Antimicrobial Peptides and Their Assemblies
Abstract
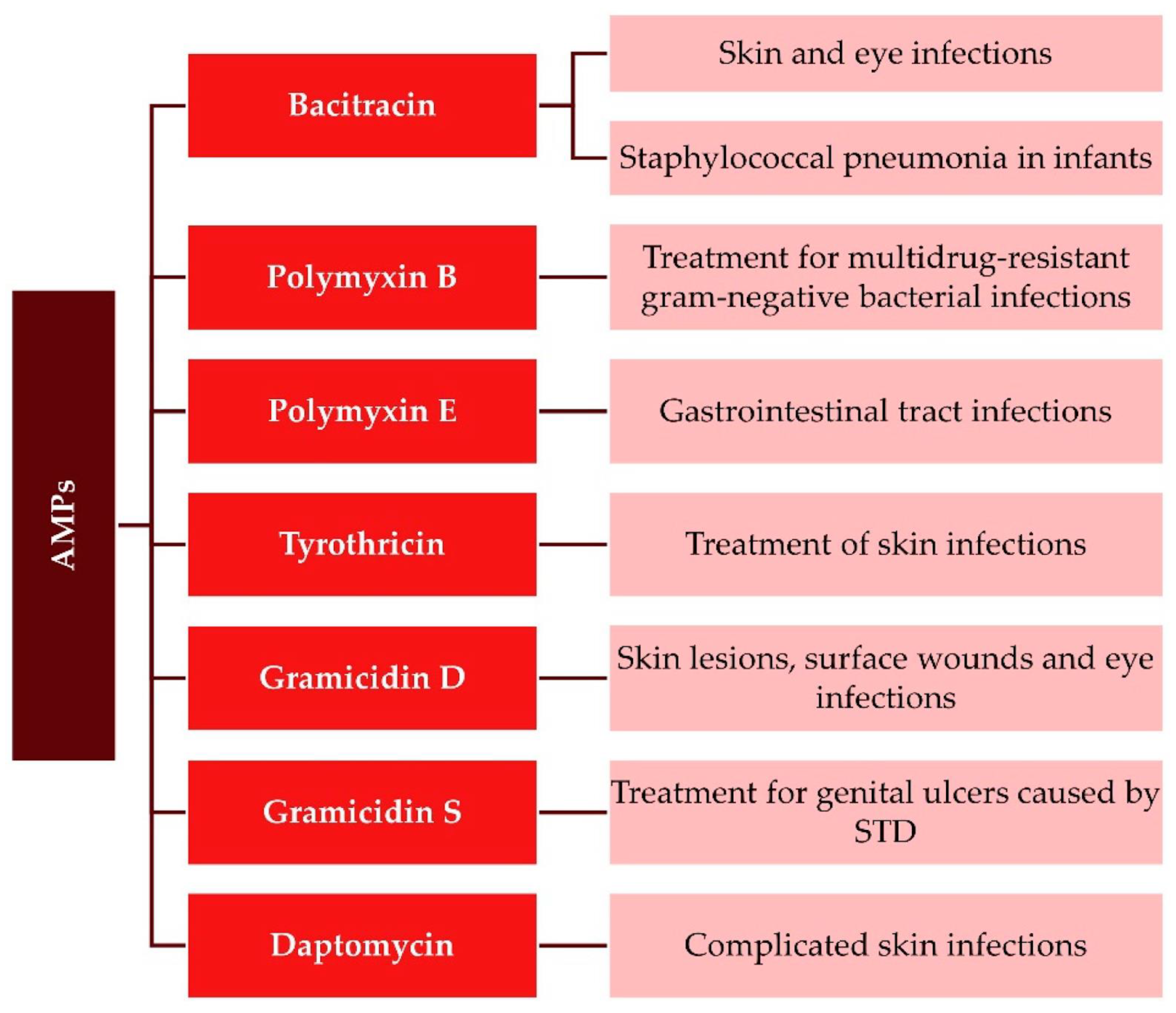
1. Introducing Antimicrobial Peptides in Pharmacology
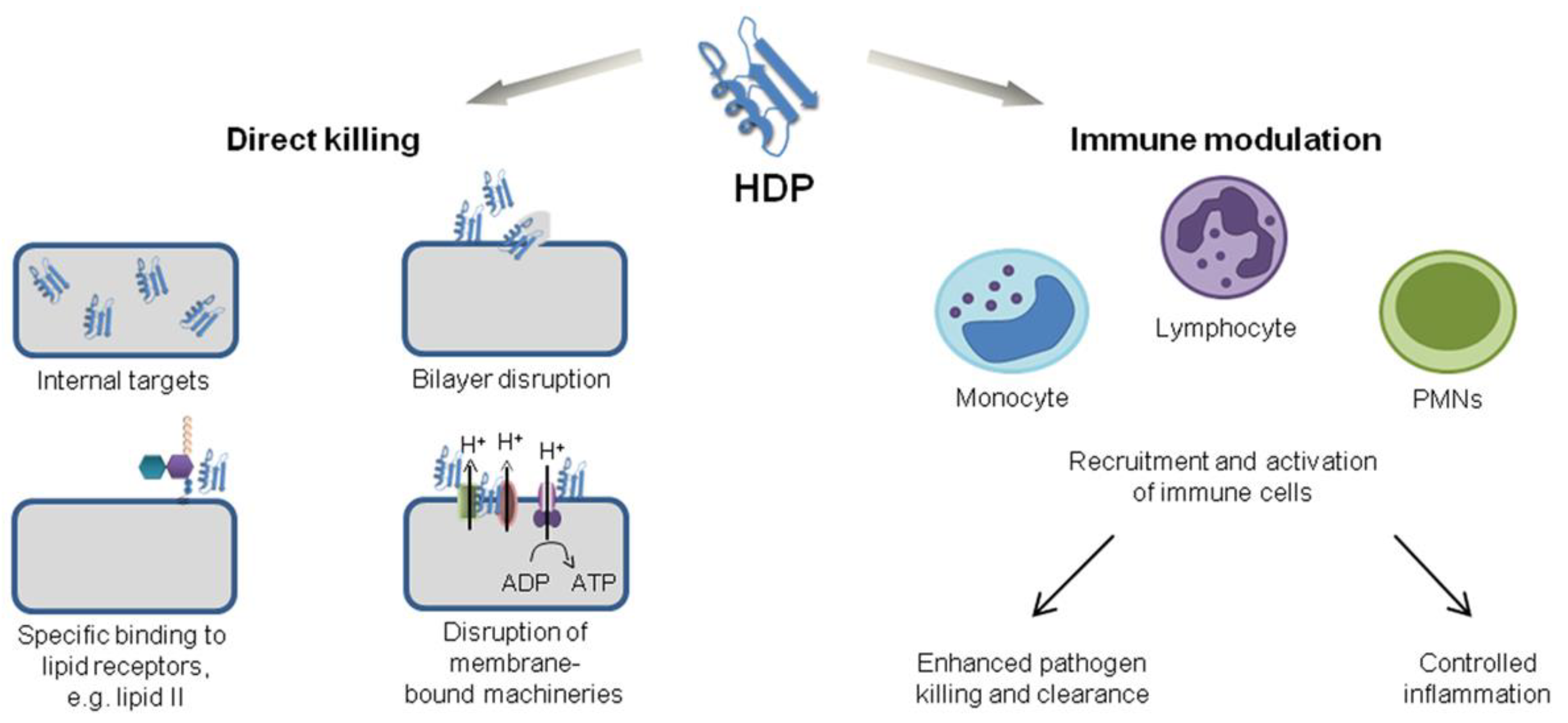
2. Chemical Structure and Mode of Action for AMPs
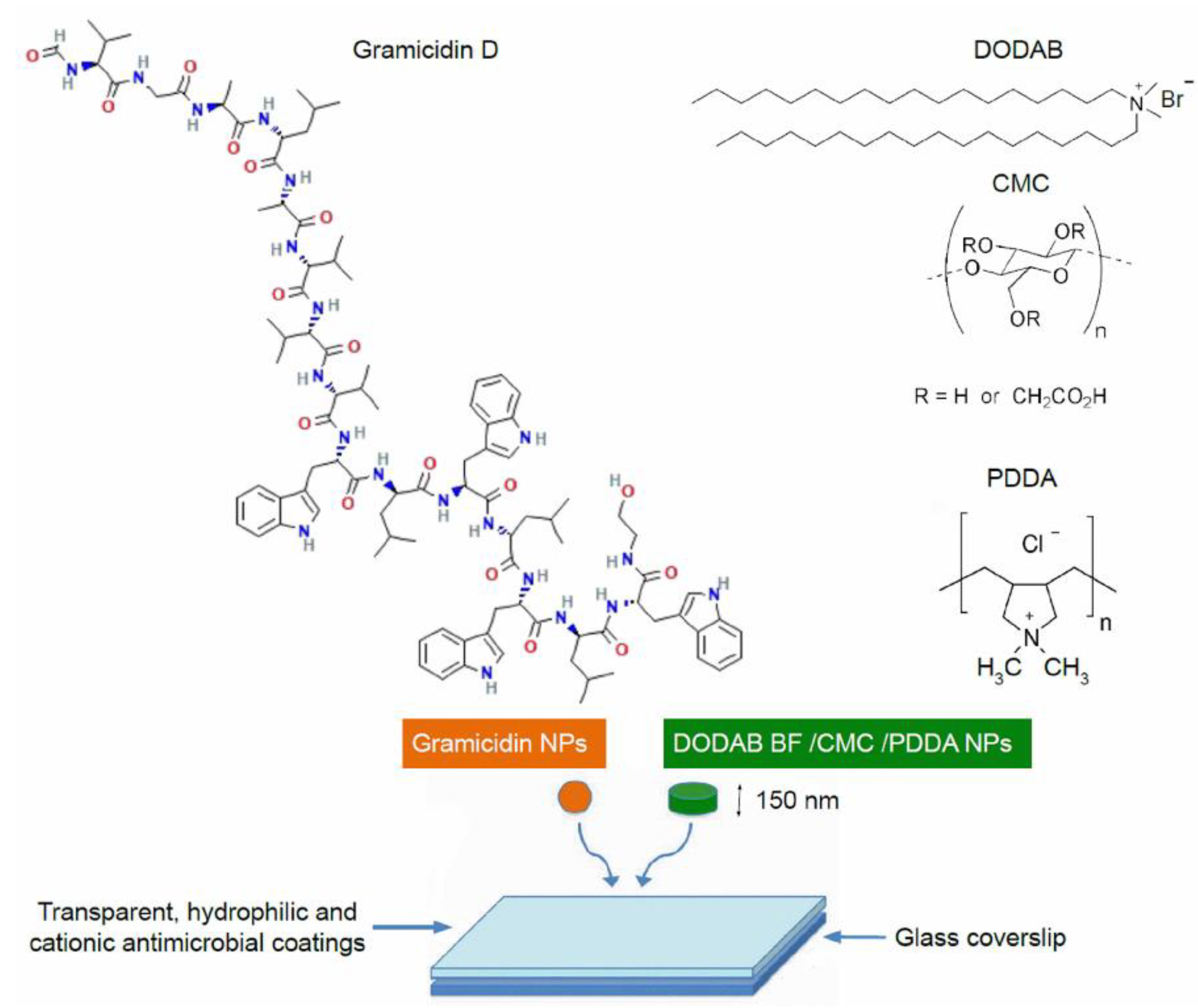
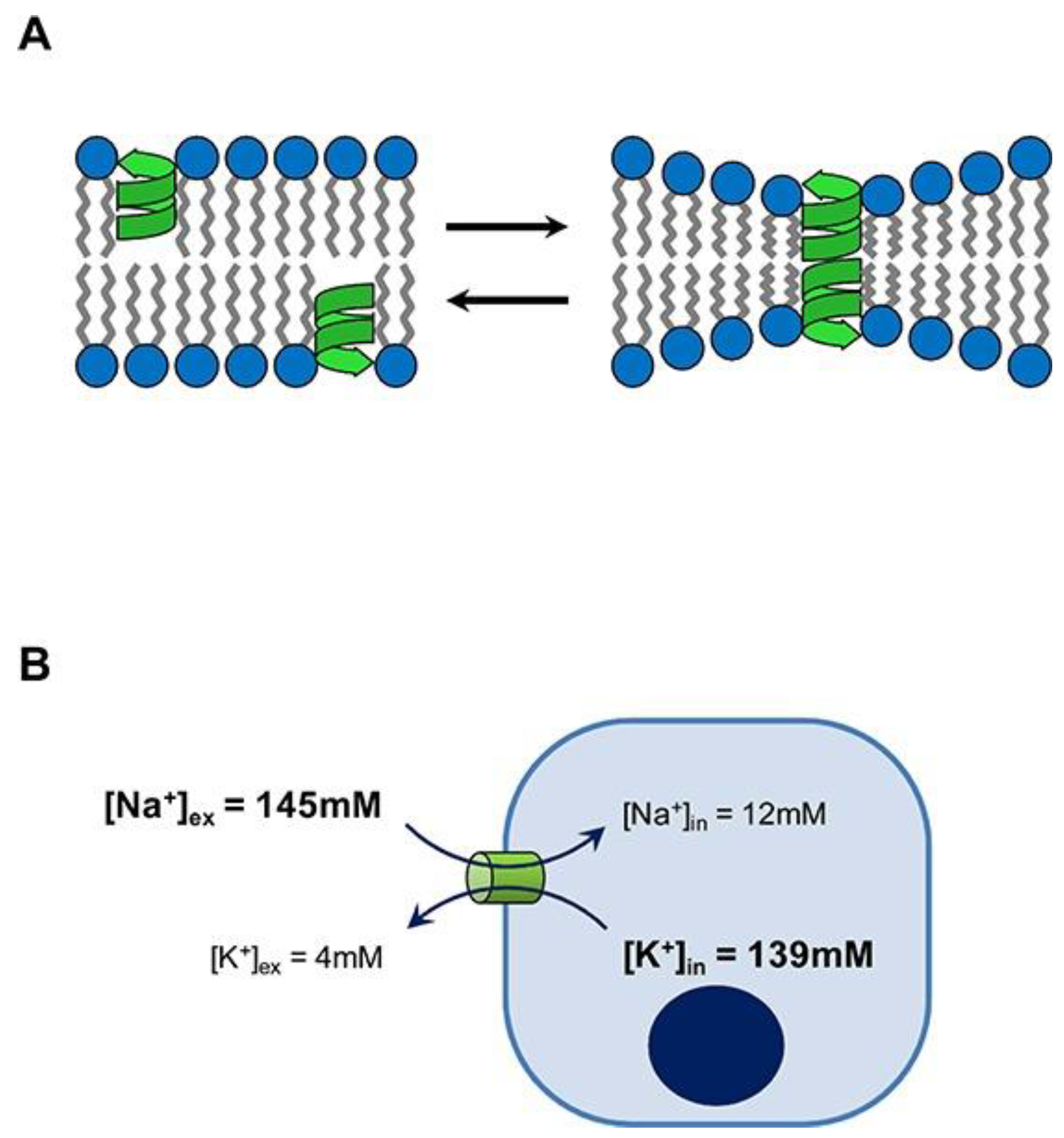
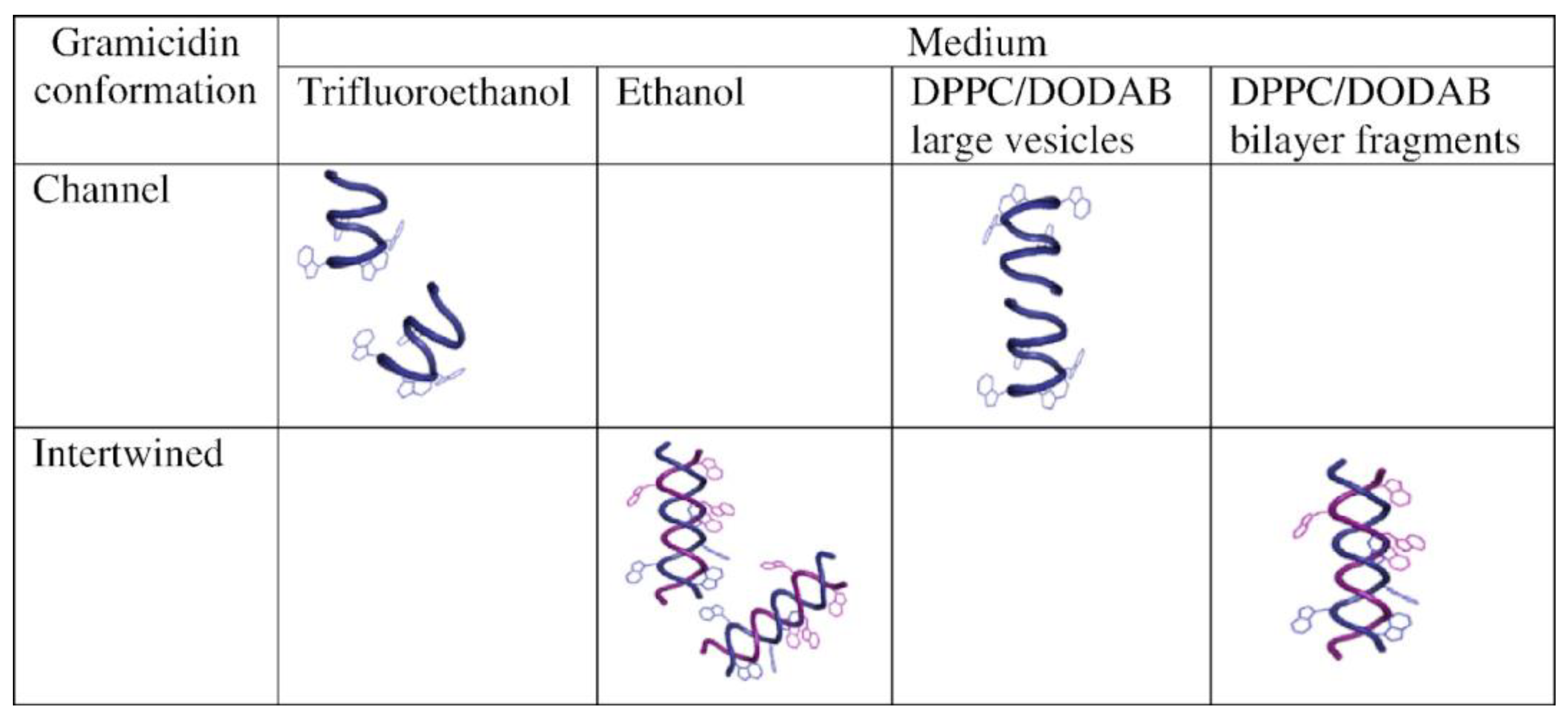
3. Gramicidin D and Its Assemblies
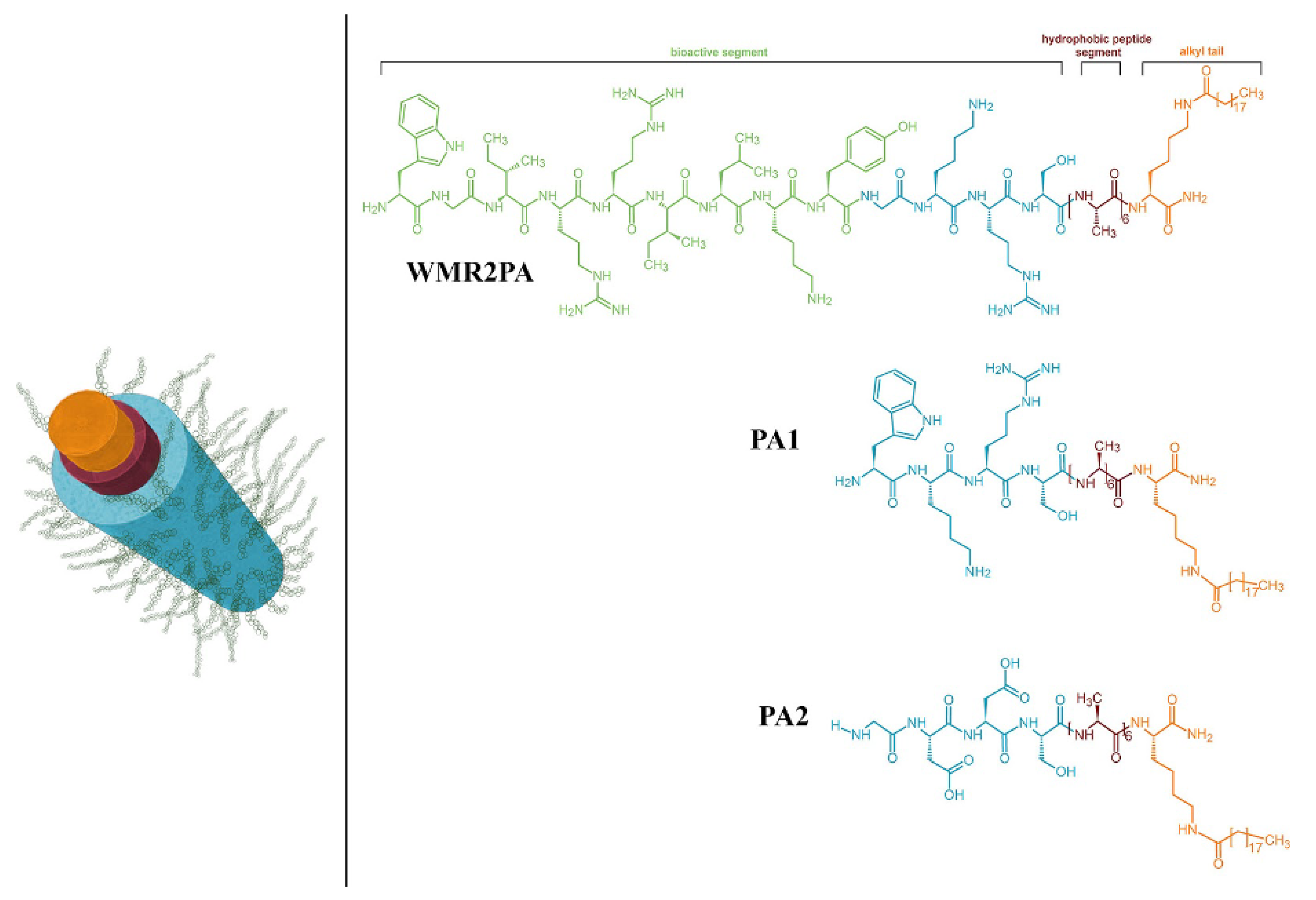
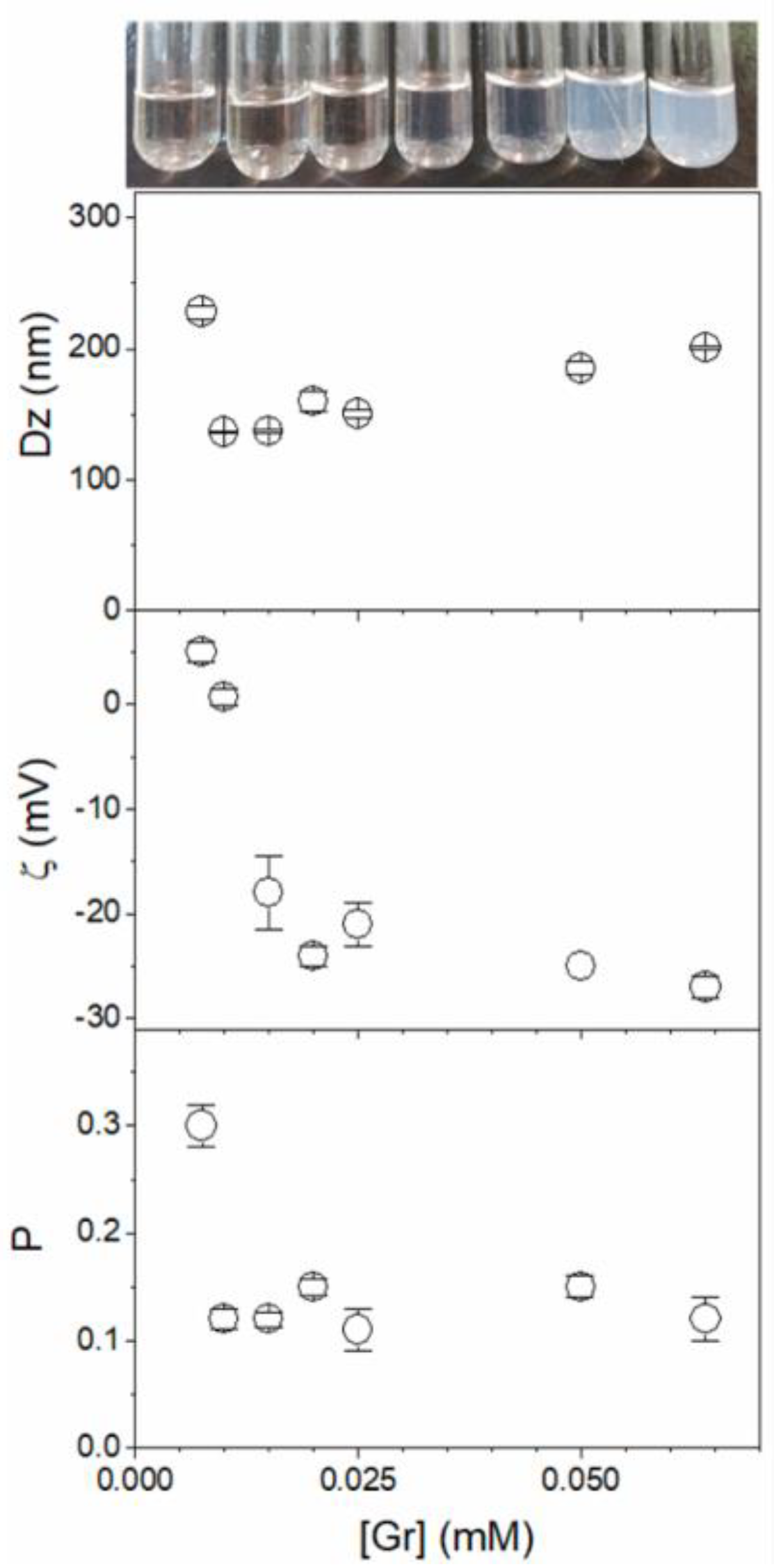
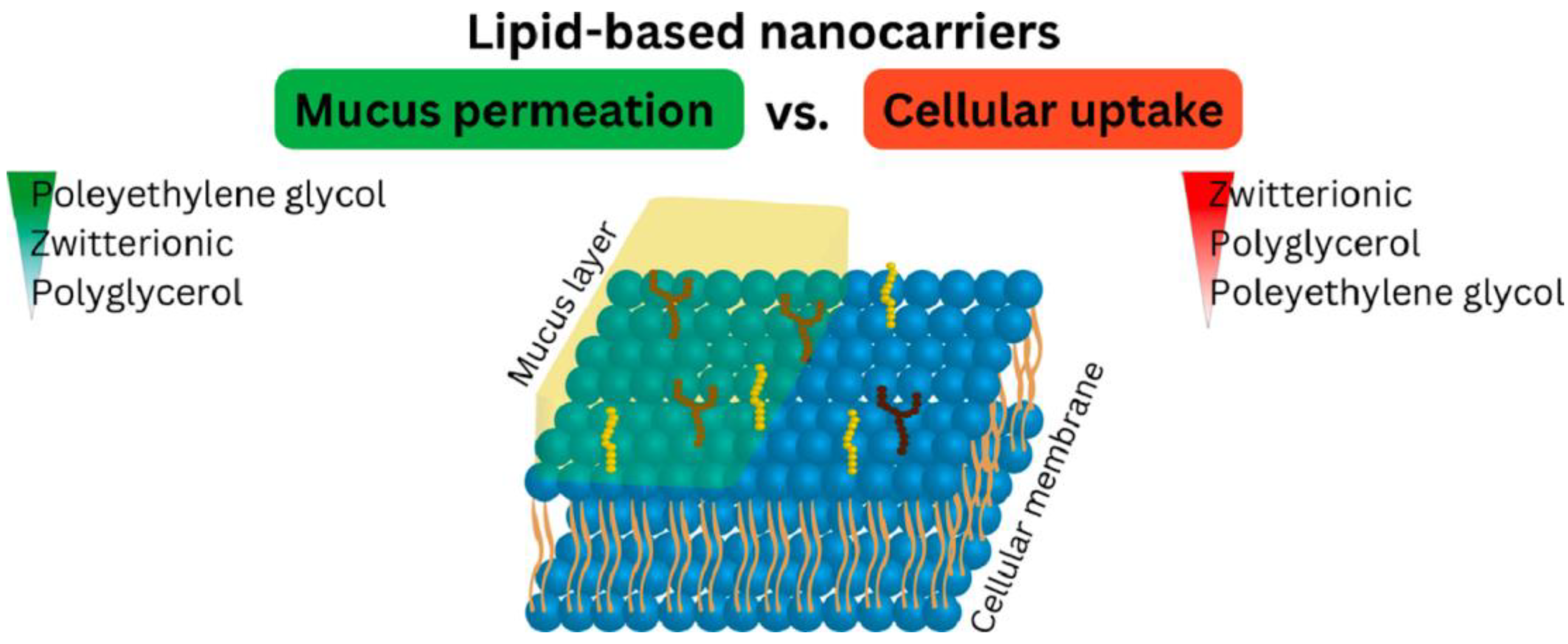
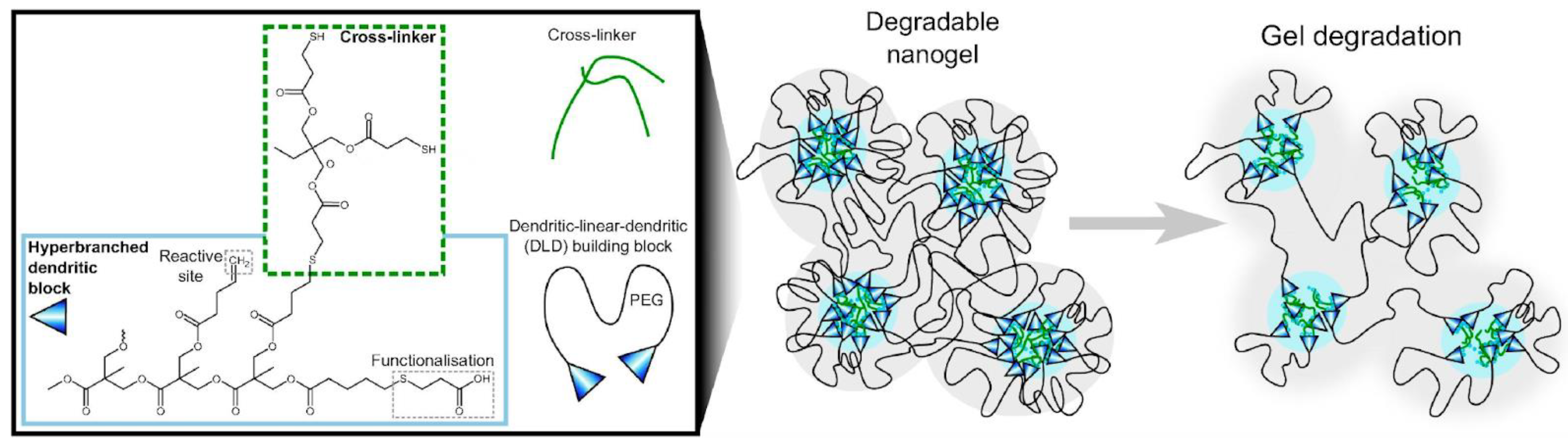
4. Recent Formulations for Antimicrobial Peptides
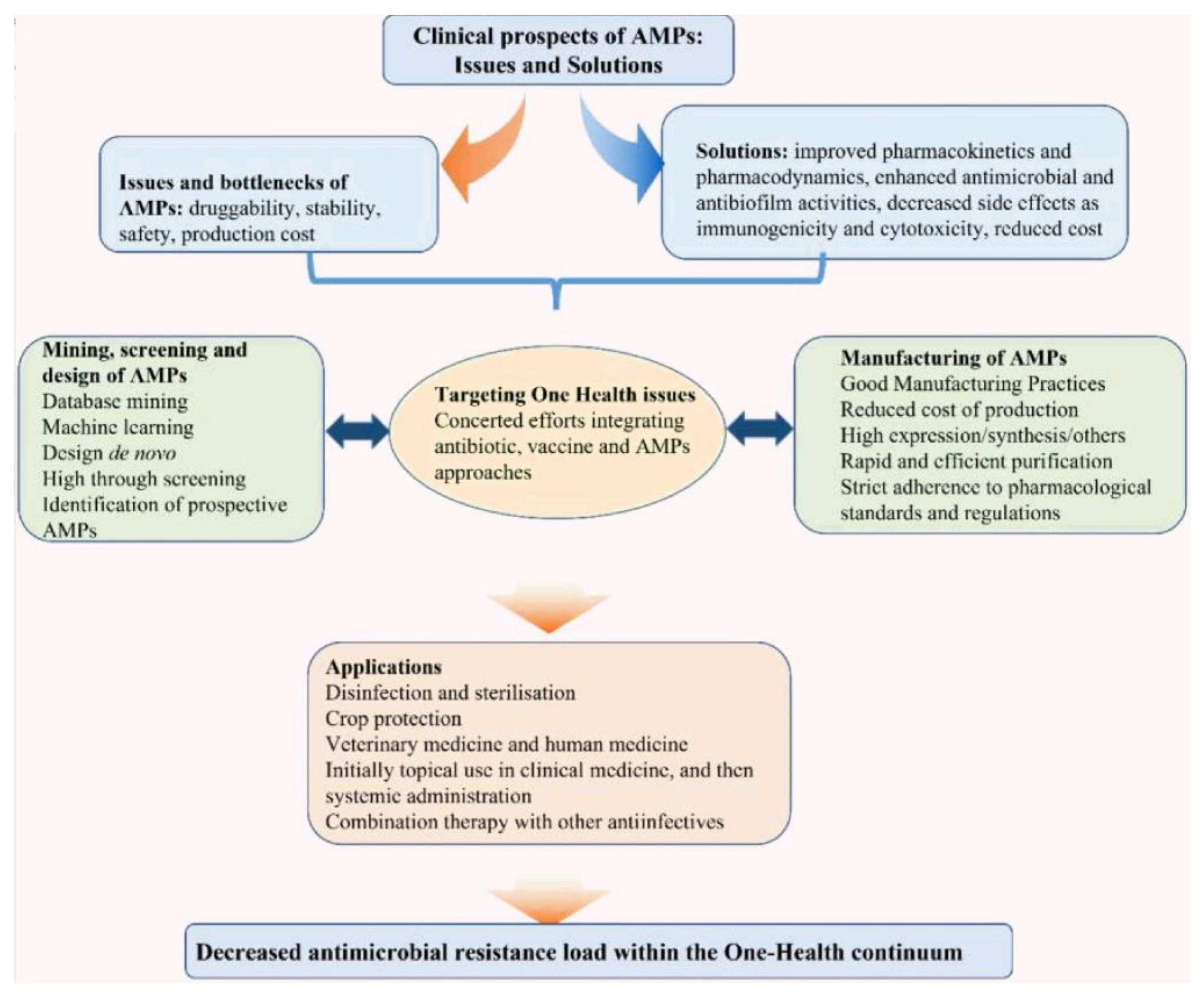
5. Conclusions
Funding
Conflicts of Interest
References
- Bechinger, B. Rationalizing the Membrane Interactions of Cationic Amphipathic Antimicrobial Peptides by Their Molecular Shape. Curr. Opin. Colloid Interface Sci. 2009, 14, 349–355. [Google Scholar] [CrossRef]
- Bechinger, B.; Lohner, K. Detergent-like Actions of Linear Amphipathic Cationic Antimicrobial Peptides. Biochim. Biophys. Acta (BBA)—Biomembr. 2006, 1758, 1529–1539. [Google Scholar] [CrossRef]
- Zhang, C.; Yang, M. Antimicrobial Peptides: From Design to Clinical Application. Antibiotics 2022, 11, 349. [Google Scholar] [CrossRef]
- Talapko, J.; Meštrović, T.; Juzbašić, M.; Tomas, M.; Erić, S.; Horvat Aleksijević, L.; Bekić, S.; Schwarz, D.; Matić, S.; Neuberg, M.; et al. Antimicrobial Peptides—Mechanisms of Action, Antimicrobial Effects and Clinical Applications. Antibiotics 2022, 11, 1417. [Google Scholar] [CrossRef]
- Browne, K.; Chakraborty, S.; Chen, R.; Willcox, M.D.; Black, D.S.; Walsh, W.R.; Kumar, N. A New Era of Antibiotics: The Clinical Potential of Antimicrobial Peptides. Int. J. Mol. Sci. 2020, 21, 7047. [Google Scholar] [CrossRef] [PubMed]
- Pavithrra, G.; Rajasekaran, R. Gramicidin Peptide to Combat Antibiotic Resistance: A Review. Int. J. Pept. Res. Ther. 2020, 26, 191–199. [Google Scholar] [CrossRef]
- Avedissian, S.N.; Liu, J.; Rhodes, N.J.; Lee, A.; Pais, G.M.; Hauser, A.R.; Scheetz, M.H. A Review of the Clinical Pharmacokinetics of Polymyxin B. Antibiotics 2019, 8, 31. [Google Scholar] [CrossRef] [PubMed]
- Field, D.; Fernandez de Ullivarri, M.; Ross, R.P.; Hill, C. After a Century of Nisin Research—Where Are We Now? FEMS Microbiol. Rev. 2023, 47, fuad023. [Google Scholar] [CrossRef]
- Shin, J.M.; Gwak, J.W.; Kamarajan, P.; Fenno, J.C.; Rickard, A.H.; Kapila, Y.L. Biomedical Applications of Nisin. J. Appl. Microbiol. 2016, 120, 1449–1465. [Google Scholar] [CrossRef]
- Vilhena, C.; Bettencourt, A. Daptomycin: A Review of Properties, Clinical Use, Drug Delivery and Resistance. Mini-Rev. Med. Chem. 2012, 12, 202–209. [Google Scholar] [CrossRef]
- Sharma, C.K.; Sharma, M. Up Scaling Strategies to Improve the Industrial Production of Bacitracin at Largescale. Mini-Rev. Med. Chem. 2017, 17, 1548–1556. [Google Scholar] [CrossRef] [PubMed]
- Guan, Q.; Huang, S.; Jin, Y.; Campagne, R.; Alezra, V.; Wan, Y. Recent Advances in the Exploration of Therapeutic Analogues of Gramicidin S, an Old but Still Potent Antimicrobial Peptide. J. Med. Chem. 2019, 62, 7603–7617. [Google Scholar] [CrossRef] [PubMed]
- Ganesan, N.; Mishra, B.; Felix, L.; Mylonakis, E. Antimicrobial Peptides and Small Molecules Targeting the Cell Membrane of Staphylococcus Aureus. Microbiol. Mol. Biol. Rev. 2023, 87, e00037222023. [Google Scholar] [CrossRef] [PubMed]
- Enayathullah, M.G.; Parekh, Y.; Banu, S.; Ram, S.; Nagaraj, R.; Kumar, B.K.; Idris, M.M. Gramicidin S and Melittin: Potential Anti-Viral Therapeutic Peptides to Treat SARS-CoV-2 Infection. Sci. Rep. 2022, 12, 3446. [Google Scholar] [CrossRef]
- Mousavi Maleki, M.S.; Sardari, S.; Ghandehari Alavijeh, A.; Madanchi, H. Recent Patents and FDA-Approved Drugs Based on Antiviral Peptides and Other Peptide-Related Antivirals. Int. J. Pept. Res. Ther. 2023, 29, 5. [Google Scholar] [CrossRef]
- Elnagdy, S.; AlKhazindar, M. The Potential of Antimicrobial Peptides as an Antiviral Therapy against COVID-19. ACS Pharmacol. Transl. Sci. 2020, 3, 780–782. [Google Scholar] [CrossRef]
- Malekkhaiat Häffner, S.; Malmsten, M. Influence of Self-Assembly on the Performance of Antimicrobial Peptides. Curr. Opin. Colloid Interface Sci. 2018, 38, 56–79. [Google Scholar] [CrossRef]
- Juhl, D.W.; Glattard, E.; Aisenbrey, C.; Bechinger, B. Antimicrobial Peptides: Mechanism of Action and Lipid-Mediated Synergistic Interactions within Membranes. Faraday Discuss. 2021, 232, 419–434. [Google Scholar] [CrossRef]
