, 653146. [Google Scholar] [CrossRef] [PubMed]
- Rathbone, M.; Pilutti, L.; Caciagli, F.; Jiang, S. Neurotrophic Effects of Extracellular Guanosine. Nucleosides Nucleotides Nucleic Acids 2008, 27, 666–672. [Google Scholar] [CrossRef] [PubMed]
- Mindt, M.; Walter, T.; Kugler, P.; Wendisch, V.F. Microbial Engineering for Production of N-Functionalized Amino Acids and Amines. Biotechnol. J. 2020, 15, 1900451. [Google Scholar] [CrossRef] [PubMed]
- Galderisi, S.; Milella, M.S.; Rossi, M.; Cicaloni, V.; Rossi, R.; Giustarini, D.; Spiga, O.; Tinti, L.; Salvini, L.; Tinti, C.; et al. Homogentisic Acid Induces Autophagy Alterations Leading to Chondroptosis in Human Chondrocytes: Implications in Alkaptonuria. Arch. Biochem. Biophys. 2022, 717, 109137. [Google Scholar] [CrossRef]
- Zhou, J.; Hou, D.; Zou, W.; Wang, J.; Luo, R.; Wang, M.; Yu, H. Comparison of Widely Targeted Metabolomics and Untargeted Metabolomics of Wild Ophiocordyceps Sinensis. Molecules 2022, 27, 3645. [Google Scholar] [CrossRef]
- Gil-Ortiz, F.; Ramón-Maiques, S.; Fernández-Murga, M.L.; Fita, I.; Rubio, V. Two Crystal Structures of Escherichia Coli N-Acetyl-l-Glutamate Kinase Demonstrate the Cycling between Open and Closed Conformations. J. Mol. Biol. 2010, 399, 476–490. [Google Scholar] [CrossRef]
- Xiong, X.; Xu, J.; Yan, X.; Wu, S.; Ma, J.; Wang, Z.; He, Q.; Gong, J.; Rao, Y. Gut Microbiome and Serum Metabolome Analyses Identify Biomarkers Associated with Sexual Maturity in Quails. Poult. Sci. 2023, 102, 102762. [Google Scholar] [CrossRef]
- Yaku, K.; Palikhe, S.; Izumi, H.; Yoshida, T.; Hikosaka, K.; Hayat, F.; Karim, M.; Iqbal, T.; Nitta, Y.; Sato, A.; et al. BST1 Regulates Nicotinamide Riboside Metabolism via Its Glycohydrolase and Base-Exchange Activities. Nat. Commun. 2021, 12, 6767. [Google Scholar] [CrossRef] [PubMed]
- Elhassan, Y.S.; Kluckova, K.; Fletcher, R.S.; Schmidt, M.S.; Garten, A.; Doig, C.L.; Cartwright, D.M.; Oakey, L.; Burley, C.V.; Jenkinson, N.; et al. Nicotinamide Riboside Augments the Aged Human Skeletal Muscle NAD+ Metabolome and Induces Transcriptomic and Anti-Inflammatory Signatures. Cell Rep. 2019, 28, 1717–1728.e6. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Sareddy, G.R.; Lu, Y.; Pratap, U.P.; Tang, F.; Greene, K.M.; Meyre, P.L.; Tekmal, R.R.; Vadlamudi, R.K.; Brann, D.W. Astrocyte-Derived Estrogen Regulates Reactive Astrogliosis and Is Neuroprotective Following Ischemic Brain Injury. J. Neurosci. 2020, 40, 9751–9771. [Google Scholar] [CrossRef] [PubMed]
- González-Mula, A.; Lang, J.; Grandclément, C.; Naquin, D.; Ahmar, M.; Soulère, L.; Queneau, Y.; Dessaux, Y.; Faure, D. Lifestyle of the Biotroph Agrobacterium Tumefaciens in the Ecological Niche Constructed on Its Host Plant. New Phytol. 2018, 219, 350–362. [Google Scholar] [CrossRef]
- Amin, N.; Tagliapietra, F.; Arango, S.; Guzzo, N.; Bailoni, L. Free and Microencapsulated Essential Oils Incubated In Vitro: Ruminal Stability and Fermentation Parameters. Animals 2021, 11, 180. [Google Scholar] [CrossRef]
