Microbial Eukaryotes in Natural and Artificial Salt Marsh Pools
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. Metabarcoding
2.3. Bioinformatics Processing and Taxonomic Assignment
2.4. Diatom Enumeration
2.5. Data Analysis
3. Results
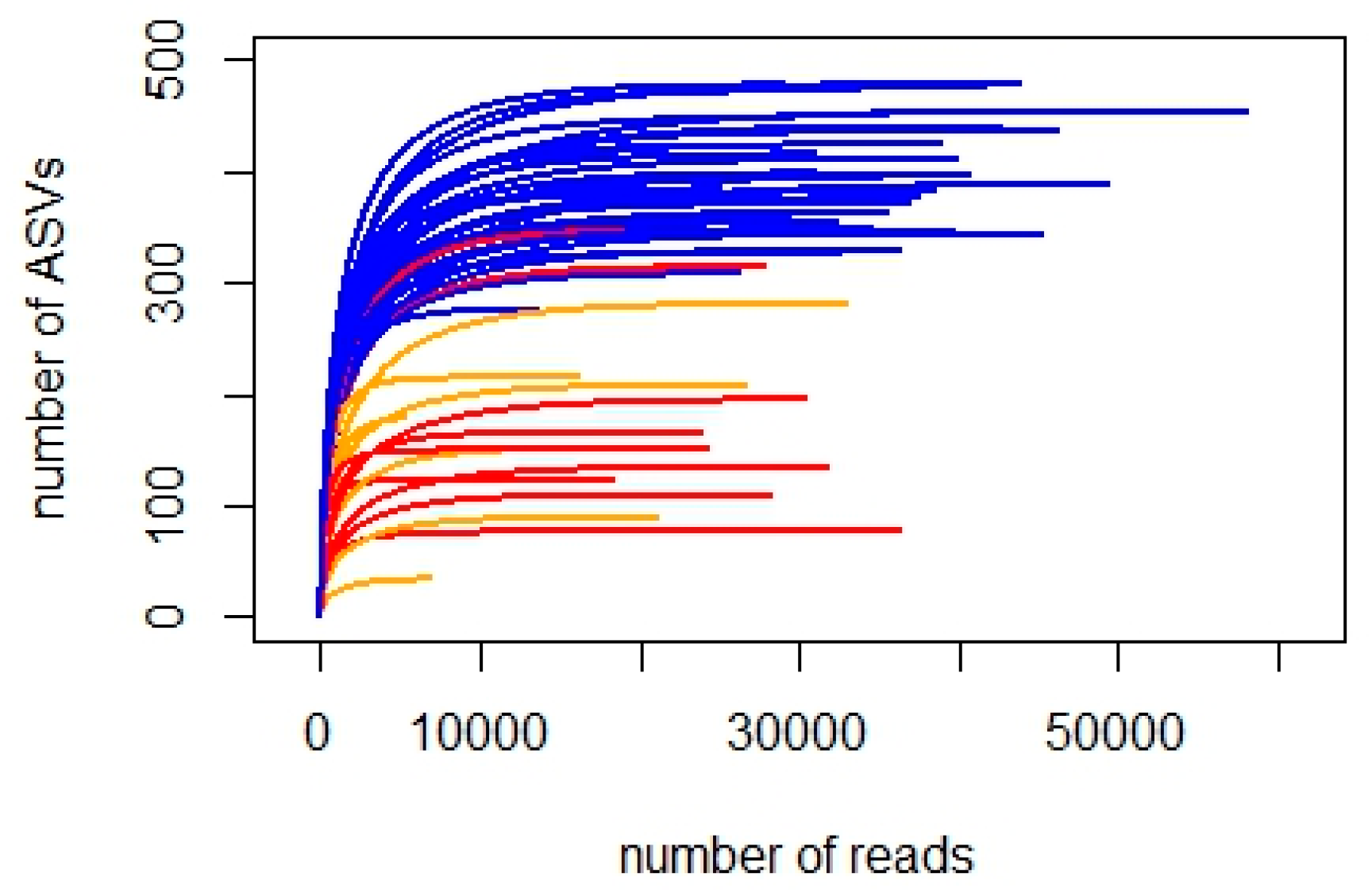
3.1. Taxonomic Diversity
3.1.1. 18S_V9 Metabarcoding
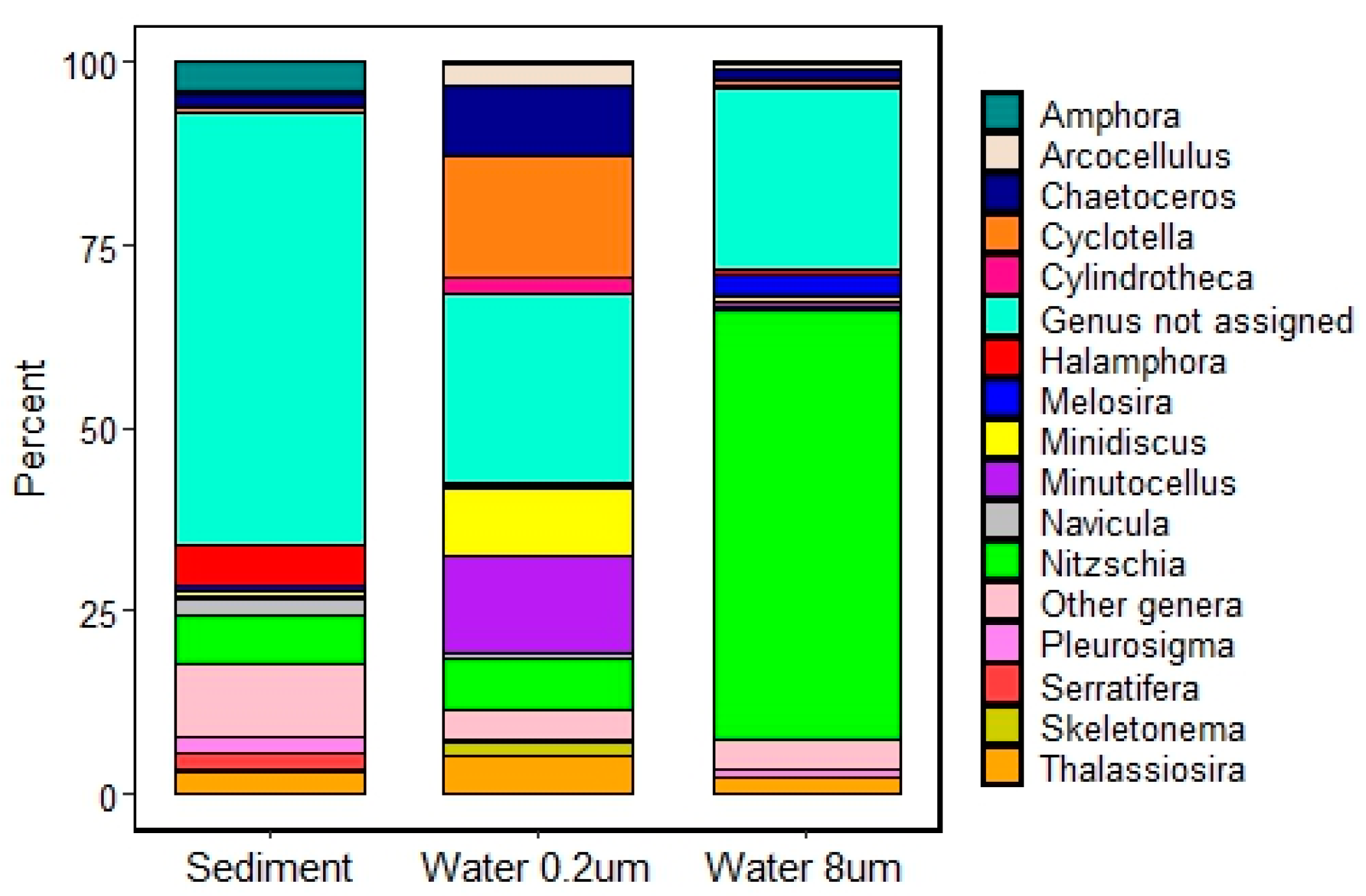
3.1.2. Diatom rbcL Metabarcoding
3.1.3. Diatom Counts
3.2. Differences between Natural and Artificial Pools
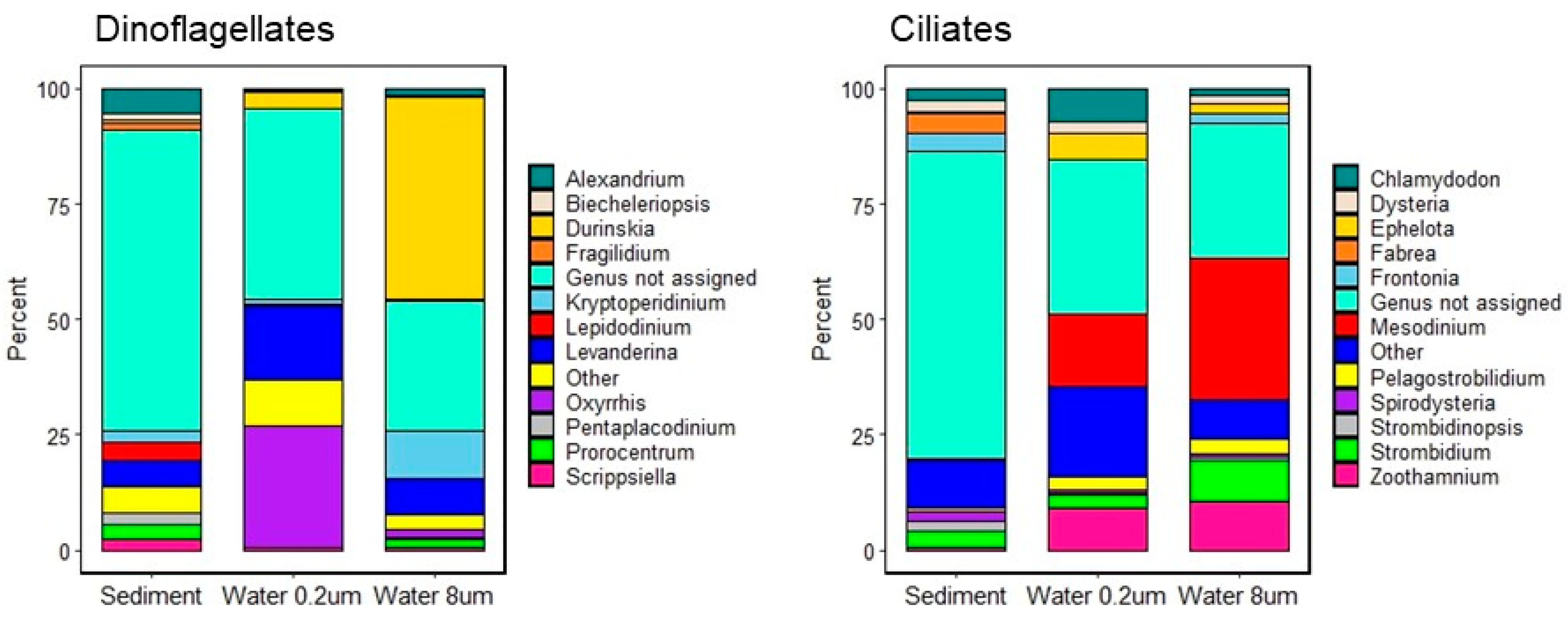
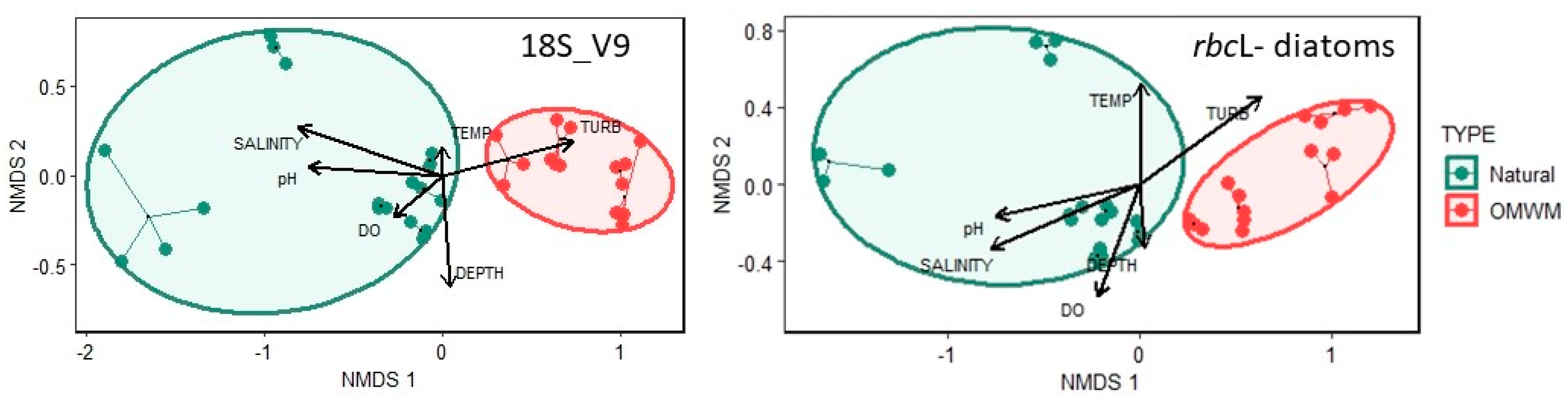
3.2.1. Assemblage Composition
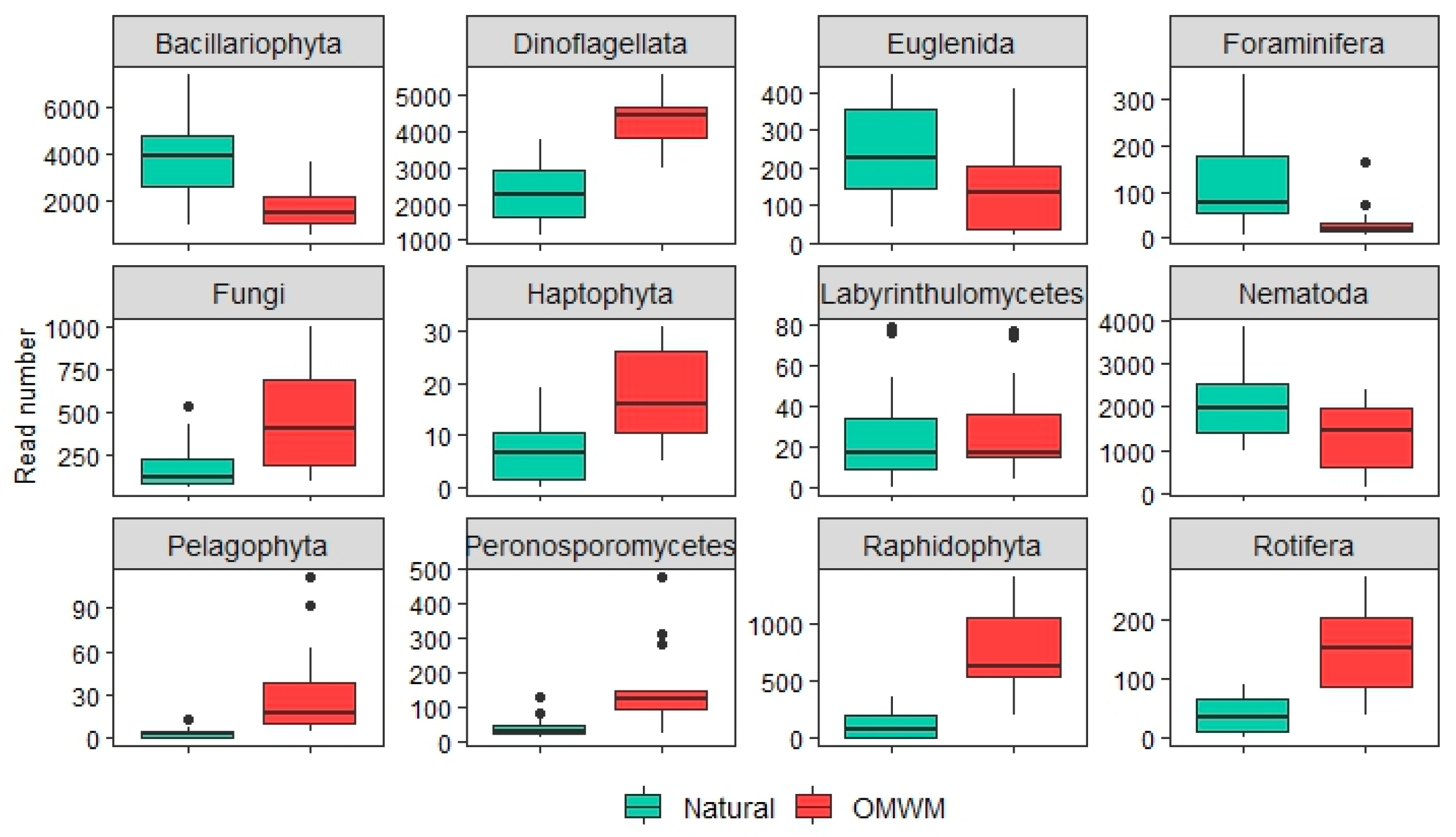
3.2.2. Differential Abundance of ASVs and Morphotaxa
4. Discussion
4.1. Assemblage Composition
4.2. Comparison of Assemblages from Natural and Artificial Pools
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Koop-Jakobsen, K.; Gutbrod, M.S. Shallow salt marsh tidal ponds—An environment with extreme oxygen dynamics. Front. Environ. Sci. 2019, 7, 137. [Google Scholar] [CrossRef]
- Able, K.W.; Smith, K.J.; Hagan, S.M. Fish composition and abundance in New Jersey salt marsh pools: Sampling technique effects. Northeast. Nat. 2005, 12, 485–502. [Google Scholar] [CrossRef]
- Bolduc, F.; Afton, A.D. Relationships between wintering waterbirds and invertebrates, sediments and hydrology of coastal marsh ponds. Waterbirds 2004, 27, 333–341. [Google Scholar] [CrossRef]
- Minello, T.J.; Able, K.W.; Weinstein, M.P.; Hays, C.G. Salt marshes as nurseries for nekton: Testing hypotheses on density, growth and survival through meta-analysis. Mar. Ecol. Prog. Ser. 2003, 246, 39–59. [Google Scholar] [CrossRef]
- Reed, H.E.; Martiny, J.B.H. Microbial composition affects the functioning of estuarine sediments. ISME J. 2013, 7, 868–879. [Google Scholar] [CrossRef] [PubMed]
- Frates, E.S.; Spietz, R.L.; Silverstein, M.R.; Girguis, P.; Hatzenpichler, R.; Marlow, J.J. Natural and anthropogenic carbon input affect microbial activity in salt marsh sediment. Front. Microbiol. 2023, 14, 1235906. [Google Scholar] [CrossRef]
- Calabon, M.S.; Jones, E.B.G.; Promputtha, I.; Hyde, K.D. Fungal biodiversity in salt marsh ecosystems. J. Fungi 2021, 7, 648. [Google Scholar] [CrossRef] [PubMed]
- Gessner, R.V.; Goos, R.D. Fungi from decomposing Spartina alterniflora. Can. J. Bot. 1973, 51, 51–55. [Google Scholar] [CrossRef]
- Rinke, M.; Maraun, M.; Scheu, S. Spatial and temporal variations in salt marsh microorganisms of the Wadden Sea. Ecol. Evol. 2022, 12, e8767. [Google Scholar] [CrossRef]
- Kearns, P.; Angell, J.; Howard, E.M.; Deegan, L.A.; Stanley, R.H.R.; Bowen, J.L. Nutrient enrichment induces dormancy and decreases diversity of active bacteria in salt marsh sediments. Nat. Commun. 2016, 7, 12881. [Google Scholar] [CrossRef]
- Kearns, P.J.; Bulseco-McKim, A.N.; Hoyt, H.; Angell, J.H.; Bowen, J.L. Nutrient enrichment alters salt marsh fungal communities and promotes putative fungal denitrifiers. Microb. Ecol. 2019, 77, 358–369. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Chaudhary, D.R.; Kang, H. Nitrogen addition differently alters GHGs production and soil microbial community of tidal salt marsh soil depending on the types of halophyte. Appl. Soil Ecol. 2020, 150, 103440. [Google Scholar] [CrossRef]
- Hanley, T.C.; Bowen, J.L.; Kearns, P.J.; Hughes, A.R. Short- and long-term effects of nutrient enrichment on salt marsh plant production and microbial community structure. J. Ecol. 2021, 109, 3779–3793. [Google Scholar] [CrossRef]