- Bechinger, B.; Salnikov, E.S. The Membrane Interactions of Antimicrobial Peptides Revealed by Solid-State NMR Spectroscopy. Chem. Phys. Lipids 2012, 165, 282–301. [Google Scholar] [CrossRef]
- Lombardi, L.; Shi, Y.; Falanga, A.; Galdiero, E.; de Alteriis, E.; Franci, G.; Chourpa, I.; Azevedo, H.S.; Galdiero, S. Enhancing the Potency of Antimicrobial Peptides through Molecular Engineering and Self-Assembly. Biomacromolecules 2019, 20, 1362–1374. [Google Scholar] [CrossRef]
- Pérez-Betancourt, Y.; Zaia, R.; Evangelista, M.F.; Ribeiro, R.T.; Roncoleta, B.M.; Mathiazzi, B.I.; Carmona-Ribeiro, A.M. Characterization and Differential Cytotoxicity of Gramicidin Nanoparticles Combined with Cationic Polymer or Lipid Bilayer. Pharmaceutics 2022, 14, 2053. [Google Scholar] [CrossRef] [PubMed]
- Cao, F.; Zhu, G.; Song, M.; Zhao, X.; Ma, G.; Zhang, M. Study on the Self-Assembly of Aromatic Antimicrobial Peptides Based on Different PAF26 Peptide Sequences. e-Polymers 2022, 22, 276–284. [Google Scholar] [CrossRef]
- Hu, X.; Liao, M.; Gong, H.; Zhang, L.; Cox, H.; Waigh, T.A.; Lu, J.R. Recent Advances in Short Peptide Self-Assembly: From Rational Design to Novel Applications. Curr. Opin. Colloid Interface Sci. 2020, 45, 1–13. [Google Scholar] [CrossRef]
- Lei, J.; Sun, L.; Huang, S.; Zhu, C.; Li, P.; He, J.; Mackey, V.; Coy, D.H.; He, Q. The Antimicrobial Peptides and Their Potential Clinical Applications. Am. J. Transl. Res. 2019, 11, 3919–3931. [Google Scholar]
- Jiang, Y.; Chen, Y.; Song, Z.; Tan, Z.; Cheng, J. Recent Advances in Design of Antimicrobial Peptides and Polypeptides toward Clinical Translation. Adv. Drug Deliv. Rev. 2021, 170, 261–280. [Google Scholar] [CrossRef]
- Sobczak, M.; Dębek, C.; Olędzka, E.; Kozłowski, R. Polymeric Systems of Antimicrobial Peptides—Strategies and Potential Applications. Molecules 2013, 18, 14122–14137. [Google Scholar] [CrossRef]
- Copling, A.; Akantibila, M.; Kumaresan, R.; Fleischer, G.; Cortes, D.; Tripathi, R.S.; Carabetta, V.J.; Vega, S.L. Recent Advances in Antimicrobial Peptide Hydrogels. Int. J. Mol. Sci. 2023, 24, 7563. [Google Scholar] [CrossRef]
- Haidari, H.; Melguizo-Rodríguez, L.; Cowin, A.J.; Kopecki, Z. Therapeutic Potential of Antimicrobial Peptides for Treatment of Wound Infection. Am. J. Physiol. Cell Physiol. 2023, 324, C29–C38. [Google Scholar] [CrossRef]
- Costa, B.; Martínez-de-Tejada, G.; Gomes, P.A.C.; Martins, M.C.L.; Costa, F. Antimicrobial Peptides in the Battle against Orthopedic Implant-Related Infections: A Review. Pharmaceutics 2021, 13, 1918. [Google Scholar] [CrossRef]
- Luo, Y.; Song, Y. Mechanism of Antimicrobial Peptides: Antimicrobial, Anti-Inflammatory and Antibiofilm Activities. Int. J. Mol. Sci. 2021, 22, 11401. [Google Scholar] [CrossRef]
- Hancock, R.E.W.; Alford, M.A.; Haney, E.F. Antibiofilm Activity of Host Defence Peptides: Complexity Provides Opportunities. Nat. Rev. Microbiol. 2021, 19, 786–797. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Huang, J.; Zhang, W.; Zheng, X.; Wang, H.; Liu, J.; Leng, H.; Yuan, W.; Song, C. Simvastatin-Hydroxyapatite Coatings Prevent Biofilm Formation and Improve Bone Formation in Implant-Associated Infections. Bioact. Mater. 2023, 21, 44–56. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Ma, L.; Sun, X.; Sloan, A.J.; O’Brien-Simpson, N.M.; Li, W. The Overview of Antimicrobial Peptide-Coated Implants against Oral Bacterial Infections. Aggregate 2023, 4, e309. [Google Scholar] [CrossRef]
- Sun, H.; Hong, Y.; Xi, Y.; Zou, Y.; Gao, J.; Du, J. Synthesis, Self-Assembly, and Biomedical Applications of Antimicrobial Peptide-Polymer Conjugates. Biomacromolecules 2018, 19, 1701–1720. [Google Scholar] [CrossRef]
- Patrulea, V.; Borchard, G.; Jordan, O. An Update on Antimicrobial Peptides (AMPs) and Their Delivery Strategies for Wound Infections. Pharmaceutics 2020, 12, 840. [Google Scholar] [CrossRef] [PubMed]
- Riool, M.; Patrulea, V.; Monteiro, C. Antimicrobial Peptide–Polymer Conjugates. Pharmaceutics 2022, 14, 2171. [Google Scholar] [CrossRef]
- Carmona-Ribeiro, A.M.; De Melo Carrasco, L.D. Novel Formulations for Antimicrobial Peptides. Int. J. Mol. Sci. 2014, 15, 18040–18083. [Google Scholar] [CrossRef]
- Carmona-Ribeiro, A.M. Self-Assembled Antimicrobial Nanomaterials. Int. J. Environ. Res. Public Health 2018, 15, 1408. [Google Scholar] [CrossRef]