- Ogunade, I.M.; McCoun, M.; Idowu, M.D.; Peters, S.O. Comparative Effects of Two Multispecies Direct-Fed Microbial Products on Energy Status, Nutrient Digestibility, and Ruminal Fermentation, Bacterial Community, and Metabolome of Beef Steers. J. Anim. Sci. 2020, 98, skaa201. [Google Scholar] [CrossRef]
- Schenck, C.A.; Maeda, H.A. Tyrosine Biosynthesis, Metabolism, and Catabolism in Plants. Phytochemistry 2018, 149, 82–102. [Google Scholar] [CrossRef]
- Meganathan, R. Ubiquinone Biosynthesis in Microorganisms. FEMS Microbiol. Lett. 2001, 203, 131–139. [Google Scholar] [CrossRef]
- Herman, M.A.; Birnbaum, M.J. Molecular Aspects of Fructose Metabolism and Metabolic Disease. Cell Metab. 2021, 33, 2329–2354. [Google Scholar] [CrossRef]
- Del Olmo, A.; Calzada, J.; Nuñez, M. Benzoic Acid and Its Derivatives as Naturally Occurring Compounds in Foods and as Additives: Uses, Exposure, and Controversy. Crit. Rev. Food Sci. Nutr. 2017, 57, 3084–3103. [Google Scholar] [CrossRef] [PubMed]
- Knarreborg, A.; Miquel, N.; Granli, T.; Jensen, B. Establishment and Application of an in Vitro Methodology to Study the Effects of Organic Acids on Coliform and Lactic Acid Bacteria in the Proximal Part of the Gastrointestinal Tract of Piglets. Anim. Feed. Sci. Technol. 2002, 99, 131–140. [Google Scholar] [CrossRef]
- Halas, D.; Hansen, C.F.; Hampson, D.J.; Mullan, B.P.; Wilson, R.H.; Pluske, J.R. Effect of Dietary Supplementation with Inulin and/or Benzoic Acid on the Incidence and Severity of Post-Weaning Diarrhoea in Weaner Pigs after Experimental Challenge with Enterotoxigenic Escherichia coli. Arch. Anim. Nutr. 2009, 63, 267–280. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.; Yang, Q.; Chen, D.; Yu, B.; He, J. Benzoic Acid Used as Food and Feed Additives Can Regulate Gut Functions. BioMed Res. Int. 2019, 2019, 5721585. [Google Scholar] [CrossRef]
- Kinscherf, T.G.; Coleman, R.H.; Barta, T.M.; Willis, D.K. Cloning and Expression of the Tabtoxin Biosynthetic Region from Pseudomonas syringae. J. Bacteriol. 1991, 173, 4124–4132. [Google Scholar] [CrossRef]
- Liu, M.; Zeng, M.; Wang, S.; Cao, B.; Guo, P.; Zhang, Y.; Jia, J.; Zhang, Q.; Zhang, B.; Wang, R.; et al. Thymidine and 2′-Deoxyuridine Reduce Microglial Activation and Improve Oxidative Stress Damage by Modulating Glycolytic Metabolism on the Aβ25-35-Induced Brain Injury. Arch. Biochem. Biophys. 2022, 729, 109377. [Google Scholar] [CrossRef]
- Sun, Z.-J.; Li, Z.-X. The Terpenoid Backbone Biosynthesis Pathway Directly Affects the Biosynthesis of Alarm Pheromone in the Aphid. Insect Mol. Biol. 2018, 27, 824–834. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Chen, X.; Li, Y.; Guo, S.; Wang, Z.; Yu, X. Advances in Pharmacological Activities of Terpenoids. Nat. Prod. Commun. 2020, 15, 1934578X2090355. [Google Scholar] [CrossRef]
