Comparison of the Proteome of Staphylococcus aureus Planktonic Culture and 3-Day Biofilm Reveals Potential Role of Key Proteins in Biofilm
Abstract
:1. Introduction
2. Materials and Methods
2.1. Microorganism and Culture Conditions
2.2. Protein Extraction and Fractionation
2.3. Protein Reduction, Alkylation, and Digestion
2.4. TMT Labeling and High pH Fractionation
2.5. Nanoflow LC-ESI-MS/MS
2.5.1. Nanoflow LC-ESI-MS/MS Using Orbitrap Elite
2.5.2. Nanoflow LC-ESI-MS/MS Using Q Exactive
2.6. Database Search, Statistical Analysis, and Bioinformatics
2.7. Validation of TMT Data with qPCR Results
3. Results
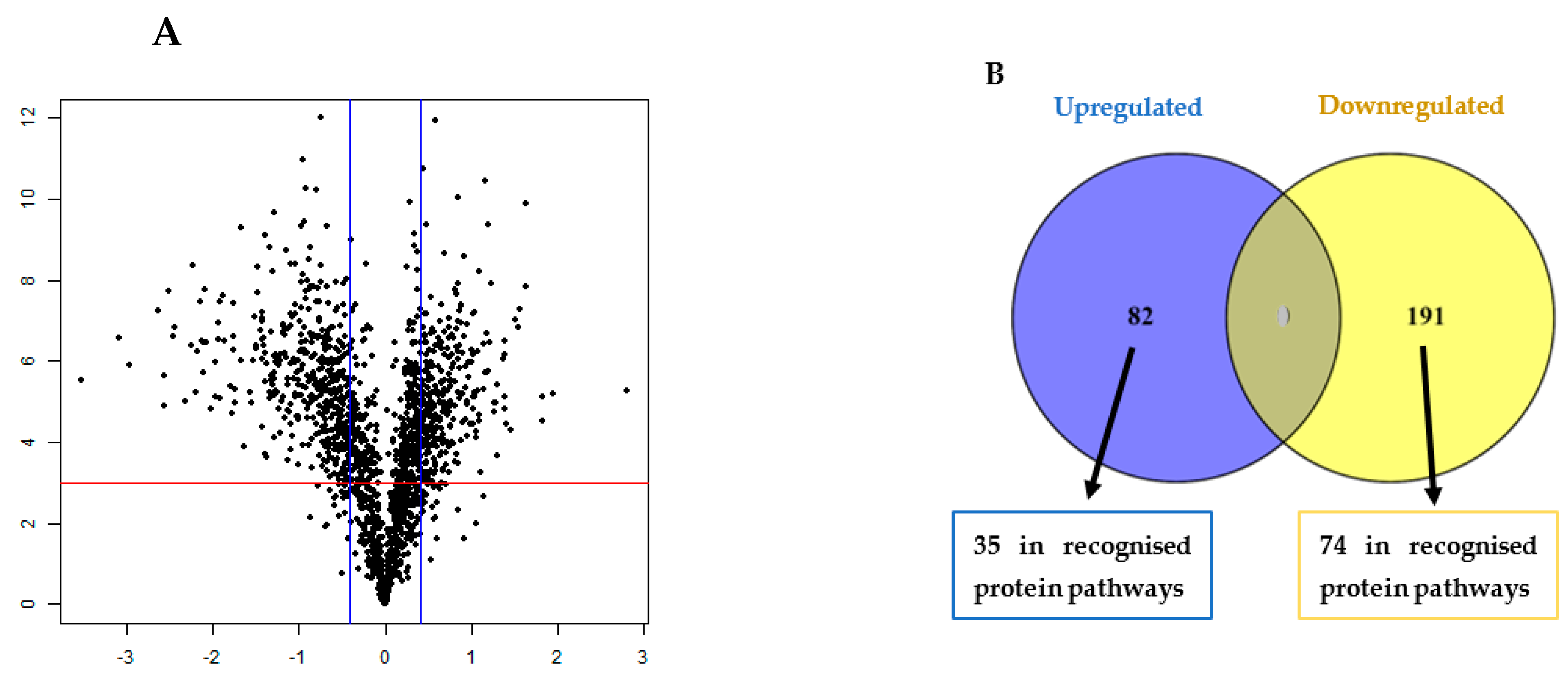
3.1. TMT Identification of Differentially Regulated Proteins in the Biofilm
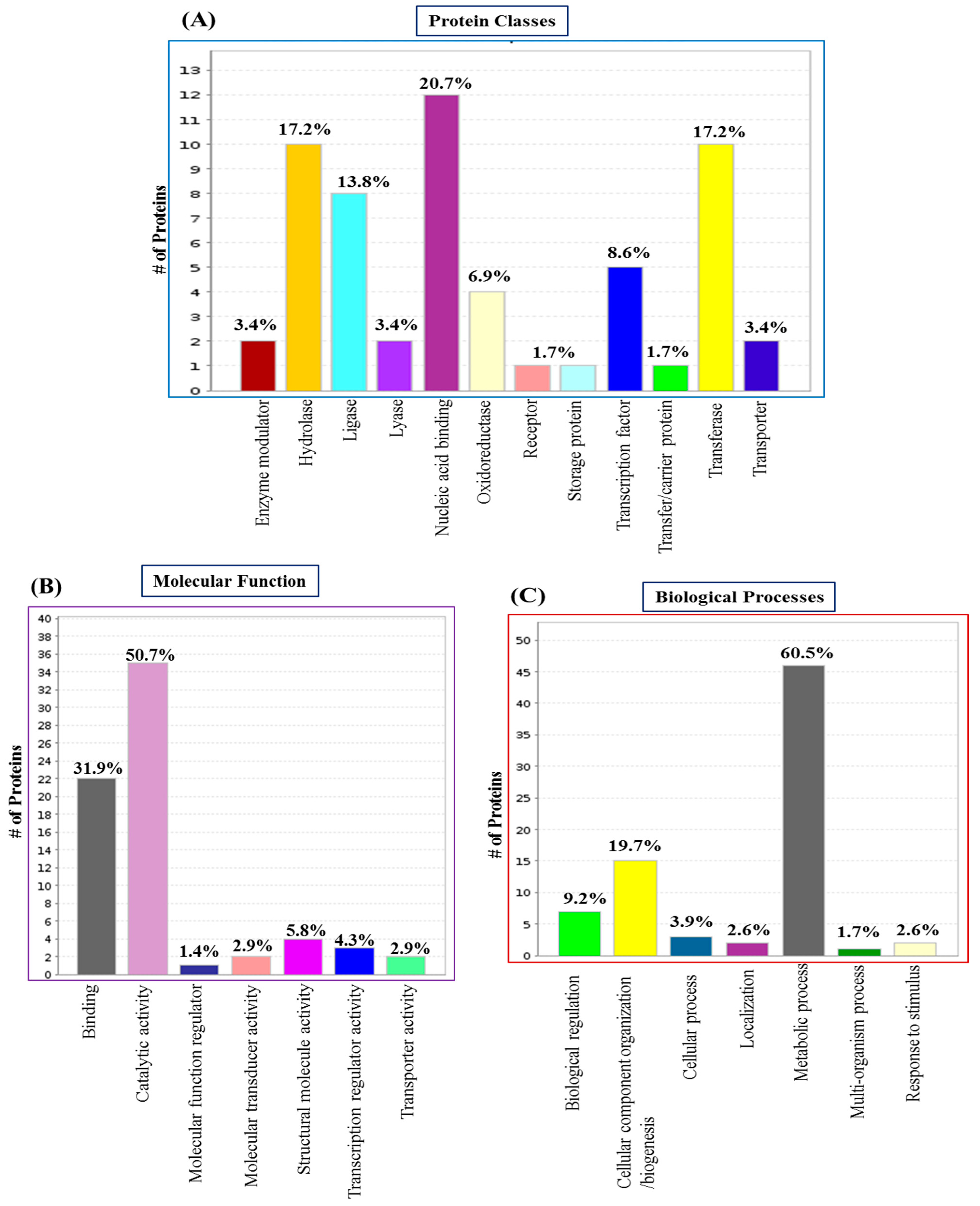
3.2. GO Analysis and Annotation of Differentially Regulated Proteins in the Biofilm
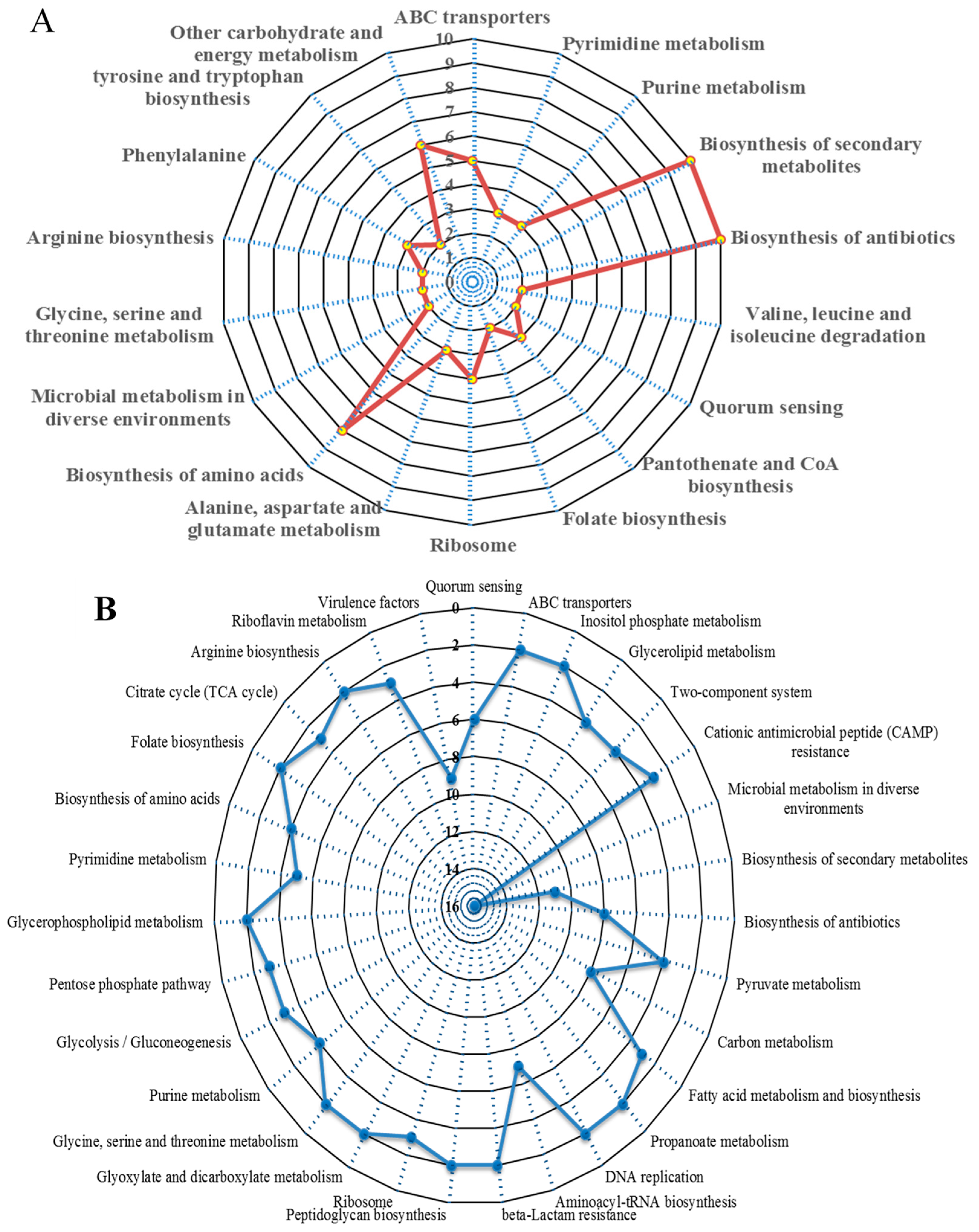
3.3. Significantly Dysregulated Proteins and Pathway Analysis in the Biofilm
3.4. Protein–Protein Interaction (PPI) Analysis in the Biofilm
3.5. Validation of TMT Data with Real-Time qPCR
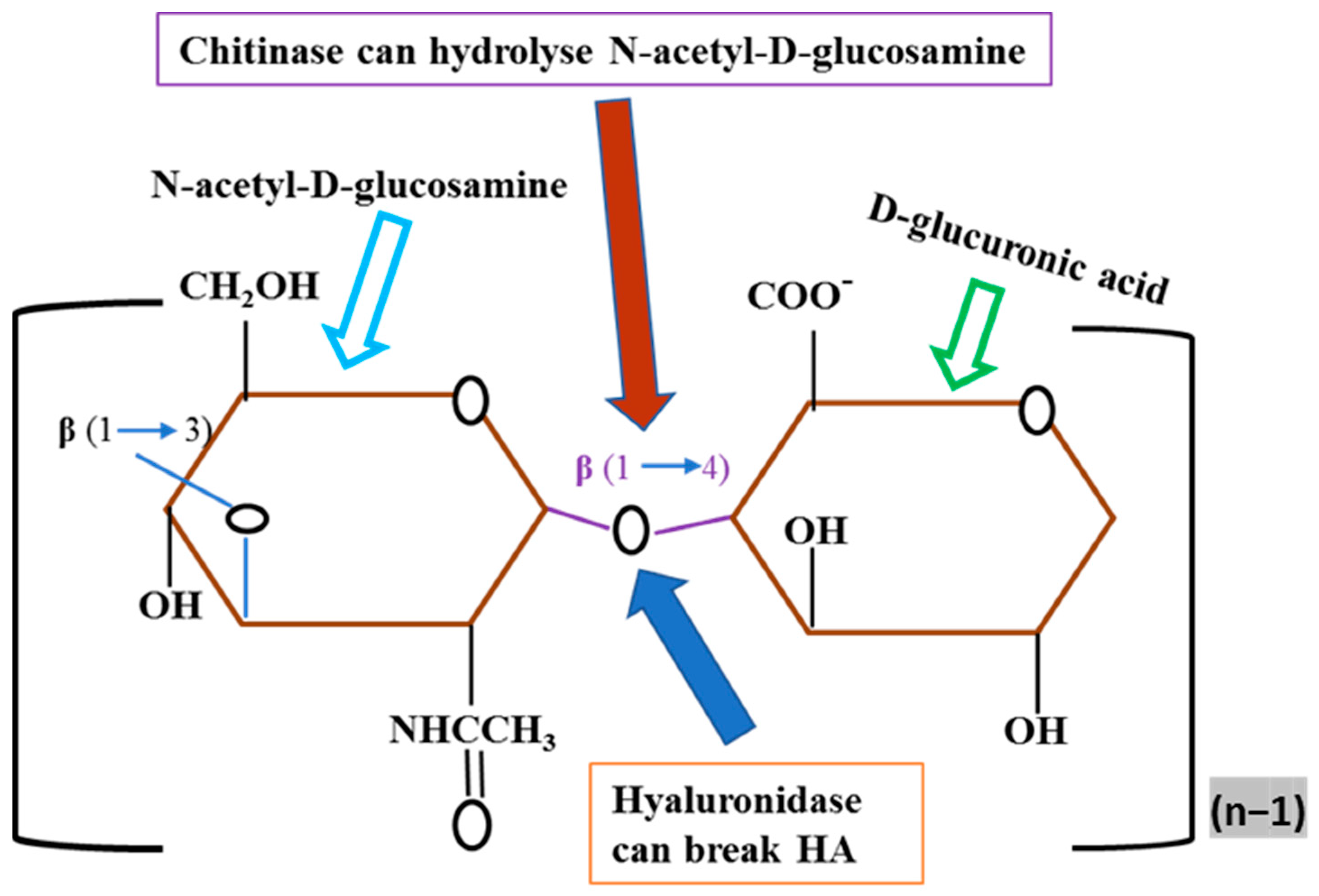
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tam, K.; Torres, V.J. Staphylococcus aureus secreted toxins and extracellular enzymes. Microbiol. Spectr. 2019, 7, 2. [Google Scholar] [CrossRef] [PubMed]
- Lister, J.L.; Horswill, A.R. Staphylococcus aureus biofilms: Recent developments in biofilm dispersal. Front. Cell. Infect. Microbiol. 2014, 4, 178. [Google Scholar] [CrossRef]
- Francis, D.; Bhairaddy, A.; Joy, A. The biofilm proteome of Staphylococcus aureus and its implications for therapeutic interventions to biofilm-associated infections. Adv. Protein Chem. Struct. Biol. 2023, 138, 327–400. [Google Scholar] [PubMed]
- Ajdic, D.; Zoghbi, Y.; Gerth, D.; Panthaki, Z.J.; Thaller, S. The relationship of bacterial biofilms and capsular contracture in breast implants. Aesthetic Surg. J. 2016, 36, 297–309. [Google Scholar] [CrossRef] [PubMed]
- Barrett, L.; Atkins, B. The clinical presentation of prosthetic joint infection. J. Antimicrob. Chemother. 2014, 69, i25–i27. [Google Scholar] [CrossRef]
- Chatterjee, S.; Maiti, P.; Dey, R.; Kundu, A.; Dey, R. Biofilms on indwelling urologic devices: Microbes and antimicrobial management prospect. Ann. Med. Health Sci. Res. 2014, 4, 100–104. [Google Scholar] [PubMed]
- Karygianni, L.; Ren, Z.; Koo, H.; Thurnheer, T. Biofilm matrixome: Extracellular components in structured microbial communities. Trends Microbiol. 2020, 28, 668–681. [Google Scholar] [CrossRef] [PubMed]
- Vickery, K. Microbial biofilms in healthcare: Formation, prevention and treatment. Materials 2019, 12, 2001. [Google Scholar] [CrossRef]
- Graf, A.C.; Leonard, A.; Schäuble, M.; Rieckmann, L.M.; Hoyer, J.; Maaß, S.; Lalk, M.; Becher, D.; Pané-Farré, J.; Riedel, K. Virulence factors produced by Staphylococcus aureus biofilms have a moonlighting function contributing to biofilm integrity. Mol. Cell. Proteom. 2019, 18, 1036–1053. [Google Scholar] [CrossRef]