- Leite, M.L.; da Cunha, N.B.; Costa, F.F. Antimicrobial Peptides, Nanotechnology, and Natural Metabolites as Novel Approaches for Cancer Treatment. Pharmacol. Ther. 2018, 183, 160–176. [Google Scholar] [CrossRef]
- Carratalá, J.V.; Serna, N.; Villaverde, A.; Vázquez, E.; Ferrer-Miralles, N. Nanostructured Antimicrobial Peptides: The Last Push towards Clinics. Biotechnol. Adv. 2020, 44, 107603. [Google Scholar] [CrossRef]
- Radaic, A.; de Jesus, M.B.; Kapila, Y.L. Bacterial Anti-Microbial Peptides and Nano-Sized Drug Delivery Systems: The State of the Art toward Improved Bacteriocins. J. Control. Release 2020, 321, 100–118. [Google Scholar] [CrossRef] [PubMed]
- Zaia, R.; Quinto, G.M.; Camargo, L.C.S.; Ribeiro, R.T.; Carmona-Ribeiro, A.M. Transient Coatings from Nanoparticles Achieving Broad-Spectrum and High Antimicrobial Performance. Pharmaceuticals 2023, 16, 816. [Google Scholar] [CrossRef] [PubMed]
- Melo, L.D.; Mamizuka, E.M.; Carmona-Ribeiro, A.M. Antimicrobial Particles from Cationic Lipid and Polyelectrolytes. Langmuir 2010, 26, 12300–12306. [Google Scholar] [CrossRef]
- de Melo Carrasco, L.D.; Sampaio, J.L.M.; Carmona-Ribeiro, A.M. Supramolecular Cationic Assemblies against Multidrug-Resistant Microorganisms: Activity and Mechanism of Action. Int. J. Mol. Sci. 2015, 16, 6337–6352. [Google Scholar] [CrossRef] [PubMed]
- Vieira, D.B.; Carmona-Ribeiro, A.M. Cationic Nanoparticles for Delivery of Amphotericin B: Preparation, Characterization and Activity in Vitro. J. Nanobiotechnology 2008, 6, 6. [Google Scholar] [CrossRef]
- Carrasco, L.D.d.M.; Bertolucci, R.J.; Ribeiro, R.T.; Sampaio, J.L.M.; Carmona-Ribeiro, A.M. Cationic Nanostructures against Foodborne Pathogens. Front. Microbiol. 2016, 7, 1804. [Google Scholar] [CrossRef]
- Carmona-Ribeiro, A.M.; Araújo, P.M. Antimicrobial Polymer−Based Assemblies: A Review. Int. J. Mol. Sci. 2021, 22, 5424. [Google Scholar] [CrossRef]
- Mookherjee, N.; Anderson, M.A.; Haagsman, H.P.; Davidson, D.J. Antimicrobial Host Defence Peptides: Functions and Clinical Potential. Nat. Rev. Drug Discov. 2020, 19, 311–332. [Google Scholar] [CrossRef]
- Iannella, H.; Luna, C.; Waterer, G. Inhaled Corticosteroids and the Increased Risk of Pneumonia: What’s New? A 2015 Updated Review. Ther. Adv. Respir. Dis. 2016, 10, 235–255. [Google Scholar] [CrossRef]
- Mant, C.T.; Jiang, Z.; Gera, L.; Davis, T.; Nelson, K.L.; Bevers, S.; Hodges, R.S. De Novo Designed Amphipathic α-Helical Antimicrobial Peptides Incorporating Dab and Dap Residues on the Polar Face To Treat the Gram-Negative Pathogen, Acinetobacter Baumannii. J. Med. Chem. 2019, 62, 3354–3366. [Google Scholar] [CrossRef]
- Mishra, B.; Reiling, S.; Zarena, D.; Wang, G. Host Defense Antimicrobial Peptides as Antibiotics: Design and Application Strategies. Curr. Opin. Chem. Biol. 2017, 38, 87–96. [Google Scholar] [CrossRef]
- Jiang, S.; Deslouches, B.; Chen, C.; Di, M.E.; Di, Y.P. Antibacterial Properties and Efficacy of a Novel SPLUNC1-Derived Antimicrobial Peptide, A4-Short, in a Murine Model of Respiratory Infection. mBio 2019, 10, e00226-19. [Google Scholar] [CrossRef] [PubMed]
- Ho, S.; Pothoulakis, C.; Koon, H.W. Antimicrobial Peptides and Colitis. Curr. Pharm. Des. 2013, 19, 40–47. [Google Scholar] [CrossRef]
- Piyadasa, H.; Hemshekhar, M.; Altieri, A.; Basu, S.; van der Does, A.M.; Halayko, A.J.; Hiemstra, P.S.; Mookherjee, N. Immunomodulatory Innate Defence Regulator (IDR) Peptide Alleviates Airway Inflammation and Hyper-Responsiveness. Thorax 2018, 73, 908–917. [Google Scholar] [CrossRef]
- Chow, L.N.Y.; Choi, K.-Y.G.; Piyadasa, H.; Bossert, M.; Uzonna, J.; Klonisch, T.; Mookherjee, N. Human Cathelicidin LL-37-Derived Peptide IG-19 Confers Protection in a Murine Model of Collagen-Induced Arthritis. Mol. Immunol. 2014, 57, 86–92. [Google Scholar] [CrossRef]
- Li, D.; Wang, W.; Shi, H.; Fu, Y.; Chen, X.; Chen, X.; Liu, Y.; Kan, B.; Wang, Y. Gene Therapy with Beta-Defensin 2 Induces Antitumor Immunity and Enhances Local Antitumor Effects. Hum. Gene Ther. 2014, 25, 63–72. [Google Scholar] [CrossRef] [PubMed]
- Felício, M.R.; Silva, O.N.; Gonçalves, S.; Santos, N.C.; Franco, O.L. Peptides with Dual Antimicrobial and Anticancer Activities. Front. Chem. 2017, 5, 5. [Google Scholar] [CrossRef] [PubMed]
