Single-Cell Transcriptome Profiling of Scale Drop Disease Virus-Infected Asian Seabass (Lates calcarifer)
Abstract
:1. Introduction
2. Materials and Methods
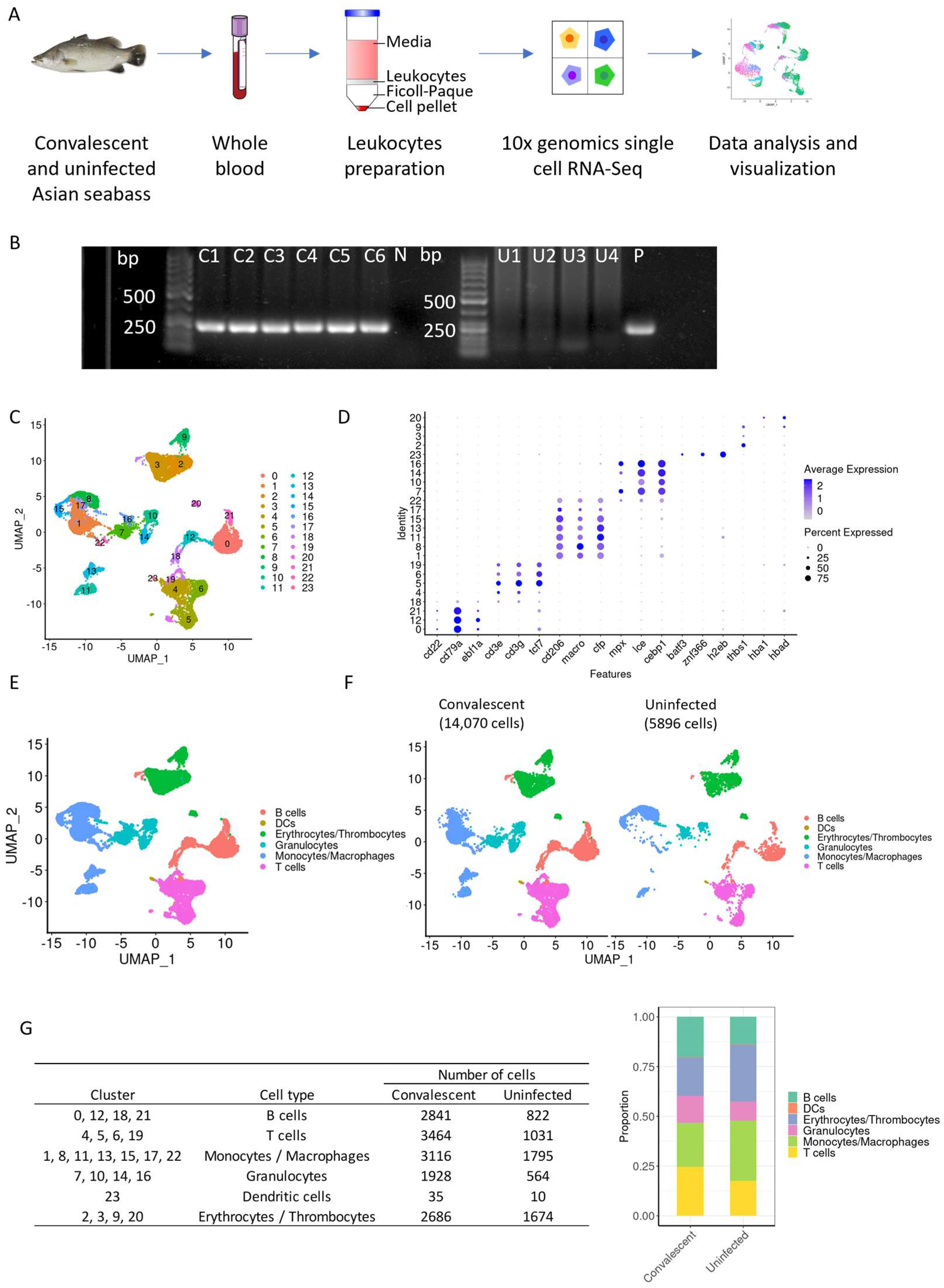
2.1. Acquisition of SDDV-Infected and -Uninfected Healthy Asian Seabass and Sample Collection
2.2. Detection of SDDV by PCR
2.3. Leukocyte Isolation from Whole Blood and Single-Cell RNA Sequencing
2.4. Single-Cell RNA Sequencing Analysis
2.5. Pseudotime Analysis
2.6. Analysis of Differentially Expressed Genes (DEGs)
3. Results
3.1. Single-Cell RNA Sequencing Reveals the Presence of 24 Distinct Clusters of Blood Leukocytes
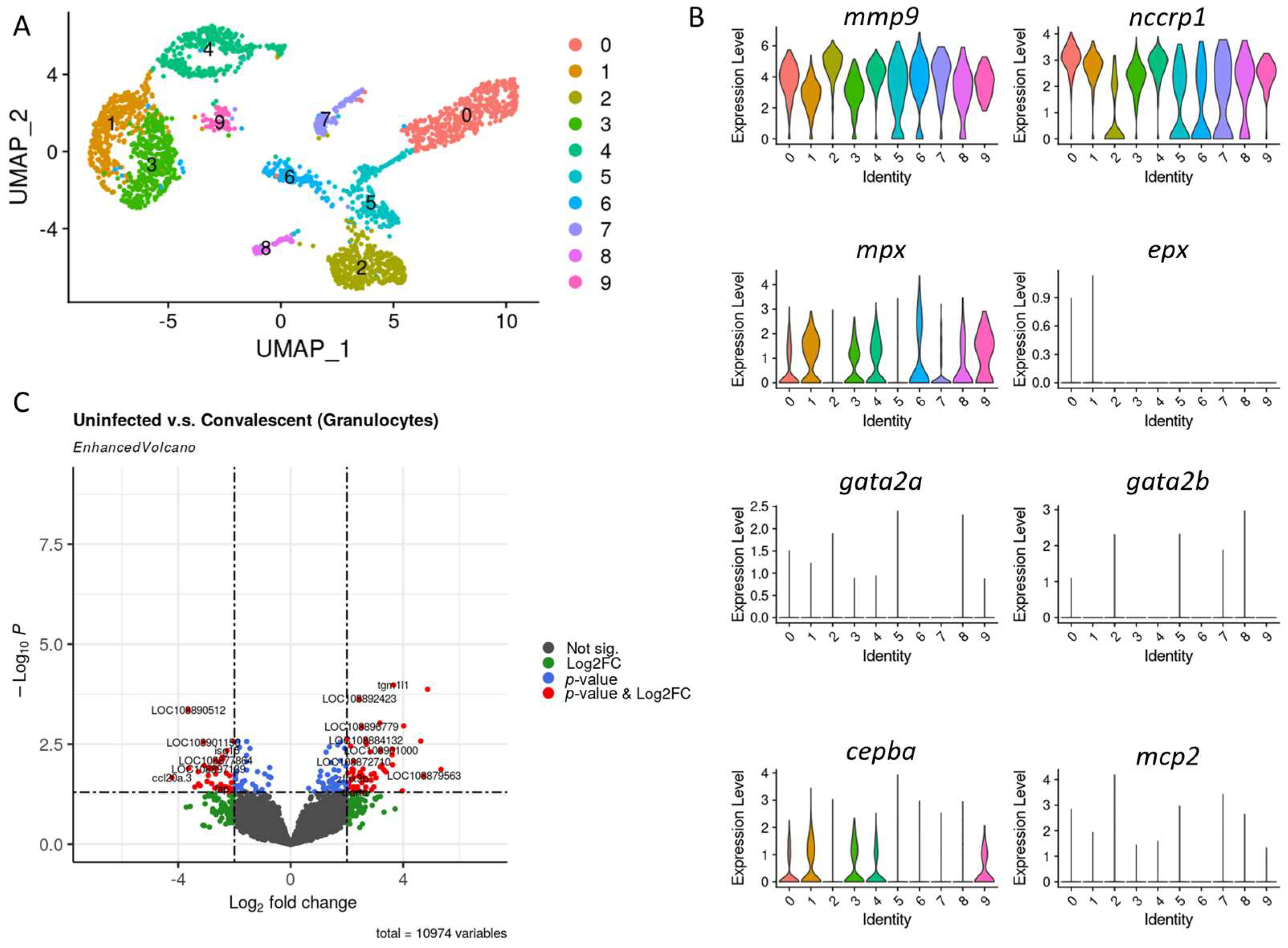
3.2. Granulocytes Profile in Convalescent and Uninfected Asian Seabass
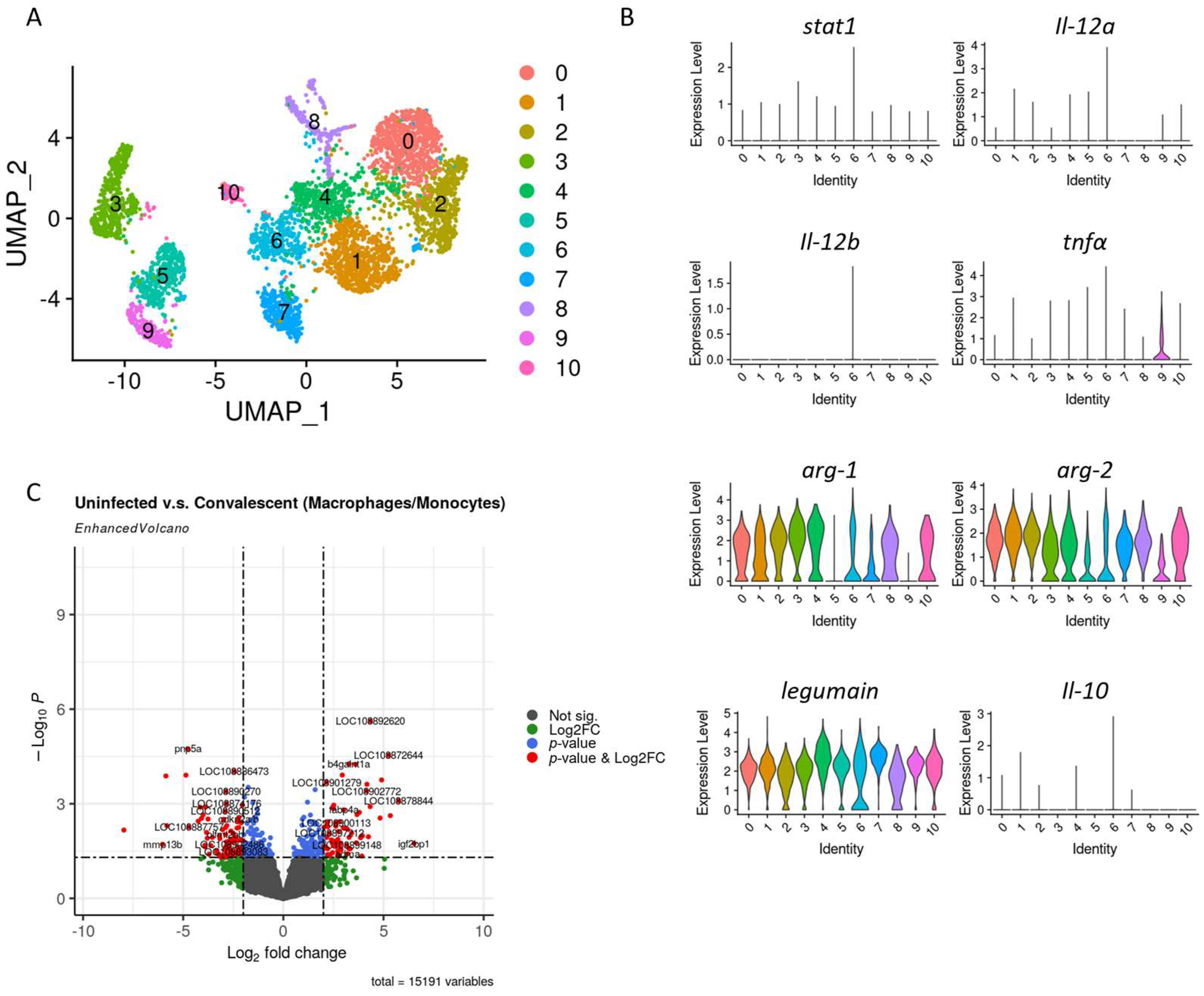
3.3. Macrophages Profile in Convalescent and Uninfected Asian Seabass
3.4. T Cells Profile in Convalescent and Uninfected Asian Seabass
3.5. B Cells Profile in Convalescent and Uninfected Asian Seabass
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gibson-Kueh, S.; Chee, D.; Chen, J.; Wang, Y.H.; Tay, S.; Leong, L.N.; Ng, M.L.; Jones, J.B.; Nicholls, P.K.; Ferguson, H.W. The pathology of ‘scale drop syndrome’ in Asian seabass, Lates calcarifer Bloch, a first description. J. Fish. Dis. 2012, 35, 19–27. [Google Scholar] [CrossRef] [PubMed]
- de Groof, A.; Guelen, L.; Deijs, M.; van der Wal, Y.; Miyata, M.; Ng, K.S.; van Grinsven, L.; Simmelink, B.; Biermann, Y.; Grisez, L.; et al. A Novel Virus Causes Scale Drop Disease in Lates calcarifer. PLoS Pathog. 2015, 11, e1005074. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.T.; Jitrakorn, S.; Kayansamruaj, P.; Pirarat, N.; Rodkhum, C.; Rattanarojpong, T.; Senapin, S.; Saksmerprome, V. Infectious spleen and kidney necrosis disease (ISKND) outbreaks in farmed barramundi (Lates calcarifer) in Vietnam. Fish. Shellfish. Immunol. 2017, 68, 65–73. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.F.; Ng, K.S.; Grisez, L.; de Groof, A.; Vogels, W.; van der Hoek, L.; Deijs, M. Novel Fish Pathogenic Virus; International Publication Number WO 2018/029301 A1; World Intellectual Property Organization International Bureau: Geneva, Switzerland, 2018. [Google Scholar]