- Engel, A.S.; Liu, C.; Paterson, A.T.; Anderson, L.C.; Turner, R.E.; Overton, E.B. Salt marsh bacterial communities before and after the Deepwater Horizon oil spill. Appl. Environ. Microbiol. 2017, 83, e00784-17. [Google Scholar] [CrossRef] [PubMed]
- An, S.U.; Cho, H.; Jung, U.J.; Kim, B.; Lee, H.; Hyun, J.H. Invasive Spartina anglica greatly alters the rates and pathways of organic carbon oxidation and associated microbial communities in an intertidal wetland of the Han River Estuary, Yellow Sea. Front. Mar. Sci. 2020, 7, 59. [Google Scholar] [CrossRef]
- Kim, J.; Chaudhary, D.R.; Lee, J.; Byun, C.; Ding, W.; Kwon, B.O.; Khim, J.S.; Kang, H. Microbial mechanism for enhanced methane emission in deep soil layer of Phragmites-introduced tidal marsh. Environ. Int. 2020, 134, 105251. [Google Scholar] [CrossRef] [PubMed]
- Simon, M.R.; Zogg, G.P.; Travis, S.E. Impacts of sea-level rise on sediment microbial community structure and function in two New England salt marshes, USA. J. Soil Sediment 2017, 17, 2847–2855. [Google Scholar] [CrossRef]
- Abbott, K.M.; Quirk, T.; Fultz, L.M. Soil microbial community development across a 32-year coastal wetland restoration time series and the relative importance of environmental factors. Sci. Total Environ. 2022, 821, 153359. [Google Scholar] [CrossRef]
- Barry, A.; Ooi, S.K.; Helton, A.M.; Steven, B.; Elphick, C.S.; Lawrence, B.A. Vegetation zonation predicts soil carbon mineralization and microbial communities in Southern New England salt marshes. Estuar. Coasts 2022, 45, 168–180. [Google Scholar] [CrossRef]
- Armitage, D.; Gallagher, K.; Youngblut, N.; Buckley, D.; Zinder, S. Millimeter-scale patterns of phylogenetic and trait diversity in a salt marsh microbial mat. Front. Microbiol. 2012, 3, 293. [Google Scholar] [CrossRef]
- Lee, H.; Heo, Y.M.; Kwon, S.L.; Yoo, Y.; Kim, D.; Lee, J.; Kwon, B.O.; Khim, J.S.; Kim, J.J. Environmental drivers affecting the bacterial community of intertidal sediments in the Yellow Sea. Sci. Total Environ. 2021, 755, 142726. [Google Scholar] [CrossRef]
- First, M.R.; Hollibaugh, J.T. Environmental factors shaping microbial community structure in salt marsh sediments. Mar. Ecol. Prog. Ser. 2010, 399, 15–26. [Google Scholar] [CrossRef]
- Barnett, R.L.; Newton, T.L.; Charman, D.J.; Gehrels, W.R. Salt-marsh testate amoebae as precise and widespread indicators of sea-level change. Earth-Sci. Rev. 2017, 164, 193–207. [Google Scholar] [CrossRef]
- Horton, B.; Sawai, Y. Diatoms as indicators of former sea levels, earthquakes, tsunamis, and hurricanes. In The Diatoms: Applications for the Environmental and Earth Sciences; Smol, J., Stoermer, E., Eds.; Cambridge University Press: Cambridge, UK, 2010; pp. 357–372. [Google Scholar]
- Kemp, A.C.; Horton, B.P.; Vann, D.R.; Engelhart, S.E.; Grand Pre, C.A.; Vane, C.H.; Nikitina, D.; Anisfeld, S.C. Quantitative vertical zonation of salt-marsh foraminifera for reconstructing former sea level; an example from New Jersey, USA. Quat. Sci. Rev. 2012, 54, 26–39. [Google Scholar] [CrossRef]
- Desianti, N.; Enache, M.D.; Griffiths, M.; Biskup, K.; Degen, A.; DaSilva, M.; Millemann, D.; Lippincott, L.; Watson, E.; Gray, A.; et al. The potential and limitations of diatoms as environmental indicators in mid-Atlantic coastal wetlands. Estuar. Coasts 2019, 42, 1440–1458. [Google Scholar] [CrossRef]
- Walker, J.S.; Khan, N.S.; Shaw, T.A.; Barber, D.C.; Switzer, A.D.; Horton, B.P. Spatial and temporal distributions of live salt-marsh foraminifera in southern New Jersey: Implications for sea-level studies. J. Foraminifer. Res. 2023, 53, 3–19. [Google Scholar] [CrossRef]