- Tornesello, A.L.; Borrelli, A.; Buonaguro, L.; Buonaguro, F.M.; Tornesello, M.L. Antimicrobial Peptides as Anticancer Agents: Functional Properties and Biological Activities. Molecules 2020, 25, 2850. [Google Scholar] [CrossRef] [PubMed]
- Wodlej, C.; Riedl, S.; Rinner, B.; Leber, R.; Drechsler, C.; Voelker, D.R.; Choi, J.-Y.; Lohner, K.; Zweytick, D. Interaction of Two Antitumor Peptides with Membrane Lipids—Influence of Phosphatidylserine and Cholesterol on Specificity for Melanoma Cells. PLoS ONE 2019, 14, e0211187. [Google Scholar] [CrossRef] [PubMed]
- Yang, N.; Aminov, R.; Franco, O.L.; de la Fuente-Nunez, C.; Wang, J. Editorial: Community Series in Antimicrobial Peptides: Molecular Design, Structure Function Relationship and Biosynthesis Optimization. Front. Microbiol. 2023, 14, 1125426. [Google Scholar] [CrossRef]
- Yang, Z.; He, S.; Wu, H.; Yin, T.; Wang, L.; Shan, A. Nanostructured Antimicrobial Peptides: Crucial Steps of Overcoming the Bottleneck for Clinics. Front. Microbiol. 2021, 12, 710199. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.-Y.; Yan, Z.-B.; Meng, Y.-M.; Hong, X.-Y.; Shao, G.; Ma, J.-J.; Cheng, X.-R.; Liu, J.; Kang, J.; Fu, C.-Y. Antimicrobial Peptides: Mechanism of Action, Activity and Clinical Potential. Mil. Med. Res. 2021, 8, 48. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, J.E.; Alford, M.A.; Yung, D.B.Y.; Molchanova, N.; Fortkort, J.A.; Lin, J.S.; Diamond, G.; Hancock, R.E.W.; Jenssen, H.; Pletzer, D.; et al. Self-Assembly of Antimicrobial Peptoids Impacts Their Biological Effects on ESKAPE Bacterial Pathogens. ACS Infect. Dis. 2022, 8, 533–545. [Google Scholar] [CrossRef] [PubMed]
- Jordan, J.B.; Easton, P.L.; Hinton, J.F. Effects of Phenylalanine Substitutions in Gramicidin A on the Kinetics of Channel Formation in Vesicles and Channel Structure in SDS Micelles. Biophys. J. 2005, 88, 224–234. [Google Scholar] [CrossRef] [PubMed]
- Molloy, E.M.; Field, D.; O’ Connor, P.M.; Cotter, P.D.; Hill, C.; Ross, R.P. Saturation Mutagenesis of Lysine 12 Leads to the Identification of Derivatives of Nisin A with Enhanced Antimicrobial Activity. PLoS ONE 2013, 8, e58530. [Google Scholar] [CrossRef]
- Rifkind, D. Prevention by Polymyxin B of Endotoxin Lethality in Mice. J. Bacteriol. 1967, 93, 1463–1464. [Google Scholar] [CrossRef]
- Shoji, H.; Ferrer, R. Potential Survival Benefit and Early Recovery from Organ Dysfunction with Polymyxin B Hemoperfusion: Perspectives from a Real-World Big Data Analysis and the Supporting Mechanisms of Action. J. Anesth. Analg. Crit. Care 2022, 2, 27. [Google Scholar] [CrossRef]
- Katagiri, D.; Ishikane, M.; Asai, Y.; Izumi, S.; Takasaki, J.; Katsuoka, H.; Kondo, I.; Ide, S.; Nakamura, K.; Nakamoto, T.; et al. Direct Hemoperfusion Using a Polymyxin B-Immobilized Polystyrene Column for COVID-19. J. Clin. Apher. 2021, 36, 313–321. [Google Scholar] [CrossRef]
- De Rosa, S.; Cutuli, S.L.; Ferrer, R.; Antonelli, M.; Ronco, C. COVID-19 EUPHAS2 Collaborative Group. Polymyxin B Hemoperfusion in Coronavirus Disease 2019 Patients with Endotoxic Shock: Case Series from EUPHAS2 Registry. Artif. Organs 2021, 45, E187–E194. [Google Scholar] [CrossRef]
- Trimble, M.J.; Mlynárčik, P.; Kolář, M.; Hancock, R.E.W. Polymyxin: Alternative Mechanisms of Action and Resistance. Cold Spring Harb. Perspect. Med. 2016, 6, a025288. [Google Scholar] [CrossRef]
- Moore, R.A.; Bates, N.C.; Hancock, R.E. Interaction of Polycationic Antibiotics with Pseudomonas Aeruginosa Lipopolysaccharide and Lipid A Studied by Using Dansyl-Polymyxin. Antimicrob. Agents Chemother. 1986, 29, 496–500. [Google Scholar] [CrossRef] [PubMed]
- Schröder, G.; Brandenburg, K.; Seydel, U. Polymyxin B Induces Transient Permeability Fluctuations in Asymmetric Planar Lipopolysaccharide/Phospholipid Bilayers. Biochemistry 1992, 31, 631–638. [Google Scholar] [CrossRef] [PubMed]
- Storm, D.R.; Rosenthal, K.S.; Swanson, P.E. Polymyxin and Related Peptide Antibiotics. Annu. Rev. Biochem. 1977, 46, 723–763. [Google Scholar] [CrossRef] [PubMed]
- Nang, S.C.; Azad, M.A.K.; Velkov, T.; Zhou, Q.T.; Li, J. Rescuing the Last-Line Polymyxins: Achievements and Challenges. Pharmacol. Rev. 2021, 73, 679–728. [Google Scholar] [CrossRef]
- Economou, N.J.; Cocklin, S.; Loll, P.J. High-Resolution Crystal Structure Reveals Molecular Details of Target Recognition by Bacitracin. Proc. Natl. Acad. Sci. USA 2013, 110, 14207–14212. [Google Scholar] [CrossRef]