- Chen, J.; Toh, X.; Ong, J.; Wang, Y.; Teo, X.H.; Lee, B.; Wong, P.S.; Khor, D.; Chong, S.M.; Chee, D.; et al. Detection and characterization of a novel marine birnavirus isolated from Asian seabass in Singapore. Virol. J. 2019, 16, 71. [Google Scholar] [CrossRef] [PubMed]
- Girisha, S.; Puneeth, T.; Nithin, M.; Kumar, B.N.; Ajay, S.; Vinay, T.; Suresh, T.; Venugopal, M.; Ramesh, K. Red sea bream iridovirus disease (RSIVD) outbreak in Asian seabass (Lates calcarifer) cultured in open estuarine cages along the west coast of India: First report. Aquaculture 2020, 520, 734712. [Google Scholar] [CrossRef]
- Senapin, S.; Dong, H.T.; Meemetta, W.; Gangnonngiw, W.; Sangsuriya, P.; Vanichviriyakit, R.; Sonthi, M.; Nuangsaeng, B. Mortality from scale drop disease in farmed Lates calcarifer in Southeast Asia. J. Fish. Dis. 2019, 42, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Kerddee, P.; Dong, H.T.; Chokmangmeepisarn, P.; Rodkhum, C.; Srisapoome, P.; Areechon, N.; Del-Pozo, J.; Kayansamruaj, P. Simultaneous detection of scale drop disease virus and Flavobacterium columnare from diseased freshwater-reared barramundi Lates calcarifer. Dis. Aquat. Organ. 2020, 140, 119–128. [Google Scholar] [CrossRef]
- Nurliyana, M.; Lukman, B.; Ina-Salwany, M.Y.; Zamri-Saad, M.; Annas, S.; Dong, H.T.; Rodkhum, C.; Amal, M.N.A. First evidence of scale drop disease virus in farmed Asian seabass (Lates calcarifer) in Malaysia. Aquaculture 2020, 528. [Google Scholar] [CrossRef]
- Carmona, S.J.; Teichmann, S.A.; Ferreira, L.; Macaulay, I.C.; Stubbington, M.J.; Cvejic, A.; Gfeller, D. Single-cell transcriptome analysis of fish immune cells provides insight into the evolution of vertebrate immune cell types. Genome Res. 2017, 27, 451–461. [Google Scholar] [CrossRef]