- Rymut, H.E.; Rund, L.A.; Southey, B.R.; Johnson, R.W.; Rodriguez-Zas, S.L. Terpenoid Backbone Biosynthesis among Pig Hippocampal Pathways Impacted by Stressors. Genes 2022, 13, 814. [Google Scholar] [CrossRef]
- Mambelli, L.I.; Teixeira, S.F.; Jorge, S.D.; Kawamura, B.; Meneguelo, R.; Barbuto, J.A.M.; De Azevedo, R.A.; Ferreira, A.K. Phosphoethanolamine Induces Caspase-Independent Cell Death by Reducing the Expression of C-RAF and Inhibits Tumor Growth in Human Melanoma Model. Biomed. Pharmacother. 2018, 103, 18–28. [Google Scholar] [CrossRef]
- Li, Y.; Hu, N.; Yang, D.; Oxenkrug, G.; Yang, Q. Regulating the Balance between the Kynurenine and Serotonin Pathways of Tryptophan Metabolism. FEBS J. 2017, 284, 948–966. [Google Scholar] [CrossRef]
- Campbell, B.M.; Charych, E.; Lee, A.W.; Möller, T. Kynurenines in CNS Disease: Regulation by Inflammatory Cytokines. Front. Neurosci. 2014, 8, 12. [Google Scholar] [CrossRef] [PubMed]
- Yue, H.; Zhao, Y.; Ma, X.; Gong, J. Ethylene Glycol: Properties, Synthesis, and Applications. Chem. Soc. Rev. 2012, 41, 4218–4244. [Google Scholar] [CrossRef]
- Farah, A.; Donangelo, C.M. Phenolic Compounds in Coffee. Braz. J. Plant Physiol. 2006, 18, 23–36. [Google Scholar] [CrossRef]
- Gutiérrez Ortiz, A.L.; Berti, F.; Solano Sánchez, W.; Navarini, L.; Colomban, S.; Crisafulli, P.; Forzato, C. Distribution of P-Coumaroylquinic Acids in Commercial Coffea Spp. of Different Geographical Origin and in Other Wild Coffee Species. Food Chem. 2019, 286, 459–466. [Google Scholar] [CrossRef] [PubMed]
- Chandel, N.S. Nucleotide Metabolism. Cold Spring Harb. Perspect. Biol. 2021, 13, a040592. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Cui, J.; Ma, H.; Lu, W.; Huang, J. Targeting Pyrimidine Metabolism in the Era of Precision Cancer Medicine. Front. Oncol. 2021, 11, 684961. [Google Scholar] [CrossRef]
- Zhang, H.L.; Chen, Y.; Xu, X.L.; Yang, Y.X. Effects of Branched-Chain Amino Acids on in Vitro Rumen Fermentation of Wheat Straw. Asian-Australas. J. Anim. Sci. 2013, 26, 523. [Google Scholar] [CrossRef]
- Xue, M.Y.; Sun, H.Z.; Wu, X.H.; Liu, J.X.; Guan, L.L. Multi-Omics Reveals That the Rumen Microbiome and Its Metabolome Together with the Host Metabolome Contribute to Individualized Dairy Cow Performance. Microbiome 2020, 8, 64. [Google Scholar] [CrossRef]
- Leandro, J.; Houten, S.M. The Lysine Degradation Pathway: Subcellular Compartmentalization and Enzyme Deficiencies. Mol. Genet. Metab. 2020, 131, 14–22. [Google Scholar] [CrossRef]
- Quinville, B.M.; Deschenes, N.M.; Ryckman, A.E.; Walia, J.S. A Comprehensive Review: Sphingolipid Metabolism and Implications of Disruption in Sphingolipid Homeostasis. Int. J. Mol. Sci. 2021, 22, 5793. [Google Scholar] [CrossRef] [PubMed]
- Arslan, E.; Koyuncu, I.; Temiz, E.; Ari, M.; Uyanikoglu, H. Metabolomic Analysis of Seminal Fluids in Infertile Individuals. Eur. Rev. Med. Pharmacol. Sci. 2023, 27, 11923–11931. [Google Scholar] [PubMed]