- Quintana, X.D.; Moreno-Amich, R. Phytoplankton composition of Empordà salt marshes, Spain and its response to freshwater flux regulation. J. Coast. Res. 2002, 36, 581–590. [Google Scholar] [CrossRef]
- Sullivan, M.J.; Currin, C.A. Community structure and functional dynamics of benthic microalgae in salt marshes. In Concepts and Controversies in Tidal Marsh Ecology; Weinstein, M.P., Kreeger, D.A., Eds.; Springer: Dordrecht, The Netherlands, 2002; pp. 81–106. [Google Scholar]
- Underwood, G.J.C. Microalgal colonization in a saltmarsh restoration scheme. Estuar. Coast. Shelf Sci. 1997, 44, 471–481. [Google Scholar] [CrossRef]
- Whitcraft, C.R.; Levin, L.A. Regulation of benthic algal and animal communities by salt marsh plants: Impact of shading. Ecology 2007, 88, 904–917. [Google Scholar] [CrossRef]
- Barnes, R.S.K.; Sattelle, D.B.; Everton, I.J.; Nicholas, W.; Scott, D.H. Intertidal sands and interstitial fauna associated with different stages of salt-marsh development. Estuar. Coast. Mar. Sci. 1976, 4, 497–511. [Google Scholar] [CrossRef]
- Elliott, P.B.; Bamforth, S.S. Interstitial protozoa and algae of Louisiana salt marshes. J. Protozool. 1975, 4, 514–519. [Google Scholar] [CrossRef]
- Xu, Y.; Yao, S.; Soetaert, K.; Fan, X. Effects of salt marsh restoration on eukaryotic microbenthic communities in the Yangtze Estuary. Mar. Ecol. Prog. Ser. 2020, 638, 39–50. [Google Scholar] [CrossRef]
- Fleeger, J.; Chandler, G.; Fitzhugh, G.; Phillips, F. Effects of tidal currents on meiofauna densities in vegetated salt marsh sediments. Mar. Ecol. Prog. Ser. 1984, 19, 49–53. [Google Scholar] [CrossRef]
- Fleeger, W.; Johnson, D.S.; Galván, K.A.; Deegan, L.A. Top-down and bottom-up control of infauna varies across the saltmarsh landscape. J. Exp. Mar. Biol. Ecol. 2008, 357, 20–34. [Google Scholar] [CrossRef][Green Version]
- Teal, J.M.; Wieser, W. The distribution and ecology of nematodes in a Georgia salt marsh. Limnol. Oceanogr. 1966, 11, 217–222. [Google Scholar] [CrossRef]
- Alteio, L.; Séneca, J.; Canarini, A.; Angel, R.; Jansa, J.; Guseva, K.; Kaiser, C.; Richter, A.; Schmidt, H. A critical perspective on interpreting amplicon sequencing data in soil ecological research. Soil Biol. Biochem. 2021, 160, 108357. [Google Scholar] [CrossRef]
- Pawlowski, J.; Kelly-Quinn, M.; Altermatt, F.; Apothéloz-Perret-Gentil, L.; Beja, P.; Boggero, A.; Borja, A.; Bouchez, A.; Cordier, T.; Domaizon, I.; et al. The future of biotic indices in the ecogenomic era: Integrating (e)DNA metabarcoding in biological assessment of aquatic ecosystems. Sci. Total Environ. 2018, 637–638, 1295–1310. [Google Scholar] [CrossRef]
- de Vargas, C.; Audic, S.; Henry, N.; Decelle, J.; Mahe, F.; Logares, R.; Lara, E.; Berney, C.; Le Bescot, N.; Probert, I.; et al. Eukaryotic plankton diversity in the sunlit ocean. Science 2015, 348, 1261605. [Google Scholar] [CrossRef]
- Dünn, M.; Arndt, H. Distribution patterns of benthic protist communities depending on depth revealed by environmental sequencing—From the sublittoral to the deep sea. Microorganisms 2023, 11, 1664. [Google Scholar] [CrossRef]
- Forster, D.; Dunthorn, M.; Mahé, F.; Dolan, J.R.; Audic, S.; Bass, D.; Bittner, L.; Boutte, C.; Christen, R.; Claverie, J.-M.; et al. Benthic protists: The under-charted majority. FEMS Microbiol. Ecol. 2016, 92, fiw120. [Google Scholar] [CrossRef]
- Massana, R.; Gobet, A.; Audic, S.; Bass, D.; Bittner, L.; Boutte, C.; Chambouvet, A.; Christen, R.; Claverie, J.-M.; Decelle, J.; et al. Protist diversity in European coastal areas. Environ. Microbiol. 2015, 17, 4035–4049. [Google Scholar] [CrossRef]