- Hernández, P.P.; Strzelecka, P.M.; Athanasiadis, E.I.; Hall, D.; Robalo, A.F.; Collins, C.M.; Boudinot, P.; Levraud, J.P.; Cvejic, A. Single-cell transcriptional analysis reveals ILC-like cells in zebrafish. Sci. Immunol. 2018, 3. [Google Scholar] [CrossRef]
- Peuß, R.; Box, A.C.; Chen, S.; Wang, Y.; Tsuchiya, D.; Persons, J.L.; Kenzior, A.; Maldonado, E.; Krishnan, J.; Scharsack, J.P.; et al. Adaptation to low parasite abundance affects immune investment and immunopathological responses of cavefish. Nat. Ecol. Evol. 2020, 4, 1416–1430. [Google Scholar] [CrossRef] [PubMed]
- Perdiguero, P.; Morel, E.; Díaz-Rosales, P.; Tafalla, C. Individual B cells transcribe multiple rearranged immunoglobulin light chains in teleost fish. iScience 2021, 24, 102615. [Google Scholar] [CrossRef] [PubMed]
- Perdiguero, P.; Morel, E.; Tafalla, C. Diversity of Rainbow Trout Blood B Cells Revealed by Single Cell RNA Sequencing. Biology 2021, 10, 511. [Google Scholar] [CrossRef] [PubMed]
- Niu, J.; Huang, Y.; Liu, X.; Zhang, Z.; Tang, J.; Wang, B.; Lu, Y.; Cai, J.; Jian, J. Single-cell RNA-seq reveals different subsets of non-specific cytotoxic cells in teleost. Genomics 2020, 112, 5170–5179. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Peng, C.; Yang, M.; Huang, F.; Duan, X.; Wang, S.; Cheng, H.; Yang, H.; Zhao, H.; Qin, Q. Single-cell RNA-seq landscape midbrain cell responses to red spotted grouper nervous necrosis virus infection. PLoS Pathog. 2021, 17, e1009665. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Huang, J.; Wang, W.; Wang, Y.; Chen, H.; Wang, Q.; Zhang, Y.; Liu, Q.; Yang, D. Multi-tissue scRNA-seq reveals immune cell landscape of turbot (Scophthalmus maximus). Fundam. Res. 2022, 2, 550–561. [Google Scholar] [CrossRef]
- Sun, X.; Li, L.; Wu, B.; Ge, J.; Zheng, Y.; Yu, T.; Zhou, L.; Zhang, T.; Yang, A.; Liu, Z. Cell type diversity in scallop adductor muscles revealed by single-cell RNA-Seq. Genomics 2021, 113, 3582–3598. [Google Scholar] [CrossRef]