- Moche, M.; Schlüter, R.; Bernhardt, J.; Plate, K.; Riedel, K.; Hecker, M.; Becher, D. Time-resolved analysis of cytosolic and surface-associated proteins of Staphylococcus aureus HG001 under planktonic and biofilm conditions. J. Proteome Res. 2015, 14, 3804–3822. [Google Scholar] [CrossRef]
- Atshan, S.S.; Shamsudin, M.N.; Sekawi, Z.; Lung, L.T.T.; Barantalab, F.; Liew, Y.K.; Alreshidi, M.A.; Abduljaleel, S.A.; Hamat, R.A. Comparative proteomic analysis of extracellular proteins expressed by various clonal types of Staphylococcus aureus and during planktonic growth and biofilm development. Front. Microbiol. 2015, 6, 524. [Google Scholar] [CrossRef] [PubMed]
- Islam, N.; Kim, Y.; Ross, J.M.; Marten, M.R. Proteomic analysis of Staphylococcus aureus biofilm cells grown under physiologically relevant fluid shear stress conditions. Proteome Sci. 2014, 12, 21. [Google Scholar] [CrossRef] [PubMed]
- Ammons, M.C.B.; Tripet, B.P.; Carlson, R.P.; Kirker, K.R.; Gross, M.A.; Stanisich, J.J.; Copié, V.R. Quantitative NMR metabolite profiling of methicillin-resistant and methicillin-susceptible Staphylococcus aureus discriminates between biofilm and planktonic phenotypes. J. Proteome Res. 2014, 13, 2973–2985. [Google Scholar] [CrossRef] [PubMed]
- Resch, A.; Leicht, S.; Saric, M.; Pásztor, L.; Jakob, A.; Götz, F.; Nordheim, A. Comparative proteome analysis of Staphylococcus aureus biofilm and planktonic cells and correlation with transcriptome profiling. Proteomics 2006, 6, 1867–1877. [Google Scholar] [CrossRef]
- Kranjec, C.; Morales Angeles, D.; Torrissen Mårli, M.; Fernández, L.; García, P.; Kjos, M.; Diep, D.B. Staphylococcal biofilms: Challenges and novel therapeutic perspectives. Antibiotics 2021, 10, 131. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.A.; Amirkhani, A.; Parvin, F.; Chowdhury, D.; Molloy, M.P.; Deva, A.K.; Vickery, K.; Hu, H. One step forward with dry surface biofilm (DSB) of Staphylococcus aureus: TMT-based quantitative proteomic analysis reveals proteomic shifts between DSB and hydrated biofilm. Int. J. Mol. Sci. 2022, 23, 12238. [Google Scholar] [CrossRef]
- Rahman, M.A.; Amirkhani, A.; Chowdhury, D.; Mempin, M.; Molloy, M.P.; Deva, A.K.; Vickery, K.; Hu, H. Proteome of Staphylococcus aureus Biofilm Changes Significantly with Aging. Int. J. Mol. Sci. 2022, 23, 6415. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Cheng, G.; Karunakaran, R.; East, A.K.; Poole, P.S. Multiplicity of sulfate and molybdate transporters and their role in nitrogen fixation in Rhizobium leguminosarum bv. viciae Rlv3841. Mol. Plant-Microbe Interact. 2016, 29, 143–152. [Google Scholar] [CrossRef]
- Vandenesch, F.; Lina, G.; Henry, T. Staphylococcus aureus hemolysins, bi-component leukocidins, and cytolytic peptides: A redundant arsenal of membrane-damaging virulence factors? Front. Cell. Infect. Microbiol. 2012, 2, 12. [Google Scholar] [CrossRef]