- Sommeria-Klein, G.; Watteaux, R.; Ibarbalz, F.M.; Pierella Karlusich, J.J.; Iudicone, D.; Bowler, C.; Morlon, H. Global drivers of eukaryotic plankton biogeography in the sunlit ocean. Science 2021, 374, 594–599. [Google Scholar] [CrossRef]
- Stern, R.F.; Horak, A.; Andrew, R.L.; Coffroth, M.-A.; Andersen, R.A.; Küpper, F.C.; Jameson, I.; Hoppenrath, M.; Véron, B.; Kasai, F.; et al. Environmental barcoding reveals massive dinoflagellate diversity in marine environments. PLoS ONE 2010, 5, e13991. [Google Scholar] [CrossRef]
- Stoeck, T.; Bass, D.; Nebel, M.; Christen, R.; Jones, M.D.M.; Breiner, H.-W.; Richards, T.A. Multiple marker parallel tag environmental DNA sequencing reveals a highly complex eukaryotic community in marine anoxic water. Mol. Ecol. 2010, 19 (Suppl. S1), 21–31. [Google Scholar] [CrossRef]
- Stoeck, T.; Kochems, R.; Forster, D.; Lejzerowicz, F.; Pawlowski, J. Metabarcoding of benthic ciliate communities shows high potential for environmental monitoring in salmon aquaculture. Ecol. Indic. 2018, 85, 153–164. [Google Scholar] [CrossRef]
- Kalu, E.I.; Reyes-Prieto, A.; Barbeau, M.A. Community dynamics of microbial eukaryotes in intertidal mudflats in the hypertidal Bay of Fundy. ISME Commun. 2023, 3, 21. [Google Scholar] [CrossRef]
- Plante, C.J.; Hill-Spanik, K.; Lowry, J. Controls on diatom biogeography on South Carolina (USA) barrier island beaches. Mar. Ecol. Prog. Ser. 2021, 661, 17–33. [Google Scholar] [CrossRef]
- Rivera-Garcia, L.G.; Hill-Spanik, K.M.; Berthrong, S.T.; Plante, C.J. Tidal stage changes in structure and diversity of intertidal benthic diatom assemblages: A case study from two contrasting Charleston harbor flats. Estuar. Coasts 2018, 41, 772–783. [Google Scholar] [CrossRef]
- Wang, M.; Yergaliyev, T.; Sun, C.; Martinez, J.G.; Wang, B. Environmental DNA metabarcoding of intertidal meiofauna sheds light on its potential for habitat discovery. Ecol. Indic. 2023, 150, 110223. [Google Scholar] [CrossRef]
- Plante, C.J.; Hill-Spanik, K.; Cook, M.; Graham, C. Environmental and spatial influences on biogeography and community structure of saltmarsh benthic diatoms. Estuar. Coasts 2021, 44, 147–161. [Google Scholar] [CrossRef]
- Hovel, K.A.; Morgan, S.G. Susceptibility of estuarine crab larvae to ultraviolet radiation. J. Exp. Mar. Biol. Ecol. 1999, 237, 107–125. [Google Scholar] [CrossRef]
- Noël, P.E.; Chmura, G.L. Spatial and environmental variability of pools on a natural and a recovering salt marsh in the Bay of Fundy. J. Coast. Res. 2011, 27, 847–856. [Google Scholar] [CrossRef]
- Smith, K.J.; Able, K.W. Dissolved oxygen dynamics in salt marsh pools and its potential impacts on fish assemblages. Mar. Ecol. Prog. Ser. 2003, 258, 223–232. [Google Scholar] [CrossRef]
- Kearns, P.J.; Holloway, D.; Angell, J.H.; Feinman, S.G.; Bowen, J.L. Effect of short-term, diel changes in environmental conditions on active microbial communities in a salt marsh pond. Aquat. Microb. Ecol. 2017, 80, 29–41. [Google Scholar] [CrossRef]
- Barnby, M.A.; Collins, J.N.; Resh, V.H. Aquatic macroinvertebrate communities of natural and ditched potholes in a San Francisco Bay salt marsh. Estuar. Coast. Shelf Sci. 1985, 20, 331–347. [Google Scholar] [CrossRef]
- Meredith, W.H.; Saveikis, D.E.; Stachecki, C.J. Guidelines for “Open Marsh Water Management” in Delaware’s salt marshes—Objectives, system designs and installation procedures. Wetlands 1985, 5, 119–133. [Google Scholar] [CrossRef]
- Powell, E.B.; Krause, J.R.; Martin, R.M.; Watson, E.B. Pond excavation reduces coastal wetland carbon dioxide assimilation. J. Geophys. Res.-Biogeosciences 2020, 1252, 1–19. [Google Scholar] [CrossRef]