- Storm, D.R. Mechanism of Bacitracin Action: A Specific Lipid-Peptide Interaction. Ann. N. Y. Acad. Sci. 1974, 235, 387–398. [Google Scholar] [CrossRef]
- Toscano, W.A.; Storm, D.R. Bacitracin. Pharmocol. Ther. 1982, 16, 199–210. [Google Scholar] [CrossRef]
- Stone, K.J.; Strominger, J.L. Mechanism of Action of Bacitracin: Complexation with Metal Ion and C55-Isoprenyl Pyrophosphate. Proc. Natl. Acad. Sci. USA 1971, 68, 3223–3227. [Google Scholar] [CrossRef]
- Huang, H.W. DAPTOMYCIN, Its Membrane-Active Mechanism vs. That of Other Antimicrobial Peptides. Biochim. Biophys. Acta Biomembr. 2020, 1862, 183395. [Google Scholar] [CrossRef]
- Reid, D.J.; Dash, T.; Wang, Z.; Aspinwall, C.A.; Marty, M.T. Investigating Daptomycin-Membrane Interactions Using Native MS and Fast Photochemical Oxidation of Peptides in Nanodiscs. Anal. Chem. 2023, 95, 4984–4991. [Google Scholar] [CrossRef]
- Acharya, Y.; Dhanda, G.; Sarkar, P.; Haldar, J. Pursuit of Next-Generation Glycopeptides: A Journey with Vancomycin. Chem. Commun. 2022, 58, 1881–1897. [Google Scholar] [CrossRef] [PubMed]
- Hassan, D.; Omolo, C.A.; Gannimani, R.; Waddad, A.Y.; Mocktar, C.; Rambharose, S.; Agrawal, N.; Govender, T. Delivery of Novel Vancomycin Nanoplexes for Combating Methicillin Resistant Staphylococcus Aureus (MRSA) Infections. Int. J. Pharm. 2019, 558, 143–156. [Google Scholar] [CrossRef] [PubMed]
- Jang, C.H.; Cho, Y.B.; Jang, Y.S.; Kim, M.S.; Kim, G.H. Antibacterial Effect of Electrospun Polycaprolactone/Polyethylene Oxide/Vancomycin Nanofiber Mat for Prevention of Periprosthetic Infection and Biofilm Formation. Int. J. Pediatr. Otorhinolaryngol. 2015, 79, 1299–1305. [Google Scholar] [CrossRef]
- McGuinness, W.A.; Malachowa, N.; DeLeo, F.R. Vancomycin Resistance in Staphylococcus Aureus. Yale J. Biol. Med. 2017, 90, 269–281. [Google Scholar]
- Acaroğlu Degitz, İ.; Hakkı Gazioğlu, B.; Burak Aksu, M.; Malta, S.; Demir Sezer, A.; Eren, T. Antibacterial and Hemolytic Activity of Cationic Polymer-Vancomycin Conjugates. Eur. Polym. J. 2020, 141, 110084. [Google Scholar] [CrossRef]
- Courvalin, P. Vancomycin Resistance in Gram-Positive Cocci. Clin. Infect. Dis. 2006, 42 (Suppl. S1), S25–S34. [Google Scholar] [CrossRef]
- Hammes, W.P.; Neuhaus, F.C. On the Mechanism of Action of Vancomycin: Inhibition of Peptidoglycan Synthesis in Gaffkya Homari. Antimicrob. Agents Chemother. 1974, 6, 722–728. [Google Scholar] [CrossRef]
- Reynolds, P.E. Structure, Biochemistry and Mechanism of Action of Glycopeptide Antibiotics. Eur. J. Clin. Microbiol. Infect. Dis. 1989, 8, 943–950. [Google Scholar] [CrossRef]
- Hallett, J.W.; Wolkowicz, M.I.; Leopold, I.H. Ophthalmic Use of Neosporin. Am. J. Ophthalmol. 1956, 41, 850–853. [Google Scholar] [CrossRef]
- Carmona-Ribeiro, A.M.; de Melo Carrasco, L.D. Cationic Antimicrobial Polymers and Their Assemblies. Int. J. Mol. Sci. 2013, 14, 9906–9946. [Google Scholar] [CrossRef]
- Sanches, L.M.; Petri, D.F.S.; de Melo Carrasco, L.D.; Carmona-Ribeiro, A.M. The Antimicrobial Activity of Free and Immobilized Poly (Diallyldimethylammonium) Chloride in Nanoparticles of Poly (Methylmethacrylate). J. Nanobiotechnol. 2015, 13, 58. [Google Scholar] [CrossRef] [PubMed]
- Galvão, C.N.; Sanches, L.M.; Mathiazzi, B.I.; Ribeiro, R.T.; Petri, D.F.S.; Carmona-Ribeiro, A.M. Antimicrobial Coatings from Hybrid Nanoparticles of Biocompatible and Antimicrobial Polymers. Int. J. Mol. Sci. 2018, 19, 2965. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, R.T.; Galvão, C.N.; Betancourt, Y.P.; Mathiazzi, B.I.; Carmona-Ribeiro, A.M. Microbicidal Dispersions and Coatings from Hybrid Nanoparticles of Poly (Methyl Methacrylate), Poly (Diallyl Dimethyl Ammonium) Chloride, Lipids, and Surfactants. Int. J. Mol. Sci. 2019, 20, 6150. [Google Scholar] [CrossRef]
- de Arauz, L.J.; Jozala, A.F.; Mazzola, P.G.; Vessoni Penna, T.C. Nisin Biotechnological Production and Application: A Review. Trends Food Sci. Technol. 2009, 20, 146–154. [Google Scholar] [CrossRef]
- Jalalifar, S.; Mirzaei, R.; Motallebirad, T.; Razavi, S.; Talebi, M. The Emerging Role of Probiotics and Their Derivatives against Biofilm-Producing MRSA: A Scoping Review. Biomed. Res. Int. 2022, 2022, 4959487. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.G.; Becattini, S.; Moody, T.U.; Shliaha, P.V.; Littmann, E.R.; Seok, R.; Gjonbalaj, M.; Eaton, V.; Fontana, E.; Amoretti, L.; et al. Microbiota-Derived Lantibiotic Restores Resistance against Vancomycin-Resistant Enterococcus. Nature 2019, 572, 665–669. [Google Scholar] [CrossRef] [PubMed]