- Shah, P.; Swiatlo, E. A multifaceted role for polyamines in bacterial pathogens. Mol. Microbiol. 2008, 68, 4–16. [Google Scholar] [CrossRef] [PubMed]
- Antognoni, F.; Del Duca, S.; Kuraishi, A.; Kawabe, E.; Fukuchi-Shimogori, T.; Kashiwagi, K.; Igarashi, K. Transcriptional inhibition of the operon for the spermidine uptake system by the substrate-binding protein PotD. J. Biol. Chem. 1999, 274, 1942–1948. [Google Scholar] [CrossRef]
- Atshan, S.S.; Shamsudin, M.N.; Lung, T.; Than, L.; Sekawi, Z.; Ghaznavi-Rad, E.; Pei Pei, C. Comparative characterisation of genotypically different clones of MRSA in the production of biofilms. BioMed Res. Int. 2012, 2012, 417247. [Google Scholar] [CrossRef]
- da Silva, W.M.; Bei, J.; Amigo, N.; Valacco, P.; Amadio, A.F.; Zhang, Q.; Wu, X.; Larzábal, M.; Chen, Z.; Cataldi, A. Quantification of Enterohemorrhagic Escherichia coli O157: H7 proteome using TMT-Based Analysis. bioRxiv 2018, 312652. [Google Scholar] [CrossRef]
- Piras, C.; Di Ciccio, P.A.; Soggiu, A.; Greco, V.; Tilocca, B.; Costanzo, N.; Ceniti, C.; Urbani, A.; Bonizzi, L.; Ianieri, A.S. aureus biofilm protein expression linked to antimicrobial resistance: A proteomic study. Animals 2021, 11, 966. [Google Scholar] [CrossRef]
- Parvin, F.; Rahman, M.A.; Deva, A.K.; Vickery, K.; Hu, H. Staphylococcus aureus cell wall phenotypic changes associated with biofilm maturation and water availability: A key contributing factor for chlorine resistance. Int. J. Mol. Sci. 2023, 24, 4983. [Google Scholar] [CrossRef]
- Theodoulou, F.L.; Kerr, I.D. ABC transporter research: Going strong 40 years on. Biochem. Soc. Trans. 2015, 43, 1033–1040. [Google Scholar] [CrossRef]
- Brady, R.A.; Leid, J.G.; Camper, A.K.; Costerton, J.W.; Shirtliff, M.E. Identification of Staphylococcus aureus proteins recognized by the antibody-mediated immune response to a biofilm infection. Infect. Immun. 2006, 74, 3415–3426. [Google Scholar] [CrossRef] [PubMed]
- Islam, N.; Ross, J.M.; Marten, M.R. Proteome analyses of Staphylococcus aureus biofilm at elevated levels of NaCl. Clin. Microbiol. 2015, 4, 219. [Google Scholar]
- Yang, J.; He, Y.; Jiang, J.; Chen, W.; Gao, Q.; Pan, L.; Shi, C. Comparative proteomic analysis by iTRAQ-2DLC-MS/MS provides insight into the key proteins involved in Cronobacter sp. biofilm formation. Food Control 2016, 63, 93–100. [Google Scholar] [CrossRef]
- Branda, S.S.; González-Pastor, J.E.; Dervyn, E.; Ehrlich, S.D.; Losick, R.; Kolter, R. Genes involved in formation of structured multicellular communities by Bacillus subtilis. J. Bacteriol. 2004, 186, 3970–3979. [Google Scholar] [CrossRef] [PubMed]
- Crowley, R.; Leigh, J.; Ward, P.; Lappin-Scott, H.; Bowler, L. Differential protein expression in Streptococcus uberis under planktonic and biofilm growth conditions. Appl. Environ. Microbiol. 2011, 77, 382–384. [Google Scholar] [CrossRef] [PubMed]