- Koiwai, K.; Koyama, T.; Tsuda, S.; Toyoda, A.; Kikuchi, K.; Suzuki, H.; Kawano, R. Single-cell RNA-seq analysis reveals penaeid shrimp hemocyte subpopulations and cell differentiation process. Elife 2021, 10, e66954. [Google Scholar] [CrossRef] [PubMed]
- Wen, S.; Lu, H.; Wang, D.; Guo, J.; Dai, W.; Wang, Z. TCF-1 maintains CD8+ T cell stemness in tumor microenvironment. J. Leukoc. Biol. 2021, 110, 585–590. [Google Scholar] [CrossRef]
- Morera, D.; Roher, N.; Ribas, L.; Balasch, J.C.; Doñate, C.; Callol, A.; Boltaña, S.; Roberts, S.; Goetz, G.; Goetz, F.W.; et al. RNA-Seq reveals an integrated immune response in nucleated erythrocytes. PLoS ONE 2011, 6, e26998. [Google Scholar] [CrossRef]
- Morera, D.; MacKenzie, S.A. Is there a direct role for erythrocytes in the immune response? Vet. Res. 2011, 42, 89. [Google Scholar] [CrossRef]
- Passantino, L.; Massaro, M.A.; Jirillo, F.; Di Modugno, D.; Ribaud, M.R.; Modugno, G.D.; Passantino, G.F.; Jirillo, E. Antigenically activated avian erythrocytes release cytokine-like factors: A conserved phylogenetic function discovered in fish. Immunopharmacol. Immunotoxicol. 2007, 29, 141–152. [Google Scholar] [CrossRef] [PubMed]
- Passantino, L.; Altamura, M.; Cianciotta, A.; Patruno, R.; Tafaro, A.; Jirillo, E.; Passantino, G.F. Fish immunology. I. Binding and engulfment of Candida albicans by erythrocytes of rainbow trout (Salmo gairdneri Richardson). Immunopharmacol. Immunotoxicol. 2002, 24, 665–678. [Google Scholar] [CrossRef] [PubMed]
- Workenhe, S.T.; Kibenge, M.J.; Wright, G.M.; Wadowska, D.W.; Groman, D.B.; Kibenge, F.S. Infectious salmon anaemia virus replication and induction of alpha interferon in Atlantic salmon erythrocytes. Virol. J. 2008, 5, 36. [Google Scholar] [CrossRef] [PubMed]
- Kyriakis, J.M. Activation of the AP-1 transcription factor by inflammatory cytokines of the TNF family. Gene Expr. 1999, 7, 217–231. [Google Scholar]
- Ainsworth, A.J. Fish granulocytes: Morphology, distribution, and function. Annu. Rev. Fish. Dis. 1992, 2, 123–148. [Google Scholar] [CrossRef]