- Clarke, J.A.; Harrington, B.A.; Hruby, T.; Waserman, F.E. The effect of ditching for mosquito control on salt marsh use by birds in Rowley, Massachusetts. J. Field Ornithol. 1984, 55, 160–180. [Google Scholar]
- James-Pirri, M.J.; Ginsberg, H.S.; Erwin, R.M.; Taylor, J. Effects of open marsh water management on numbers of larval salt marsh mosquitoes. J. Med. Entomol. 2009, 46, 1392–1399. [Google Scholar] [CrossRef]
- Rochlin, I.; James-Pirri, M.J.; Adamowicz, S.C.; Dempsey, M.E.; Iwanejko, T.; Ninivaggi, D.V. The effects of integrated marsh management (IMM) on salt marsh vegetation, nekton, and birds. Estuar. Coasts 2012, 35, 727–742. [Google Scholar] [CrossRef]
- Kermarrec, L.; Franc, A.; Rimet, F.; Chaumeil, P.; Humbert, J.F.; Bouchez, A. Next-generation sequencing to inventory taxonomic diversity in eukaryotic communities: A test for freshwater diatoms. Mol. Ecol. Resour. 2013, 13, 607–619. [Google Scholar] [CrossRef]
- Pérez-Burillo, J.; Valoti, G.; Witkowski, A.P.; Prado, P.; Mann, D.G.; Trobajo, R. Assessment of marine benthic diatom communities: Insights from a combined morphological–metabarcoding approach in Mediterranean shallow coastal waters. Mar. Pollut. Bull. 2022, 174, 113183. [Google Scholar] [CrossRef]
- Turk Dermastia., T.; Vascotto, I.; Francé, J.; Stanković, D.; Mozetič, P. Evaluation of the rbcL marker for metabarcoding of marine diatoms and inference of population structure of selected genera. Front. Microbiol. 2023, 14, 1071379. [Google Scholar] [CrossRef]
- Vasselon, V.; Rimet, F.; Tapolczai, K.; Bouchez, A. Assessing ecological status with diatoms DNA metabarcoding: Scaling-up on a WFD monitoring network (Mayotte island, France). Ecol. Indic. 2017, 82, 1–12. [Google Scholar] [CrossRef]
- Thompson, L.R.; Sanders, J.G.; McDonald, D.; Amir, A.; Jansson, J.K.; Gilbert, J.A.; Knight, R. The Earth Microbiome Project Consortium. A communal catalogue reveals Earth’s multiscale microbial diversity. Nature 2017, 551, 457–463. [Google Scholar] [CrossRef]
- Gohl, D.; Vangay, P.; Garbe, J.; MacLean, A.; Hauge, A.; Becker, A.; Gould, T.J.; Clayton, J.B.; Johnson, T.J.; Hunter, R.; et al. Systematic improvement of amplicon marker gene methods for increased accuracy in microbiome studies. Nat. Biotechnol. 2016, 34, 942–949. [Google Scholar] [CrossRef]
- R Core Team. R. A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2023; Available online: https://www.R-project.org/ (accessed on 1 October 2023).
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Morien, E.; Parfrey, L.W. SILVA v128 and v132 dada2 Formatted 18s ‘train sets’ (1.0) [Data Set]. Zenodo. 2018. Available online: https://zenodo.org/records/1447330 (accessed on 1 October 2023).
- Guillou, L.; Bachar, D.; Audic, S.; Bass, D.; Berney, C.; Bittner, L.; Boutte, C.; Burgaud, G.; de Vargas, C.; Decelle, J.; et al. The Protist ribosomal reference database (PR2): A catalog of unicellular eukaryote small sub-unit rRNA sequences with curated taxonomy. Nucleic Acids Res. 2013, 41, D597–D604. [Google Scholar] [CrossRef]
- Rimet, F.; Gusev, E.; Kahlert, M.; Kelly, M.G.; Kulikovskiy, M.; Maltsev, Y.; Mann, D.G.; Pfannkuchen, M.; Trobajo, R.; Vasselon, V.; et al. Diat.Barcode, an open-access curated barcode library for diatoms. Sci. Rep. 2019, 9, 1. [Google Scholar] [CrossRef]
- Desianti, N.; Potapova, M. Diatom Flora of the New Jersey Coastal Wetlands. Report for the New Jersey Department of Environmental Protection. 2019. Available online: https://hdl.handle.net/10929/68423 (accessed on 1 September 2023).