- Hinsa, S.M.; Espinosa-Urgel, M.; Ramos, J.L.; O’toole, G.A. Transition from reversible to irreversible attachment during biofilm formation by Pseudomonas fluorescens WCS365 requires an ABC transporter and a large secreted protein. Mol. Microbiol. 2003, 49, 905–918. [Google Scholar] [CrossRef] [PubMed]
- Vanderlinde, E.M.; Harrison, J.J.; Muszyński, A.; Carlson, R.W.; Turner, R.J.; Yost, C.K. Identification of a novel ABC transporter required for desiccation tolerance, and biofilm formation in Rhizobium leguminosarum bv. viciae 3841. FEMS Microbiol. Ecol. 2010, 71, 327–340. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Long, F.; Chen, Y.; Knøchel, S.; She, Q.; Shi, X. A putative ABC transporter is involved in negative regulation of biofilm formation by Listeria monocytogenes. Appl. Environ. Microbiol. 2008, 74, 7675–7683. [Google Scholar] [CrossRef] [PubMed]
- Beenken, K.E.; Dunman, P.M.; McAleese, F.; Macapagal, D.; Murphy, E.; Projan, S.J.; Blevins, J.S.; Smeltzer, M.S. Global gene expression in Staphylococcus aureus biofilms. J. Bacteriol. 2004, 186, 4665–4684. [Google Scholar] [CrossRef]
- Vytvytska, O.; Nagy, E.; Blüggel, M.; Meyer, H.E.; Kurzbauer, R.; Huber, L.A.; Klade, C.S. Identification of vaccine candidate antigens of Staphylococcus aureus by serological proteome analysis. Proteomics 2002, 2, 580–590. [Google Scholar] [CrossRef]
- Speziale, P.; Pietrocola, G. The multivalent role of fibronectin-binding proteins A and B (FnBPA and FnBPB) of Staphylococcus aureus in host infections. Front. Microbiol. 2020, 11, 572022. [Google Scholar] [CrossRef]
- Hofbauer, B.; Vomacka, J.; Stahl, M.; Korotkov, V.S.; Jennings, M.C.; Wuest, W.M.; Sieber, S.A. Dual Inhibitor of Staphylococcus aureus Virulence and Biofilm Attenuates Expression of Major Toxins and Adhesins. Biochemistry 2018, 57, 1814–1820. [Google Scholar] [CrossRef]
- Kot, B.; Sytykiewicz, H.; Sprawka, I. Expression of the Biofilm-Associated Genes in Methicillin-Resistant Staphylococcus aureus in Biofilm and Planktonic Conditions. Int. J. Mol. Sci. 2018, 19, 3487. [Google Scholar] [CrossRef]
- Paharik, A.E.; Horswill, A.R. The Staphylococcal Biofilm: Adhesins, Regulation, and Host Response. Microbiol. Spectr. 2016, 4, 2. [Google Scholar] [CrossRef] [PubMed]
- Ibberson, C.B.; Parlet, C.P.; Kwiecinski, J.; Crosby, H.A.; Meyerholz, D.K.; Horswill, A.R. Hyaluronan modulation impacts Staphylococcus aureus biofilm infection. Infect. Immun. 2016, 84, 1917–1929. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.A.; Choi, Y.H.; Pradeep, G.; Yoo, J.C. An ammonium sulfate sensitive chitinase from Streptomyces sp. CS501. Arch. Pharmacal Res. 2014, 37, 1522–1529. [Google Scholar] [CrossRef] [PubMed]
- Boles, B.R.; Horswill, A.R. Agr-mediated dispersal of Staphylococcus aureus biofilms. PLoS Pathog. 2008, 4, e1000052. [Google Scholar] [CrossRef]
- Periasamy, S.; Joo, H.-S.; Duong, A.C.; Bach, T.-H.L.; Tan, V.Y.; Chatterjee, S.S.; Cheung, G.Y.; Otto, M. How Staphylococcus aureus biofilms develop their characteristic structure. Proc. Natl. Acad. Sci. USA 2012, 109, 1281–1286. [Google Scholar] [CrossRef] [PubMed]