- Elfiky, S.A.; Mahmoud Ahmed, S.; Elmenshawy, A.M.; Sultan, G.M.; Asser, S.L. Study of the Gut Microbiome as a Novel Target for Prevention of Hospital-Associated Infections in Intensive Care Unit Patients. Acute Crit. Care 2023, 38, 76–85. [Google Scholar] [CrossRef]
- Petrof, E.O.; Dhaliwal, R.; Manzanares, W.; Johnstone, J.; Cook, D.; Heyland, D.K. Probiotics in the Critically Ill: A Systematic Review of the Randomized Trial Evidence. Crit. Care Med. 2012, 40, 3290–3302. [Google Scholar] [CrossRef]
- Chernyshova, D.N.; Tyulin, A.A.; Ostroumova, O.S.; Efimova, S.S. Discovery of the Potentiator of the Pore-Forming Ability of Lantibiotic Nisin: Perspectives for Anticancer Therapy. Membranes 2022, 12, 1166. [Google Scholar] [CrossRef]
- Dijksteel, G.S.; Ulrich, M.M.W.; Middelkoop, E.; Boekema, B.K.H.L. Review: Lessons Learned From Clinical Trials Using Antimicrobial Peptides (AMPs). Front. Microbiol. 2021, 12, 616979. [Google Scholar] [CrossRef]
- Nayab, S.; Aslam, M.A.; Rahman, S.U.; Sindhu, Z.U.D.; Sajid, S.; Zafar, N.; Razaq, M.; Kanwar, R.; Amanullah. A Review of Antimicrobial Peptides: Its Function, Mode of Action and Therapeutic Potential. Int. J. Pept. Res. Ther. 2022, 28, 46. [Google Scholar] [CrossRef]
- Mukhopadhyay, S.; Bharath Prasad, A.S.; Mehta, C.H.; Nayak, U.Y. Antimicrobial Peptide Polymers: No Escape to ESKAPE Pathogens—A Review. World J. Microbiol. Biotechnol. 2020, 36, 131. [Google Scholar] [CrossRef]
- Ulm, H.; Wilmes, M.; Shai, Y.; Sahl, H.-G. Antimicrobial Host Defensins—Specific Antibiotic Activities and Innate Defense Modulation. Front. Immunol. 2012, 3, 249. [Google Scholar] [CrossRef] [PubMed]
- Kelkar, D.A.; Chattopadhyay, A. The Gramicidin Ion Channel: A Model Membrane Protein. Biochim. Biophys. Acta 2007, 1768, 2011–2025. [Google Scholar] [CrossRef] [PubMed]
- Koeppe, R.E.; Anderson, O.S. Engineering the Gramicidin Channel. Annu. Rev. Biophys. Biomol. Struct. 1996, 25, 231–258. [Google Scholar] [CrossRef]
- Sarges, R.; Witkop, B. Gramicidin A. V. The Structure of Valine- and Isoleucine-gramicidin A. J. Am. Chem. Soc. 1965, 87, 2011–2020. [Google Scholar] [CrossRef]
- Urban, B.W.; Hladky, S.B.; Haydon, D.A. Ion Movements in Gramicidin Pores. An Example of Single-File Transport. Biochim. Biophys. Acta (BBA)—Biomembr. 1980, 602, 331–354. [Google Scholar] [CrossRef]
- Mukherjee, S.; Chattopadhyay, A. Motionally Restricted Tryptophan Environments at the Peptide-Lipid Interface of Gramicidin Channels. Biochemistry 1994, 33, 5089–5097. [Google Scholar] [CrossRef]
- David, J.M.; Rajasekaran, A.K. Gramicidin A: A New Mission for an Old Antibiotic. J. Kidney Cancer VHL 2015, 2, 15–24. [Google Scholar] [CrossRef]
- Haoyang, W.-W.; Xiao, Q.; Ye, Z.; Fu, Y.; Zhang, D.-W.; Li, J.; Xiao, L.; Li, Z.-T.; Hou, J.-L. Gramicidin A-Based Unimolecular Channel: Cancer Cell-Targeting Behavior and Ion Transport-Induced Apoptosis. Chem. Commun. 2021, 57, 1097–1100. [Google Scholar] [CrossRef]
- David, J.M.; Owens, T.A.; Barwe, S.P.; Rajasekaran, A.K. Gramicidin A Induces Metabolic Dysfunction and Energy Depletion Leading to Cell Death in Renal Cell Carcinoma Cells. Mol. Cancer Ther. 2013, 12, 2296–2307. [Google Scholar] [CrossRef] [PubMed]
- Raileanu, M.; Popescu, A.; Bacalum, M. Antimicrobial Peptides as New Combination Agents in Cancer Therapeutics: A Promising Protocol against HT-29 Tumoral Spheroids. Int. J. Mol. Sci. 2020, 21, 6964. [Google Scholar] [CrossRef]
- Xue, Y.-W.; Itoh, H.; Dan, S.; Inoue, M. Gramicidin A Accumulates in Mitochondria, Reduces ATP Levels, Induces Mitophagy, and Inhibits Cancer Cell Growth. Chem. Sci. 2022, 13, 7482–7491. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Wang, Y.; Yang, Y.; Yu, K.; Cao, X.; Su, F.; Xu, H.; Peng, Y.; Hu, Y.; Qian, F.; et al. Gramicidin Inhibits Human Gastric Cancer Cell Proliferation, Cell Cycle and Induced Apoptosis. Biol. Res. 2019, 52, 57. [Google Scholar] [CrossRef] [PubMed]
- Bubin, R.; Uljanovs, R.; Strumfa, I. Cancer Stem Cells in Pancreatic Ductal Adenocarcinoma. Int. J. Mol. Sci. 2023, 24, 7030. [Google Scholar] [CrossRef]
- Wang, R.-Q.; Geng, J.; Sheng, W.-J.; Liu, X.-J.; Jiang, M.; Zhen, Y.-S. The Ionophore Antibiotic Gramicidin A Inhibits Pancreatic Cancer Stem Cells Associated with CD47 Down-Regulation. Cancer Cell Int. 2019, 19, 145. [Google Scholar] [CrossRef]