- Zheng, C.; Ma, J.; Xu, J.; Wu, L.; Wu, Y.; Liu, Y.; Shen, B. The Optimal Regimen, Efficacy and Safety of Tranexamic Acid and Aminocaproic Acid to Reduce Bleeding for Patients after Total Hip Arthroplasty: A Systematic Review and Bayesian Network Meta-Analysis. Thromb. Res. 2023, 221, 120–129. [Google Scholar] [CrossRef] [PubMed]
- Shim, Y.M.; Paige, M. Leukotriene A4 Hydrolase—An Evoloving Therapeutic Target. In Inflammatory Diseases-Immunopathology, Clinical and Pharmacological Bases; Khatami, M., Ed.; InTech: London, UK, 2012; pp. 253–278. [Google Scholar]
- Adams, J.M.; Rege, S.V.; Liu, A.T.; Vu, N.V.; Raina, S.; Kirsher, D.Y.; Nguyen, A.L.; Harish, R.; Szoke, B.; Leone, D.P.; et al. Leukotriene A4 Hydrolase Inhibition Improves Age-Related Cognitive Decline via Modulation of Synaptic Function. Sci. Adv. 2023, 9, eadf8764. [Google Scholar] [CrossRef]
- de Araújo, S.S.; Neves, C.M.L.; Guimarães, S.L.; Whitman, C.P.; Johnson, W.H.; Aparicio, R.; Nagem, R.A.P. Structural and Kinetic Characterization of Recombinant 2-Hydroxymuconate Semialdehyde Dehydrogenase from Pseudomonas Putida G7. Arch. Biochem. Biophys. 2015, 579, 8–17. [Google Scholar] [CrossRef] [PubMed]
- Sala-trepat, J.M.; Evans, W.C. The Meta Cleavage of Catechol by Azotobacter Species: 4-Oxalocrotonate Pathway. Eur. J. Biochem. 1971, 20, 400–413. [Google Scholar] [CrossRef]
- Bohdanowicz, M.; Grinstein, S. Role of Phospholipids in Endocytosis, Phagocytosis, and Macropinocytosis. Physiol. Rev. 2013, 93, 69–106. [Google Scholar] [CrossRef]
- van Meer, G.; de Kroon, A.I. Lipid Map of the Mammalian Cell. J. Cell Sci. 2011, 124, 5–8. [Google Scholar] [CrossRef]
- Kumar, A.; Shrinet, J.; Sunil, S. Chikungunya Virus Infection in Aedes Aegypti Is Modulated by L-Cysteine, Taurine, Hypotaurine and Glutathione Metabolism. PLoS Neglected Trop. Dis. 2023, 17, e0011280. [Google Scholar] [CrossRef]
- Penttilä, K.E. Role of Cysteine and Taurine in Regulating Glutathione Synthesis by Periportal and Perivenous Hepatocytes. Biochem. J. 1990, 269, 659–664. [Google Scholar] [CrossRef]

| Cycles | A | B | C | D | E | F |
|---|---|---|---|---|---|---|
| 1 | CON | EOB1 | EOB2 | EOB3 | EOB4 | EOB5 |
| 2 | EOB5 | CON | EOB1 | EOB2 | EOB3 | EOB4 |
| 3 | EOB4 | EOB5 | CON | EOB1 | EOB2 | EOB3 |
| 4 | EOB3 | EOB4 | EOB5 | CON | EOB1 | EOB2 |
| 5 | EOB2 | EOB3 | EOB4 | EOB5 | CON | EOB1 |
| 6 | EOB1 | EOB2 | EOB3 | EOB4 | EOB5 | CON |
| (As Submitted, %) | Dry Matter Basis (%, Unless Stated) | |
|---|---|---|
| DM | 86.49 | --- |
| Moisture | 13.51 | |
| CP | 9.18 | 10.61 |
| Fat | 1.19 | 1.38 |
| Ash | 6.66 | 7.71 |
| NDF | 57.75 | 66.77 |
| ADF | 34.86 | 40.30 |
| ADL | 22.61 | 19.55 |
| Unavailable protein | 1.44 | 1.66 |
| Adjusted crude protein | 8.66 | 10.01 |
| Total digestible nutrients | 55.64 | 64.32 |
| Nitrate ion | 0.09 | 0.10 |
| Non-fiber carbohydrates | 15.36 | 17.76 |
| Calcium | 0.26 | 0.30 |
| Phosphorus | 0.23 | 0.27 |
| Sulfur | 0.14 | 0.16 |
| Sodium | 0.05 | 0.06 |
| Magnesium | 0.13 | 0.15 |
| Potassium | 1.83 | 2.12 |
| Copper (ppm) | 13.00 | 15.01 |