- Resch, A.; Rosenstein, R.; Nerz, C.; Götz, F. Differential gene expression profiling of Staphylococcus aureus cultivated under biofilm and planktonic conditions. Appl. Environ. Microbiol. 2005, 71, 2663–2676. [Google Scholar] [CrossRef]
- Kantyka, T.; Plaza, K.; Koziel, J.; Florczyk, D.; Stennicke, H.R.; Thogersen, I.B.; Enghild, J.J.; Silverman, G.A.; Pak, S.C.; Potempa, J. Inhibition of Staphylococcus aureus cysteine proteases by human serpin potentially limits staphylococcal virulence. Biol. Chem. 2011, 392, 483–489. [Google Scholar] [CrossRef]
- Martínez-García, S.; Peralta, H.; Betanzos-Cabrera, G.; Chavez-Galan, L.; Rodríguez-Martínez, S.; Cancino-Diaz, M.E.; Cancino-Diaz, J.C. Proteomic comparison of biofilm vs. planktonic Staphylococcus epidermidis cells suggests key metabolic differences between these conditions. Res. Microbiol. 2021, 172, 103796. [Google Scholar] [CrossRef]
- Chen, X.; Wu, H.; Cao, Y.; Yao, X.; Zhao, L.; Wang, T.; Yang, Y.; Lv, D.; Chai, Y.; Cao, Y. Ion-pairing chromatography on a porous graphitic carbon column coupled with time-of-flight mass spectrometry for targeted and untargeted profiling of amino acid biomarkers involved in Candida albicans biofilm formation. Mol. BioSystems 2014, 10, 74–85. [Google Scholar] [CrossRef]
- Wall, E.A.; Caufield, J.H.; Lyons, C.E.; Manning, K.A.; Dokland, T.; Christie, G.E. Specific N-terminal cleavage of ribosomal protein L 27 in Staphylococcus aureus and related bacteria. Mol. Microbiol. 2015, 95, 258–269. [Google Scholar] [CrossRef]
- Weiss, A.; Moore, B.D.; Tremblay, M.H.; Chaput, D.; Kremer, A.; Shaw, L.N. The ω subunit governs RNA polymerase stability and transcriptional specificity in Staphylococcus aureus. J. Bacteriol. 2017, 199, e00459-16. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Li, H.; Qi, T.; Yan, X.; Wang, B.; Guan, J.; Li, Y. Comparative transcriptomics analyses of the different growth states of multidrug-resistant Acinetobacter baumannii. Biomed. Pharmacother. 2017, 85, 564–574. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.D.; Franklin, M.J.; Park, C.-H.; McFeters, G.A.; Stewart, P.S. Gene expression and protein levels of the stationary phase sigma factor, RpoS, in continuously-fed Pseudomonas aeruginosa biofilms. FEMS Microbiol. Lett. 2001, 199, 67–71. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Yao, Z.; Sun, L.; Hu, W.; Cao, J.; Lin, W.; Lin, X. Proteomics analysis reveals a potential antibiotic cocktail therapy strategy for Aeromonas hydrophila infection in biofilm. J. Proteome Res. 2016, 15, 1810–1820. [Google Scholar] [CrossRef] [PubMed]
- Weber, H.; Engelmann, S.; Becher, D.; Hecker, M. Oxidative stress triggers thiol oxidation in the glyceraldehyde-3-phosphate dehydrogenase of Staphylococcus aureus. Mol. Microbiol. 2004, 52, 133–140. [Google Scholar] [CrossRef]