- Maham, S.; Awan, S.N.; Adnan, F.; Kakar, S.J.; Mian, A.; Khan, D. PB1837: Antibiotics; a Possible Alternate Treatment Option for Myeloid Leukemia. Hemasphere 2023, 7 (Suppl. S3), e1313111. [Google Scholar] [CrossRef]
- Yang, X.; Hua, C.; Lin, L.; Ganting, Z. Antimicrobial Peptides as Potential Therapy for Gastrointestinal Cancers. Naunyn-Schmiedeberg’s Arch. Pharmacol. 2023, 396, 2831–2841. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Ruan, P.; Jiang, G.; Zhang, W. Screening and in Vitro Biological Evaluation of Novel Multiple Tyrosine Kinases Inhibitors as Promising Anticancer Agents. Anticancer Agents Med. Chem. 2023. [Google Scholar] [CrossRef]
- Gong, X.; Zou, L.; Wang, M.; Zhang, Y.; Peng, S.; Zhong, M.; Zhou, J.; Li, X.; Ma, X. Gramicidin Inhibits Cholangiocarcinoma Cell Growth by Suppressing EGR4. Artif. Cells Nanomed. Biotechnol. 2020, 48, 53–59. [Google Scholar] [CrossRef]
- Yang, M.; Liu, S.; Zhang, C. Antimicrobial Peptides with Antiviral and Anticancer Properties and Their Modification and Nanodelivery Systems. Curr. Res. Biotechnol. 2023, 5, 100121. [Google Scholar] [CrossRef]
- Carvalho, C.A.; Olivares-Ortega, C.; Soto-Arriaza, M.A.; Carmona-Ribeiro, A.M. Interaction of Gramicidin with DPPC/DODAB Bilayer Fragments. Biochim. Biophys. Acta 2012, 1818, 3064–3071. [Google Scholar] [CrossRef] [PubMed]
- Carmona Ribeiro, A.M.; Chaimovich, H. Preparation and Characterization of Large Dioctadecyldimethylammonium Chloride Liposomes and Comparison with Small Sonicated Vesicles. Biochim. Biophys. Acta (BBA)—Biomembr. 1983, 733, 172–179. [Google Scholar] [CrossRef]
- Carmona-Ribeiro, A.M. Bilayer-Forming Synthetic Lipids: Drugs or Carriers? Curr. Med. Chem. 2003, 10, 2425–2446. [Google Scholar] [CrossRef] [PubMed]
- Carmona-Ribeiro, A.M.; Castuma, C.E.; Sesso, A.; Schreier, S. Bilayer Structure and Stability in Dihexadecyl Phosphate Dispersions. J. Phys. Chem. 1991, 95, 5361–5366. [Google Scholar] [CrossRef]
- Andersson, M.; Hammarstroem, L.; Edwards, K. Effect of Bilayer Phase Transitions on Vesicle Structure, and Its Influence on the Kinetics of Viologen Reduction. J. Phys. Chem. 1995, 99, 14531–14538. [Google Scholar] [CrossRef]
- Carmona-Ribeiro, A.M. Lipid Bilayer Fragments and Disks in Drug Delivery. Curr. Med. Chem. 2006, 13, 1359–1370. [Google Scholar] [CrossRef]
- Carmona-Ribeiro, A.M. Synthetic Amphiphile Vesicles. Chem. Soc. Rev. 1992, 21, 209–214. [Google Scholar] [CrossRef]
- Carmona-Ribeiro, A.M.; Midmore, B.R. Surface Potential in Charged Synthetic Amphiphile Vesicles. J. Phys. Chem. 1992, 96, 3542–3547. [Google Scholar] [CrossRef]
- Carmona-Ribeiro, A.M. Bilayer Vesicles and Liposomes as Interface Agents. Chem. Soc. Rev. 2001, 30, 241–247. [Google Scholar] [CrossRef]
- Campanhã, M.T.; Mamizuka, E.M.; Carmona-Ribeiro, A.M. Interactions between Cationic Liposomes and Bacteria: The Physical-Chemistry of the Bactericidal Action. J. Lipid Res. 1999, 40, 1495–1500. [Google Scholar] [CrossRef] [PubMed]
- Martins, L.M.S.; Mamizuka, E.M.; Carmona-Ribeiro, A.M. Cationic Vesicles as Bactericides. Langmuir 1997, 13, 5583–5587. [Google Scholar] [CrossRef]
- Sicchierolli, S.M.; Mamizuka, E.M.; Carmona-Ribeiro, A.M. Bacteria Flocculation and Death by Cationic Vesicles. Langmuir 1995, 11, 2991–2995. [Google Scholar] [CrossRef]
- Campanhã, M.T.N.; Mamizuka, E.M.; Carmona-Ribeiro, A.M. Interactions between Cationic Vesicles and Candida Albicans. J. Phys. Chem. B 2001, 105, 8230–8236. [Google Scholar] [CrossRef]
- Ragioto, D.A.; Carrasco, L.D.; Carmona-Ribeiro, A.M. Novel Gramicidin Formulations in Cationic Lipid as Broad-Spectrum Microbicidal Agents. Int. J. Nanomed. 2014, 9, 3183–3192. [Google Scholar] [CrossRef]
- Carmonaribeiro, A.; Carrasco, L. Fungicidal Assemblies and Their Mode of Action. OA Biotechnol. 2013, 2, 25. [Google Scholar] [CrossRef]
- Xavier, G.R.S.; Carmona-Ribeiro, A.M. Cationic Biomimetic Particles of Polystyrene/Cationic Bilayer/Gramicidin for Optimal Bactericidal Activity. Nanomaterials 2017, 7, 422. [Google Scholar] [CrossRef]
- Carmona-Ribeiro, A.M. Biomimetic Systems in Nanomedicine. In Handbook of Nanobiomedical Research: Fundamentals, Applications and Recent Developments; World Scientific: Singapore, 2014; pp. 401–456. [Google Scholar] [CrossRef]