- Krammer, K.; Lange-Bertalot, H. Bacillariophyceae. 2. Teil: Bacillariaceae, Epithemiaceae, Surirellaceae. In Susswasserflora von Mitteleuropa; Ettl, H., Gerloff, J., Heynig, H., Mollenhauer, D., Eds.; Gustav Fisher Verlag: Jena, Germany, 1988; Band 2/2; pp. 1–596. [Google Scholar]
- Spaulding, S.; Potapova, M.G.; Bishop, I.W.; Lee, S.S.; Gasperak, T.S.; Jovanoska, E.; Furey, P.C.; Edlund, M.B. Diatoms.org: Supporting taxonomists, connecting communities. Diatom Res. 2021, 36, 291–304. [Google Scholar] [CrossRef]
- Stepanek, J.G.; Kociolek, J.P. Amphora and Halamphora from coastal and inland waters of the United States and Japan; Bibliotheca Diatomologica; Schweizerbart Science Publishers: Stuttgart, Germany, 2018; Volume 66, pp. 1–260. [Google Scholar]
- Witkowski, A.; Lange-Bertalot, H.; Metzeltin, D. Diatom Flora of Marine Coasts I; Iconographia Diatomologica; A.R.G. Gantner Verlag: Ruggell, Liechtenstein, 2000; Volume 7, pp. 1–925. [Google Scholar]
- Oksanen, J.; Simpson, G.; Blanchet, F.; Kindt, R.; Legendre, P.; Minchin, P.; O’Hara, R.; Solymos, P.; Stevens, M.; Szoecs, E.; et al. Vegan: Community Ecology Package. R Package Version 2.6-4. 2022. Available online: https://CRAN.R-project.org/package=vegan (accessed on 1 October 2023).
- Fernandes, A.D.; Macklaim, J.M.; Linn, T.G.; Reid, G.; Gloor, G.B. ANOVA-Like Differential Expression (Aldex) Analysis for Mixed Population Rna-Seq. PLoS ONE 2013, 8, e67019. [Google Scholar] [CrossRef] [PubMed]
- Gloor, G.B.; Macklaim, J.M.; Fernandes, A.D. Displaying variation in large datasets: A visual summary of effect sizes. J. Comput. Graph. Stat. 2016, 25, 971–979. [Google Scholar] [CrossRef]
- De Cáceres, M.; Legendre, P. Associations between species and groups of sites: Indices and statistical inference. Ecology 2009, 90, 3566–3574. [Google Scholar] [CrossRef] [PubMed]
- Anderson, M.J. Permutational Multivariate Analysis of Variance (PERMANOVA). In Wiley StatsRef: Statistics Reference Online; Balakrishnan, N.T., Colton, T., Everitt, B., Piegorsch, W., Ruggeri, F., Teugels, J.L., Eds.; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2017; pp. 1–15. [Google Scholar] [CrossRef]
- Nearing, J.T.; Douglas, G.M.; Hayes, M.G.; MacDonald, J.; Desai, D.K.; Allward, N.; Jones, C.M.A.; Wright, R.J.; Dhanani, A.S.; Comeau, A.M.; et al. Microbiome differential abundance methods produce different results across 38 datasets. Nat. Commun. 2022, 13, 342. [Google Scholar] [CrossRef] [PubMed]
- Piredda, R.; Tomasino, M.P.; D’Erchia, A.M.; Manzari, C.; Pesole, G.; Montresor, M.; Kooistra, W.H.; Sarno, D.; Zingone, A. Diversity and temporal patterns of planktonic protist assemblages at a Mediterranean Long Term Ecological Research site. FEMS Microbiol. Ecol. 2017, 93, fiw200. [Google Scholar] [CrossRef] [PubMed]
- Sildever, S.; Laas, P.; Kolesova, N.; Lips, I.; Lips, U.; Nagai, S. Plankton biodiversity and species co-occurrence based on environmental DNA—A multiple marker study. Metabarcoding Metagenom. 2021, 5, e72371. [Google Scholar] [CrossRef]
- Marangoni, R.; Paris, D.; Melck, D.; Fulgentini, L.; Colombetti, G.; Motta, A. In vivo NMR metabolic profiling of Fabrea salina reveals sequential defense mechanisms against ultraviolet radiation. Biophys. J. 2011, 100, 215–224. [Google Scholar] [CrossRef] [PubMed]
- Repak, A.J.; Anderson, O.R. The fine structure of the encysting salt marsh heterotrich ciliate Fabrea salina. J. Morphol. 1990, 205, 335–341. [Google Scholar] [CrossRef]
- Zhang, B.; Hou, L.; Qi, H.; Hou, L.; Zhang, T.; Zhao, F.; Miao, M. An extremely streamlined macronuclear genome in the free-living protozoan Fabrea salina. Mol. Biol. Evol. 2022, 39, msac062. [Google Scholar] [CrossRef]
- Fan, X.; Lin, X.; Liu, W.; Xu, Y.; Al-Farraj, S.A.; Al-Rasheid, K.A.S.; Warren, A. Morphology of three new marine Frontonia species (Ciliophora; Peniculida) with note on the phylogeny of this genus. Eur. J. Protistol. 2013, 49, 312–323. [Google Scholar] [CrossRef]
- Liu, W.; Xu, D.; Ma, H.; Al-Farraj, S.A.; Warren, A.; Yi, Z. Taxonomy and molecular systematics of three oligotrich (s.l.) ciliates including descriptions of two new species, Strombidium guangdongense sp. nov. and Strombidinopsis sinicum sp. nov. (Protozoa, Ciliophora). Syst. Biodivers. 2016, 14, 452–465. [Google Scholar] [CrossRef]
- Fenchel, T.; Kristensen, L.D.; Rasmussen, L. Water column anoxia: Vertical zonation of planktonic protozoa. Mar. Ecol. Prog. Ser. 1990, 62, 1–10. [Google Scholar] [CrossRef]
- Fenchel, T.; Bernard, C. A purple protist. Nature 1993, 362, 300. [Google Scholar] [CrossRef] [PubMed]
- Johnson, M.D.; Stoecker, D.K.; Marshall, H.G. Seasonal dynamics of Mesodinium rubrum in Chesapeake Bay. J. Plankton Res. 2013, 35, 877–893. [Google Scholar] [CrossRef]
- Ren, L. Baseline Characterization of Phytoplankton and Harmful Algal Blooms in Barnegat Bay-Little Egg Harbor, New Jersey (Year One). Final Report; New Jersey Department of Environmental Protection, Division of Science, Research and Environmental Health: Trenton, NJ, USA, 2013; pp. 1–44. [Google Scholar]
- Snyder, R.A.; Moss, J.A.; Santoferrara, L.; Head, M.; Jeffrey, W.H. Ciliate microzooplankton from the Northeastern Gulf of Mexico. ICES J. Mar. Sci. 2021, 78, 3356–3371. [Google Scholar] [CrossRef]
- Zamora-Terol, S.; Novotny, A.; Winder, M. Reconstructing marine plankton food web interactions using DNA metabarcoding. Mol Ecol. 2020, 29, 3380–3395. [Google Scholar] [CrossRef] [PubMed]
- Carty, S.; Cox, E.R. Kansodinium gen. nov. and Durinskia gen. nov.: Two genera of freshwater dinoflagellates (Pyrrhophyta). Phycologia 1986, 25, 197–204. Phycologia 1986, 25, 197–204. [Google Scholar] [CrossRef]
- Moestrup, Ø.; Hakanen, P.; Hansen, G.; Daugbjerg, N.; Ellegaard, M. On Levanderina fissa gen. & comb. nov. (Dinophyceae) (syn. Gymnodinium fissum, Gyrodinium instriatum, Gyr. uncatenum), a dinoflagellate with a very unusual sulcus. Phycologia 2014, 53, 265–292. [Google Scholar] [CrossRef]
- Tillmann, U.; Wietkamp, S.; Kretschmann, J.; Chacón, J.; Gottschling, M. Spatial fragmentation in the distribution of diatom endosymbionts from the taxonomically clarified dinophyte Kryptoperidinium triquetrum (=Kryptoperidinium foliaceum, Peridiniales). Sci. Rep. 2023, 13, 8593. [Google Scholar] [CrossRef]
- Lundholm, N.; Churro, C.; Escalera, L.; Fraga, S.; Hoppenrath, M.; Iwataki, M.; Larsen, J.; Mertens, K.; Moestrup, Ø.; Tillmann, U.; et al. 2009 Onwards. IOC-UNESCO Taxonomic Reference List of Harmful Micro Algae. Available online: https://www.marinespecies.org/hab (accessed on 1 October 2023).