- Oliveira, L.; Madureira, P.; Andrade, E.B.; Bouaboud, A.; Morello, E.; Ferreira, P.; Poyart, C.; Trieu-Cuot, P.; Dramsi, S. Group B streptococcus GAPDH is released upon cell lysis, associates with bacterial surface, and induces apoptosis in murine macrophages. PLoS ONE 2012, 7, e29963. [Google Scholar] [CrossRef] [PubMed]
- Planchon, S.; Desvaux, M.; Chafsey, I.; Chambon, C.; Leroy, S.; Hébraud, M.; Talon, R. Comparative subproteome analyses of planktonic and sessile Staphylococcus xylosus C2a: New insight in cell physiology of a coagulase-negative Staphylococcus in biofilm. J. Proteome Res. 2009, 8, 1797–1809. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yi, L.; Wu, Z.; Shao, J.; Liu, G.; Fan, H.; Zhang, W.; Lu, C. Comparative proteomic analysis of Streptococcus suis biofilms and planktonic cells that identified biofilm infection-related immunogenic proteins. PLoS ONE 2012, 7, e33371. [Google Scholar] [CrossRef] [PubMed]
- Foulston, L.; Elsholz, A.K.; DeFrancesco, A.S.; Losick, R. The extracellular matrix of Staphylococcus aureus biofilms comprises cytoplasmic proteins that associate with the cell surface in response to decreasing pH. MBio 2014, 5, e01667-14. [Google Scholar] [CrossRef]
- Gil, C.; Solano, C.; Burgui, S.; Latasa, C.; García, B.; Toledo-Arana, A.; Lasa, I.; Valle, J. Biofilm Matrix Exoproteins Induce a Protective Immune Response against Staphylococcus aureus Biofilm Infection. Infect. Immun. 2014, 82, 1017–1029. [Google Scholar] [CrossRef]
- Zhang, J.; Meng, L.; Zhang, Y.; Sang, L.; Liu, Q.; Zhao, L.; Liu, F.; Wang, G. GapB is involved in biofilm formation dependent on LrgAB but not the SinI/R system in Bacillus cereus 0-9. Front. Microbiol. 2020, 11, 591926. [Google Scholar] [CrossRef] [PubMed]
- Stewart, P.S.; Zhang, T.; Xu, R.; Pitts, B.; Walters, M.C.; Roe, F.; Kikhney, J.; Moter, A. Reaction–diffusion theory explains hypoxia and heterogeneous growth within microbial biofilms associated with chronic infections. Npj Biofilms Microbiomes 2016, 2, 16012. [Google Scholar] [CrossRef] [PubMed]
- Walters, M.C.; Roe, F.; Bugnicourt, A.; Franklin, M.J.; Stewart, P.S. Contributions of antibiotic penetration, oxygen limitation, and low metabolic activity to tolerance of Pseudomonas aeruginosa biofilms to ciprofloxacin and tobramycin. Antimicrob. Agents Chemother. 2003, 47, 317–323. [Google Scholar] [CrossRef]
- Borriello, G.; Werner, E.; Roe, F.; Kim, A.M.; Ehrlich, G.D.; Stewart, P.S. Oxygen limitation contributes to antibiotic tolerance of Pseudomonas aeruginosa in biofilms. Antimicrob. Agents Chemother. 2004, 48, 2659–2664. [Google Scholar] [CrossRef]
- Zheng, Z.; Stewart, P.S. Growth limitation of Staphylococcus epidermidis in biofilms contributes to rifampin tolerance. Biofilms 2004, 1, 31–35. [Google Scholar] [CrossRef]
- Eng, R.; Padberg, F.; Smith, S.; Tan, E.; Cherubin, C. Bactericidal effects of antibiotics on slowly growing and nongrowing bacteria. Antimicrob. Agents Chemother. 1991, 35, 1824–1828. [Google Scholar] [CrossRef] [PubMed]