- Ragini, S. Macrophage: A Key Player of Teleost Immune System. In Macrophages; Chapter 2; Vijay, K., Ed.; IntechOpen: Rijeka, Croatia, 2022. [Google Scholar]
- Nakanishi, T.; Shibasaki, Y.; Matsuura, Y. T Cells in Fish. Biology 2015, 4, 640–663. [Google Scholar] [CrossRef]
- Ashfaq, H.; Soliman, H.; Saleh, M.; El-Matbouli, M. CD4: A vital player in the teleost fish immune system. Vet. Res. 2019, 50, 1. [Google Scholar] [CrossRef] [PubMed]
- Nakanishi, T.; Toda, H.; Shibasaki, Y.; Somamoto, T. Cytotoxic T cells in teleost fish. Dev. Comp. Immunol. 2011, 35, 1317–1323. [Google Scholar] [CrossRef]
- Mittrücker, H.-W.; Visekruna, A.; Huber, M. Heterogeneity in the Differentiation and Function of CD8+ T Cells. Arch. Immunol. Et. Ther. Exp. 2014, 62, 449–458. [Google Scholar] [CrossRef]
- Guslund, N.C.; Solbakken, M.H.; Brieuc, M.S.O.; Jentoft, S.; Jakobsen, K.S.; Qiao, S.W. Single-Cell Transcriptome Profiling of Immune Cell Repertoire of the Atlantic Cod Which Naturally Lacks the Major Histocompatibility Class II System. Front. Immunol. 2020, 11, 559555. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Gao, A.; Li, L.; Chen, J.; Li, J.; Ye, J. A Single-Cell Transcriptome Profiling of Anterior Kidney Leukocytes From Nile Tilapia (Oreochromis niloticus). Front. Immunol. 2021, 12, 783196. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Qiao, Y.; Xu, W.; Gong, L.; He, R.; Qi, W.; Gao, Q.; Cai, H.; Grossart, H.P.; Yan, Q. Full-Length Transcriptome: A Reliable Alternative for Single-Cell RNA-Seq Analysis in the Spleen of Teleost Without Reference Genome. Front. Immunol. 2021, 12, 737332. [Google Scholar] [CrossRef] [PubMed]
- Star, B.; Nederbragt, A.J.; Jentoft, S.; Grimholt, U.; Malmstrøm, M.; Gregers, T.F.; Rounge, T.B.; Paulsen, J.; Solbakken, M.H.; Sharma, A.; et al. The genome sequence of Atlantic cod reveals a unique immune system. Nature 2011, 477, 207–210. [Google Scholar] [CrossRef] [PubMed]