| Manganese (ppm) | 42.00 | 48.15 |
| Iron (ppm) | 338.00 | 390.86 |
| Zinc (ppm) | 29.00 | 33.87 |
| Treatments | TVFA | Acetate | Propionate | Butyrate | Isobutyrate | Valerate | Isovalerate | A:PR |
|---|---|---|---|---|---|---|---|---|
| CONTROL | 95.4 | 0.711 | 0.178 | 0.099 | 0.0046 b | 0.0062 ab | 0.0014 b | 3.99 |
| EOB1 | 93.4 | 0.713 | 0.178 | 0.098 | 0.0039 b | 0.0058 bc | 0.0011 b | 4.02 |
| EOB2 | 88.1 | 0.709 | 0.180 | 0.096 | 0.0063 a | 0.0064 a | 0.0019 a | 3.94 |
| EOB3 | 93.3 | 0.710 | 0.179 | 0.099 | 0.0046 b | 0.0061 ab | 0.0013 b | 3.99 |
| EOB4 | 94.6 | 0.719 | 0.176 | 0.094 | 0.0040 b | 0.0055 c | 0.0011 b | 4.12 |
| EOB5 | 86.1 | 0.714 | 0.177 | 0.094 | 0.0065 a | 0.0062 ab | 0.0020 a | 4.05 |
| SEM | 3.460 | 0.0317 | 0.0020 | 0.0175 | 0.0001 | 0.0002 | 0.0001 | 0.0635 |
| p-value | 0.312 | 0.196 | 0.665 | 0.171 | 0.000 | 0.011 | 0.0001 | 0.473 |
| Treatments | CH4 | CO2 | NH3 | H2S | Rumen pH | NH3-N (mg/dL) |
|---|---|---|---|---|---|---|
| CONTROL | 11.6 | 22.3 | 54.8 | 234 | 6.48 | 12.3 |
| EOB1 | 11.6 | 19.9 | 45.2 | 101 | 6.60 | 14.1 |
| EOB2 | 11.4 | 12.0 | 70.6 | 299 | 6.62 | 14.7 |
| EOB3 | 9.29 | 18.3 | 57.3 | 180 | 6.55 | 16.1 |
| EOB4 | 10.8 | 21.3 | 62.1 | 222 | 6.57 | 14.6 |
| EOB5 | 8.31 | 16.9 | 50.3 | 179 | 6.59 | 12.4 |
| SEM | 1.613 | 3.318 | 10.169 | 65.278 | 0.0771 | 0.5773 |
| p-value | 0.610 | 0.771 | 0.570 | 0.404 | 0.0700 | 1.364 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Okedoyin, D.O.; Alabi, J.O.; Anotaenwere, C.C.; Wuaku, M.; Gray, D.; Adelusi, O.O.; Ike, K.A.; Dele, P.A.; Oderinwale, O.A.; Idowu, M.D.; et al. Metabolomic Profiling, Volatile Fatty Acids, and Greenhouse Gas Emissions of Beef Cattle Infused with Different Essential Oil Blends. Ruminants 2024, 4, 329-351. https://doi.org/10.3390/ruminants4030024
Okedoyin DO, Alabi JO, Anotaenwere CC, Wuaku M, Gray D, Adelusi OO, Ike KA, Dele PA, Oderinwale OA, Idowu MD, et al. Metabolomic Profiling, Volatile Fatty Acids, and Greenhouse Gas Emissions of Beef Cattle Infused with Different Essential Oil Blends. Ruminants. 2024; 4(3):329-351. https://doi.org/10.3390/ruminants4030024
Chicago/Turabian StyleOkedoyin, Deborah O., Joel O. Alabi, Chika C. Anotaenwere, Michael Wuaku, DeAndrea Gray, Oludotun O. Adelusi, Kelechi A. Ike, Peter A. Dele, Olatunde A. Oderinwale, Modoluwamu D. Idowu, and et al. 2024. "Metabolomic Profiling, Volatile Fatty Acids, and Greenhouse Gas Emissions of Beef Cattle Infused with Different Essential Oil Blends" Ruminants 4, no. 3: 329-351. https://doi.org/10.3390/ruminants4030024
APA StyleOkedoyin, D. O., Alabi, J. O., Anotaenwere, C. C., Wuaku, M., Gray, D., Adelusi, O. O., Ike, K. A., Dele, P. A., Oderinwale, O. A., Idowu, M. D., Ogunade, I. M., & Anele, U. Y. (2024). Metabolomic Profiling, Volatile Fatty Acids, and Greenhouse Gas Emissions of Beef Cattle Infused with Different Essential Oil Blends. Ruminants, 4(3), 329-351. https://doi.org/10.3390/ruminants4030024