- Evans, D.; Allison, D.; Brown, M.; Gilbert, P. Susceptibility of Pseudomonas aeruginosa and Escherichia coli biofilms towards ciprofloxacin: Effect of specific growth rate. J. Antimicrob. Chemother. 1991, 27, 177–184. [Google Scholar] [CrossRef] [PubMed]
- Eschbach, M.; Schreiber, K.; Trunk, K.; Buer, J.; Jahn, D.; Schobert, M. Long-term anaerobic survival of the opportunistic pathogen Pseudomonas aeruginosa via pyruvate fermentation. J. Bacteriol. 2004, 186, 4596–4604. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Weiss, E.C.; Otto, M.; Fey, P.D.; Smeltzer, M.S.; Somerville, G.A. Staphylococcus aureus biofilm metabolism and the influence of arginine on polysaccharide intercellular adhesin synthesis, biofilm formation, and pathogenesis. Infect. Immun. 2007, 75, 4219–4226. [Google Scholar] [CrossRef]
- Sambanthamoorthy, K.; Schwartz, A.; Nagarajan, V.; Elasri, M.O. The role of msa in Staphylococcus aureus biofilm formation. BMC Microbiol. 2008, 8, 221. [Google Scholar] [CrossRef]
- Klein, M.I.; Xiao, J.; Lu, B.; Delahunty, C.M.; Yates, J.R., III; Koo, H. Streptococcus mutans Protein Synthesis during Mixed-Species Biofilm Development by High-Throughput Quantitative Proteomics. PLoS ONE 2012, 7, e45795. [Google Scholar] [CrossRef] [PubMed]
- Brady, R.A.; May, G.A.; Leid, J.G.; Prior, M.L.; Costerton, J.W.; Shirtliff, M.E. Resolution of Staphylococcus aureus Biofilm Infection Using Vaccination and Antibiotic Treatment. Infect. Immun. 2011, 79, 1797. [Google Scholar] [CrossRef] [PubMed]
- Chew, J.; Zilm, P.S.; Fuss, J.M.; Gully, N.J. A proteomic investigation of Fusobacterium nucleatum alkaline-induced biofilms. BMC Microbiol. 2012, 12, 189. [Google Scholar] [CrossRef] [PubMed]
- Perez-Riverol, Y.; Bai, J.; Bandla, C.; García-Seisdedos, D.; Hewapathirana, S.; Kamatchinathan, S.; Kundu, D.J.; Prakash, A.; Frericks-Zipper, A.; Eisenacher, M. The PRIDE database resources in 2022: A hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 2022, 50, D543–D552. [Google Scholar] [CrossRef] [PubMed]

| Function | Accession ID | Uniprot ID | Virulence Factors | Related Genes | Fold Change | Protein Pathway | Subcellular Localization |
|---|---|---|---|---|---|---|---|
| Adherence | AIO22275.1 | Q7A382 | Clumping factor B | clfB | −2.96 | S aureus infection | Cell wall |
| AIO22136.1 | Q7A3J7 | Fibronectin-binding protein A | fnbA SA2291 | −2.60 | Bacterial invasion of epithelial cells | Cell wall | |
| AIO19779.1 | A0A0H2X057 | Immunoglobulin G binding protein A | spa SACOL0095 | −4.71 | S aureus infection | Cell wall | |
| AIO20229.1 | Q5HIB2 | Serine-aspartate repeat-containing protein E | sdrE SACOL0610 | −3.82 | S aureus infection | Cell wall | |
| AIO20228.1 | Q7A780 | Serine-aspartate repeat-containing protein D | sdrD SA0520 | −4.49 | S aureus infection | Cell wall | |
| Toxins | AIO20763.1 | A0A0H3JMC2 | Alpha-hemolysin | SA1007 | −13.83 | Extracellular | |