- Malmstrøm, M.; Matschiner, M.; Tørresen, O.K.; Star, B.; Snipen, L.G.; Hansen, T.F.; Baalsrud, H.T.; Nederbragt, A.J.; Hanel, R.; Salzburger, W.; et al. Evolution of the immune system influences speciation rates in teleost fishes. Nat. Genet. 2016, 48, 1204–1210. [Google Scholar] [CrossRef] [PubMed]
- Pilström, L.; Warr, G.W.; Strömberg, S. Why is the antibody response of Atlantic cod so poor? The search for a genetic explanation. Fish. Sci. 2005, 71, 961–971. [Google Scholar] [CrossRef]
- Lund, S.G.; Phillips, M.C.; Moyes, C.D.; Tufts, B.L. The effects of cell ageing on protein synthesis in rainbow trout (Oncorhynchus mykiss) red blood cells. J. Exp. Biol. 2000, 203, 2219–2228. [Google Scholar] [CrossRef] [PubMed]
- Zexia, G.; Weimin, W.; Yi, Y.; Abbas, K.; Dapeng, L.; Guiwei, Z.; Diana, J.S. Morphological studies of peripheral blood cells of the Chinese sturgeon, Acipenser sinensis. Fish. Physiol. Biochem. 2007, 33, 213–222. [Google Scholar] [CrossRef]
- Lugo-Villarino, G.; Balla, K.M.; Stachura, D.L.; Bañuelos, K.; Werneck, M.B.F.; Traver, D. Identification of dendritic antigen-presenting cells in the zebrafish. Proc. Natl. Acad. Sci. USA 2010, 107, 15850–15855. [Google Scholar] [CrossRef]
- Shao, T.; Zhu, L.-Y.; Nie, L.; Shi, W.; Dong, W.-R.; Xiang, L.-X.; Shao, J.-Z. Characterization of surface phenotypic molecules of teleost dendritic cells. Dev. Comp. Immunol. 2015, 49, 38–43. [Google Scholar] [CrossRef]
- Zoccola, E.; Delamare-Deboutteville, J.; Barnes, A.C. Identification of Barramundi (Lates calcarifer) DC-SCRIPT, a Specific Molecular Marker for Dendritic Cells in Fish. PLoS ONE 2015, 10, e0132687. [Google Scholar] [CrossRef]
- Wang, J.; Wang, T.; Benedicenti, O.; Collins, C.; Wang, K.; Secombes, C.J.; Zou, J. Characterisation of ZBTB46 and DC-SCRIPT/ZNF366 in rainbow trout, transcription factors potentially involved in dendritic cell maturation and activation in fish. Dev. Comp. Immunol. 2018, 80, 2–14. [Google Scholar] [CrossRef]