- Anderson, D.M.; Fensin, E.; Gobler, C.J.; Hoeglund, A.E.; Hubbard, K.A.; Kulis, D.M.; Landsberg, J.H.; Lefebvre, K.A.; Provoost, P.; Richlen, M.L.; et al. Marine harmful algal blooms (HABs) in the United States: History, current status and future trends. Harmful Algae 2021, 102, 101975. [Google Scholar] [CrossRef]
- Lowe, C.D.; Day, A.; Kemp, S.J.; Montagnes, D.J.S. There are high levels of functional and genetic diversity in Oxyrrhis marina. J. Eukaryot. Microbiol. 2005, 52, 250–257. [Google Scholar] [CrossRef]
- Käse, L.; Metfies, K.; Neuhaus, S.; Boersma, M.; Wiltshire, K.H.; Kraberg, A.C. Host-parasitoid associations in marine planktonic time series: Can metabarcoding help reveal them? PLoS ONE 2021, 16, e0244817. [Google Scholar] [CrossRef] [PubMed]
- Olsen, P.S.; Mahoney, J.B. Phytoplankton in the Barnegat Bay-Little Egg harbor estuarine system: Species composition and picoplankton bloom development. J. Coast. Res. 2001, SI 32, 115–143. [Google Scholar]
- Robicheau, B.M.; Tolman, J.; Bertrand, E.M.; LaRoche, J. Highly-resolved interannual phytoplankton community dynamics of the coastal Northwest Atlantic. ISME Commun. 2022, 2, 38. [Google Scholar] [CrossRef] [PubMed]
- Huapaya, K.; Echeveste, P. Physiological responses of Humboldt current system diatoms to Fe and Cu co-limitation. Mar. Environ. Res. 2023, 187, 105937. [Google Scholar] [CrossRef] [PubMed]
- McQuoid, M.R.; Hobson, L.A. Diatom resting stages. J. Phycol. 1996, 32, 889–902. [Google Scholar] [CrossRef]
- Sullivan, M.J. Diatom communities from a Delaware salt marsh. J. Phycol. 1975, 11, 384–390. [Google Scholar] [CrossRef]
- Tomas, C.R. Identifying Marine Phytoplankton; Academic Press: San Diego, CA, USA, 1997; pp. 1–858. [Google Scholar]
- Tragin, M.; Vaulot, D. Green microalgae in marine coastal waters: The Ocean Sampling Day (OSD) dataset. Sci. Rep. 2018, 8, 14020. [Google Scholar] [CrossRef] [PubMed]
- Bass, D.; Cavalier-Smith, T. Phylum-specific environmental DNA analysis reveals remarkably high global biodiversity of Cercozoa (Protozoa). Int. J. Syst. Evol. Microbiol. 2004, 54, 2393–2404. [Google Scholar] [CrossRef]
- Suter, E.A.; Pachiadaki, M.; Taylor, G.T.; Edgcomb, V.P. Eukaryotic parasites are integral to a productive microbial food web in oxygen-depleted waters. Front. Microbiol. 2022, 12, 764605. [Google Scholar] [CrossRef]
- More, K.; Simpson, A.G.B.; Hess, S. Two new marine species of Placopus (Vampyrellida, Rhizaria) that perforate the theca of Tetraselmis (Chlordendrales, Viridiplantae). J. Eukaryot. Microbiol. 2019, 66, 560–573. [Google Scholar] [CrossRef]
- Yamaguchi, A.; Yubuki, N.; Leander, B.S. Morphostasis in a novel eukaryote illuminates the evolutionary transition from phagotrophy to phototrophy: Description of Rapaza viridis n. gen. et sp. (Euglenozoa, Euglenida). BMC Evol. Biol. 2012, 12, 29. [Google Scholar] [CrossRef]
- Frontalini, F.; Cordier, T.; Balassi, E.; Armynot du Chatelet, E.; Cermakova, K.; Apothéloz-Perret-Gentil, L.; Martins, M.V.A.; Bucci, C.; Scantamburlo, E.; Treglia, M.; et al. Benthic foraminiferal metabarcoding and morphology–based assessment around three offshore gas platforms: Congruence and complementarity. Environ. Int. 2020, 144, 106049. [Google Scholar] [CrossRef]
- Morales, S.E.; Biswas, A.; Herndl, G.J.; Baltar, F. Global structuring of phylogenetic and functional diversity of pelagic fungi by depth and temperature. Front. Mar. Sci. 2019, 6, 131. [Google Scholar] [CrossRef]
- Richards, T.A.; Jones, M.D.; Leonard, G.; Bass, D. Marine fungi: Their ecology and molecular diversity. Ann. Rev. Mar. Sci. 2012, 4, 495–522. [Google Scholar] [CrossRef]
- Lee, J.J.; Muller, W.A. Trophic dynamics and niches of salt marsh foraminifera. Am. Zool. 1973, 13, 215–223. [Google Scholar] [CrossRef]
- Burkholder, J.M.; Glibert, M.P.; Skelton, H.M. Mixotrophy, a major mode of nutrition for harmful algal species in eutrophic waters. Harmful Algae 2008, 8, 77–93. [Google Scholar] [CrossRef]
- Mitra, A.; Caron, D.A.; Faure, E.; Flynn, K.J.; Leles, S.G.; Hansen, P.J.; McManus, G.B.; Not, F.; do Rosario Gomes, H.; Santoferrara, L.F.; et al. The Mixoplankton Database (MDB): Diversity of photo-phago-trophic plankton in form, function, and distribution across the global ocean. J. Eukaryot. Microbiol. 2023, 70, e12972. [Google Scholar] [CrossRef]
- Glibert, P.M.; Legrand, C. The diverse nutrient strategies of harmful algae: Focus on osmotrophy. Ecol. Stud. 2006, 189, 163–175. [Google Scholar] [CrossRef]
- Lomas, M.W.; Glibert, P.M.; Clougherty, D.A.; Huber, D.E.; Jones, J.; Alexander, J.; Haramoto, E. Elevated organic nutrient ratios associated with brown tide blooms of Aureococcus anophagefferens (Pelagophyceae). J. Plankton Res. 2001, 23, 1339–1344. [Google Scholar] [CrossRef]
- Gann, E.R.; Truchon, A.R.; Papoulis, S.E.; Dyhrman, S.T.; Gobler, C.J.; Wilhelm, S.W. Aureococcus anophagefferens (Pelagophyceae) genomes improve evaluation of nutrient acquisition strategies involved in brown tide dynamics. J. Phycol. 2022, 58, 146–160. [Google Scholar] [CrossRef] [PubMed]