| AIO22369.1 | Q5HEI1 | Phospholipase C (EC 3.1.4.3) (beta-hemolysin) (beta-toxin) (sphingomyelinase) (SMase) | hlb SACOL2003 | −12.33 | Quorum sensing, inositol phosphate metabolism, glycerophospholipid metabolism, biosynthesis of secondary metabolites | Extracellular | |
| AIO21667.1 | P0A0M2 | Delta-hemolysin (delta-lysin) (delta-toxin) | hld SA1841.1 SAS065 | −34.02 | Quorum sensing | Extracellular | |
| AIO22060.1 | Q7A3S2 | Gamma-hemolysin component C | hlgC SA2208 | −3.79 | Staphylococcus aureus infection | Extracellular | |
| AIO20093.1 | A0A0H3JSX3 | Exotoxin 11 (superantigen-like protein) | set11 | −3.63 | Staphylococcus aureus infection | Extracellular | |
| Antiphagocytosis (capsule) | AIO19823.1 | A0A0H3JKC9 | Capsular polysaccharide synthesis enzyme Cap5G | capG | 2.018 | Amino sugar and nucleotide sugar metabolism | Cytoplasmic |
| Exo-enzyme | AIO21508.1 | Q5HEW4 | Serine protease SplE | splE | −4.18 | Quorum sensing | Extracellular |
| AIO21601.1 | P65826 | Cysteine proteinase A | scpA | −3.73 | Extracellular | ||
| AIO20644.1 | Q5HH36 | Cysteine proteinase B | sspB | −11.44 | Extracellular | ||
| AIO19987.1 | Q7A7P2 | lipase | geh | −4.12 | Extracellular | ||
| AIO21839.1 | A0A0H3JN21 | Hyaluronate lyase | hysA | 2.50 | Extracellular | ||
| AIO20645.1 | Q5HH35 | Glutamyl endopeptidase | sspA | −6.52 | Quorum sensing | Extracellular | |
| OOC94232.1 | A0A0H2WZZ4 | Aureolysin | aur | −3.22 | Staphylococcus aureus infection, cationic antimicrobial peptide (CAMP) resistance | Extracellular | |
| AIO19888.1 | A0A0H3JNG8 | Staphylocoagulase | coa | 2.10 | Extracellular | ||
| Secretion system (type VII secretion system) | AIO19949.1 | Q7A7S3 | Type VII secretion protein EsaA | esaA SA0272 | −2.29 | CM * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rahman, M.A.; Amirkhani, A.; Chowdhury, D.; Vickery, K.; Hu, H. Comparison of the Proteome of Staphylococcus aureus Planktonic Culture and 3-Day Biofilm Reveals Potential Role of Key Proteins in Biofilm. Hygiene 2024, 4, 238-257. https://doi.org/10.3390/hygiene4030020
Rahman MA, Amirkhani A, Chowdhury D, Vickery K, Hu H. Comparison of the Proteome of Staphylococcus aureus Planktonic Culture and 3-Day Biofilm Reveals Potential Role of Key Proteins in Biofilm. Hygiene. 2024; 4(3):238-257. https://doi.org/10.3390/hygiene4030020
Chicago/Turabian StyleRahman, Md. Arifur, Ardeshir Amirkhani, Durdana Chowdhury, Karen Vickery, and Honghua Hu. 2024. "Comparison of the Proteome of Staphylococcus aureus Planktonic Culture and 3-Day Biofilm Reveals Potential Role of Key Proteins in Biofilm" Hygiene 4, no. 3: 238-257. https://doi.org/10.3390/hygiene4030020
APA StyleRahman, M. A., Amirkhani, A., Chowdhury, D., Vickery, K., & Hu, H. (2024). Comparison of the Proteome of Staphylococcus aureus Planktonic Culture and 3-Day Biofilm Reveals Potential Role of Key Proteins in Biofilm. Hygiene, 4(3), 238-257. https://doi.org/10.3390/hygiene4030020