- Tian, H.; Xing, J.; Tang, X.; Sheng, X.; Chi, H.; Zhan, W. Single-cell transcriptome uncovers heterogeneity and immune responses of leukocytes after vaccination with inactivated Edwardsiella tarda in flounder (Paralichthys olivaceus). Aquaculture 2023, 566, 739238. [Google Scholar] [CrossRef]
- Wiegertjes, G.F.; Wentzel, A.S.; Spaink, H.P.; Elks, P.M.; Fink, I.R. Polarization of immune responses in fish: The ‘macrophages first’ point of view. Mol. Immunol. 2016, 69, 146–156. [Google Scholar] [CrossRef]
- Desai, A. 15-Pgdh Inhibition Alternatively Activates Macrophages to Promote Hematopoietic Function during Aging. Blood 2023, 142 (Suppl. 1), 5610. [Google Scholar] [CrossRef]
- Clementi, G.; Caruso, A.; Cutuli, V.M.; Prato, A.; Mangano, N.G.; Amico-Roxas, M. Antiinflammatory activity of adrenomedullin in the acetic acid peritonitis in rats. Life Sci. 1999, 65, Pl203–Pl208. [Google Scholar] [CrossRef]
- Isumi, Y.; Kubo, A.; Katafuchi, T.; Kangawa, K.; Minamino, N. Adrenomedullin suppresses interleukin-1beta-induced tumor necrosis factor-alpha production in Swiss 3T3 cells. FEBS Lett. 1999, 463, 110–114. [Google Scholar] [CrossRef] [PubMed]
- Hansen, J.D.; Landis, E.D.; Phillips, R.B. Discovery of a unique Ig heavy-chain isotype (IgT) in rainbow trout: Implications for a distinctive B cell developmental pathway in teleost fish. Proc. Natl. Acad. Sci. USA 2005, 102, 6919–6924. [Google Scholar] [CrossRef]
- Parra, D.; Takizawa, F.; Sunyer, J.O. Evolution of B Cell Immunity. Annu. Rev. Anim. Biosci. 2013, 1, 65–97. [Google Scholar] [CrossRef] [PubMed]
- Granja, A.G.; Tafalla, C. Different IgM+ B cell subpopulations residing within the peritoneal cavity of vaccinated rainbow trout are differently regulated by BAFF. Fish. Shellfish. Immunol. 2019, 85, 9–17. [Google Scholar] [CrossRef]
- Castro, R.; Bromage, E.; Abós, B.; Pignatelli, J.; Gonzalez Granja, A.; Luque, A.; Tafalla, C. CCR7 is mainly expressed in teleost gills, where it defines an IgD+ IgM− B lymphocyte subset. J. Immunol. 2014, 192, 1257–1266. [Google Scholar] [CrossRef] [PubMed]