- Gobler, C.G.; Boneillo, G.E.; Debenham, C.J.; Caron, D.A. Nutrient limitation, organic matter cycling, and plankton dynamics during an Aureococcus anophagefferens bloom. Aquat. Microb. Ecol. 2004, 35, 31–43. [Google Scholar] [CrossRef]
- Gobler, C.J.; Koch, F.; Kang, Y.; Berry, D.L.; Tang, Y.Z.; Lasi, M.; Walters, L.; Hall, L.; Miller, J.D. Expansion of harmful brown tides caused by the pelagophyte, Aureoumbra lagunensis DeYoe et Stockwell, to the US east coast. Harmful Algae 2013, 27, 29–41. [Google Scholar] [CrossRef]
- Keppler, C.J.; Lewitus, A.J.; Ringwood, A.H.; Hoguet, J.; Staton, T. Sublethal cellular effects of short-term raphidophyte and brevetoxin exposures on the eastern oyster Crassostrea virginica. Mar. Ecol. Prog. Ser. 2006, 312, 141–147. [Google Scholar] [CrossRef]
- Dick, M.W. The Peronosporomycetes. In Systematics and Evolution. The Mycota; McLaughlin, D.J., McLaughlin, E.G., Lemke, P.A., Eds.; Springer: Berlin/Heidelberg, Germany, 2001; Volume 7A, pp. 39–72. [Google Scholar]
- Fabian, J.; Zlatanovic, S.; Mutz, M.; Premke, K. Fungal–bacterial dynamics and their contribution to terrigenous carbon turnover in relation to organic matter quality. ISME J. 2017, 11, 415–425. [Google Scholar] [CrossRef]
- Xie, N.; Wang, Z.; Hunt, D.E.; Johnson, Z.I.; He, Y.; Wang, G. Niche partitioning of labyrinthulomycete protists across sharp coastal gradients and their putative relationships with bacteria and fungi. Front. Microbiol. 2022, 13, 906864. [Google Scholar] [CrossRef]
- Hellebust, J.A.; Lewin, J. Heterotrophic nutrition. In The Biology of Diatoms; Werner, D., Ed.; University of California Press: Berkeley, CA, USA, 1977; pp. 169–197. [Google Scholar]
- Anderson, O.R.; Gorrell, T.; Bergen, A.; Kruzansky, R.; Levandowsky, M. Naked amoebas and bacteria in an oil-impacted salt marsh community. Microb. Ecol. 2000, 42, 474–481. [Google Scholar] [CrossRef]
- Lewitus, A.J.; Schmidt, L.B.; Mason, L.J.; Kempton, J.W.; Wilde, S.B.; Wolny, J.L.; Williams, B.J.; Hayes, K.C.; Hymel, S.N.; Keppler, C.J.; et al. Harmful algal blooms in South Carolina residential and golf course ponds. Pop. Environ. 2003, 24, 387–413. [Google Scholar] [CrossRef]
- Sieburth, J.M.; Johnson, P.W.; Hargraves, P.E. Ultrastructure and ecology of Aureococcus anophagefferens gen. et sp. nov. (Chrysophyceae)—The dominant picoplankter during a bloom in Narragansett Bay, Rhode Island, summer 1985. J. Phycol. 1988, 24, 416–425. [Google Scholar] [CrossRef]


| Pool Type | Depth, m | Salinity, psu | pH | Dissolved Oxygen, mg L−1 | Temperature, °C |
|---|---|---|---|---|---|
| Natural | 0.2–0.4–0.6 | 31–31–32 | 7.2–7.5–7.8 | 2.7–4.2–6.2 | 20–21–22 |
| OMWM | 0.3–0.4–0.5 | 28–29–30 | 7.0–7.2–7.4 | 1.9–3.9–5.9 | 21–21–22 |
| Group | Sediment | Water | ||||||
|---|---|---|---|---|---|---|---|---|
| Assemblage Composition (PERMANOVA) | Read Abundance (t-Test) | Assemblage Composition (PERMANOVA) | Read Abundance (t-Test) | |||||
| F | p-Value | T | p-Value | F | p-Value | T | p-Value | |
| All 18S ASVs | 10.4 | 0.001 | na | na | 4.0 | 0.001 | na | na |
| Bacillariophyta_18S | 13.1 | 0.001 | 5.3 | 0.001 | 5.0 | 0.001 | 4.0 | 0.001 |
| Dinoflagellata | 14.6 | 0.001 | −7.1 | 0.001 | 3.7 | 0.001 | 0.1 | ns |
| Ciliophora | 6.4 | 0.001 | 2.0 | 0.054 | 3.4 | 0.001 | 3.6 | 0.004 |
| Nematoda | 7.0 | 0.001 | 2.6 | 0.013 | na | na | 1.3 | ns |
| Rotifera | 6.8 | 0.001 | −5.3 | 0.001 | na | na | −1.2 | ns |
| Foraminifera | 4.6 | 0.001 | 3.4 | 0.002 | na | na | −0.5 | ns |
| Platyhelminthes | na | na | −2.3 | 0.033 | na | na | 0.7 | ns |
| Fungi | 3.6 | 0.003 | −3.5 | 0.003 | 1.4 | 0.113 | 2.6 | 0.024 |
| Pelagophyta | na | na | −3.4 | 0.004 | na | na | −1.0 | ns |
| Raphidophyta | na | na | −7.1 | 0.001 | na | na | −1.0 | ns |
| Euglenida | 6.1 | 0.001 | 2.5 | 0.017 | 1.8 | 0.005 | 1.5 | ns |
| Chlorophyta | 9.5 | 0.001 | −2.5 | 0.024 | 8.0 | 0.002 | −2.4 | 0.046 |
| Apicomplexa | 10.7 | 0.001 | −0.5 | ns | na | na | 2.2 | 0.046 |
| Cercozoa | 14.9 | 0.001 | −0.3 | ns | 5.4 | 0.001 | 0.0 | ns |
| Peronosporomycetes | 4.8 | 0.001 | −3.8 | 0.002 | 1.6 | 0.041 | 0.0 | ns |
| Labyrinthulomycetes | 4.2 | 0.001 | −3.4 | 0.004 | 3.0 | 0.001 | 1.9 | ns |
| Haptophyta | na | na | −4.2 | 0.001 | na | na | 0.3 | ns |
| Chrysophyta | na | na | −3.3 | 0.005 | na | na | −0.2 | ns |
| Bacillariophyta_rbcL | 11.6 | 0.001 | na | na | 4.2 | 0.001 | na | na |
| Bacillariophyta_counts | 1.8 | 0.047 | na | na | na | na | na | na |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Potapova, M.; Markarian, D.; King, A.; Aycock, L. Microbial Eukaryotes in Natural and Artificial Salt Marsh Pools. Coasts 2024, 4, 287-305. https://doi.org/10.3390/coasts4020015
Potapova M, Markarian D, King A, Aycock L. Microbial Eukaryotes in Natural and Artificial Salt Marsh Pools. Coasts. 2024; 4(2):287-305. https://doi.org/10.3390/coasts4020015
Chicago/Turabian StylePotapova, Marina, Daiana Markarian, Abigail King, and Laura Aycock. 2024. "Microbial Eukaryotes in Natural and Artificial Salt Marsh Pools" Coasts 4, no. 2: 287-305. https://doi.org/10.3390/coasts4020015
APA StylePotapova, M., Markarian, D., King, A., & Aycock, L. (2024). Microbial Eukaryotes in Natural and Artificial Salt Marsh Pools. Coasts, 4(2), 287-305. https://doi.org/10.3390/coasts4020015