- Perdiguero, P.; Martín-Martín, A.; Benedicenti, O.; Díaz-Rosales, P.; Morel, E.; Muñoz-Atienza, E.; García-Flores, M.; Simón, R.; Soleto, I.; Cerutti, A. Teleost IgD+ IgM− B cells mount clonally expanded and mildly mutated intestinal IgD responses in the absence of lymphoid follicles. Cell Rep. 2019, 29, 4223–4235.e5. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Zhang, X.J.; Chen, D.D.; Oriol Sunyer, J.; Zhang, Y.A. Molecular characterization and expression analysis of three subclasses of IgT in rainbow trout (Oncorhynchus mykiss). Dev. Comp. Immunol. 2017, 70, 94–105. [Google Scholar] [CrossRef]
- Tian, H.-F.; Xing, J.; Tang, X.-Q.; Chi, H.; Sheng, X.-Z.; Zhan, W.-B. Cluster of differentiation antigens: Essential roles in the identification of teleost fish T lymphocytes. Mar. Life Sci. Technol. 2022, 4, 303–316. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Loh, Z.; Lim, T.W.; Howland, S.W.; Awate, S.; Renia, L.; Chen, J.; Ren, E.C. Single-Cell Transcriptome Profiling of Scale Drop Disease Virus-Infected Asian Seabass (Lates calcarifer). Aquac. J. 2024, 4, 28-43. https://doi.org/10.3390/aquacj4020003
Loh Z, Lim TW, Howland SW, Awate S, Renia L, Chen J, Ren EC. Single-Cell Transcriptome Profiling of Scale Drop Disease Virus-Infected Asian Seabass (Lates calcarifer). Aquaculture Journal. 2024; 4(2):28-43. https://doi.org/10.3390/aquacj4020003
Chicago/Turabian StyleLoh, Zhixuan, Ting Wei Lim, Shanshan Wu Howland, Sunita Awate, Laurent Renia, Jinmiao Chen, and Ee Chee Ren. 2024. "Single-Cell Transcriptome Profiling of Scale Drop Disease Virus-Infected Asian Seabass (Lates calcarifer)" Aquaculture Journal 4, no. 2: 28-43. https://doi.org/10.3390/aquacj4020003
APA StyleLoh, Z., Lim, T. W., Howland, S. W., Awate, S., Renia, L., Chen, J., & Ren, E. C. (2024). Single-Cell Transcriptome Profiling of Scale Drop Disease Virus-Infected Asian Seabass (Lates calcarifer). Aquaculture Journal, 4(2), 28-43. https://doi.org/10.3390/aquacj4020003