- Carmona-Ribeiro, A.M. Biomimetic Nanoparticles: Preparation, Characterization and Biomedical Applications. Int. J. Nanomed. 2010, 5, 249–259. [Google Scholar] [CrossRef]
- Carmona-Ribeiro, A.M. Biomimetic Lipid Polymer Nanoparticles for Drug Delivery. In Nanoparticles in Biology and Medicine: Methods and Protocols; Ferrari, E., Soloviev, M., Eds.; Methods in Molecular Biology; Springer: New York, NY, USA, 2020; pp. 45–60. [Google Scholar] [CrossRef]
- Carmona-Ribeiro, A.M. Lipid-Based Biomimetics in Drug and Vaccine Delivery. In Biomimetics Learning from Nature; IntechOpen: London, UK, 2010. [Google Scholar] [CrossRef][Green Version]
- Carmona-Ribeiro, A.M. Biomimetic Nanomaterials from the Assembly of Polymers, Lipids, and Surfactants. In Surfactants and Detergents; IntechOpen: London, UK, 2019. [Google Scholar] [CrossRef]
- Zou, P.; Chen, W.-T.; Sun, T.; Gao, Y.; Li, L.-L.; Wang, H. Recent Advances: Peptides and Self-Assembled Peptide-Nanosystems for Antimicrobial Therapy and Diagnosis. Biomater. Sci. 2020, 8, 4975–4996. [Google Scholar] [CrossRef]
- Sahandi Zangabad, P.; Abousalman Rezvani, Z.; Tong, Z.; Esser, L.; Vasani, R.B.; Voelcker, N.H. Recent Advances in Formulations for Long-Acting Delivery of Therapeutic Peptides. ACS Appl. Bio Mater. 2023, 6, 3532–3554. [Google Scholar] [CrossRef]
- Lau, J.L.; Dunn, M.K. Therapeutic Peptides: Historical Perspectives, Current Development Trends, and Future Directions. Bioorg. Med. Chem. 2018, 26, 2700–2707. [Google Scholar] [CrossRef] [PubMed]
- Barfar, A.; Javadzadeh, Y.; Alizadeh, H.; Masoomzadeh, S. Oral Insulin Delivery: A Review On Recent Advancements and Novel Strategies. Curr. Drug Deliv. 2023. [Google Scholar] [CrossRef]
- Uhl, P.; Grundmann, C.; Sauter, M.; Storck, P.; Tursch, A.; Özbek, S.; Leotta, K.; Roth, R.; Witzigmann, D.; Kulkarni, J.A.; et al. Coating of PLA-Nanoparticles with Cyclic, Arginine-Rich Cell Penetrating Peptides Enables Oral Delivery of Liraglutide. Nanomedicine 2020, 24, 102132. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Xu, W.; Wu, Y.; Niu, B.; Zhang, X. Macroporous Organosilicon Nanocomposites Co-Deliver Bcl2-Converting Peptide and Chemotherapeutic Agent for Synergistic Treatment against Multidrug Resistant Cancer. Cancer Lett. 2020, 469, 340–354. [Google Scholar] [CrossRef]
- Yu, Q.; Deng, T.; Lin, F.-C.; Zhang, B.; Zink, J.I. Supramolecular Assemblies of Heterogeneous Mesoporous Silica Nanoparticles to Co-Deliver Antimicrobial Peptides and Antibiotics for Synergistic Eradication of Pathogenic Biofilms. ACS Nano 2020, 14, 5926–5937. [Google Scholar] [CrossRef]
- Moorcroft, S.C.T.; Roach, L.; Jayne, D.G.; Ong, Z.Y.; Evans, S.D. Nanoparticle-Loaded Hydrogel for the Light-Activated Release and Photothermal Enhancement of Antimicrobial Peptides. ACS Appl. Mater. Interfaces 2020, 12, 24544–24554. [Google Scholar] [CrossRef]
- Dutta, K.; Das, R.; Ling, J.; Monibas, R.M.; Carballo-Jane, E.; Kekec, A.; Feng, D.D.; Lin, S.; Mu, J.; Saklatvala, R.; et al. In Situ Forming Injectable Thermoresponsive Hydrogels for Controlled Delivery of Biomacromolecules. ACS Omega 2020, 5, 17531–17542. [Google Scholar] [CrossRef]
- Wang, P.; Zhuo, X.; Chu, W.; Tang, X. Exenatide-Loaded Microsphere/Thermosensitive Hydrogel Long-Acting Delivery System with High Drug Bioactivity. Int. J. Pharm. 2017, 528, 62–75. [Google Scholar] [CrossRef]
- Xiong, Z.; Cui, W.; Sun, T.; Teng, Y.; Qu, Y.; Yang, L.; Zhou, J.; Chen, K.; Yao, S.; Shao, Z.; et al. Sustained Delivery of PlGF-2123-144*-Fused BMP2-Related Peptide P28 from Small Intestinal Submucosa/Polylactic Acid Scaffold Material for Bone Tissue Regeneration. RSC Adv. 2020, 10, 7289–7300. [Google Scholar] [CrossRef]
- Luo, J.; Zhang, H.; Zhu, J.; Cui, X.; Gao, J.; Wang, X.; Xiong, J. 3-D Mineralized Silk Fibroin/Polycaprolactone Composite Scaffold Modified with Polyglutamate Conjugated with BMP-2 Peptide for Bone Tissue Engineering. Colloids Surf. B Biointerfaces 2018, 163, 369–378. [Google Scholar] [CrossRef]
- Tran, H.; ElSayed, M.E.H. Progress and Limitations of Oral Peptide Delivery as a Potentially Transformative Therapy. Expert. Opin. Drug Deliv. 2022, 19, 163–178. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Li, X.; Zhao, Y.; Zheng, Q.; Wu, Q.; Yu, Y. Nanocarrier System: An Emerging Strategy for Bioactive Peptide Delivery. Front. Nutr. 2022, 9, 1050647. [Google Scholar] [CrossRef] [PubMed]
- Haddadzadegan, S.; Dorkoosh, F.; Bernkop-Schnürch, A. Oral Delivery of Therapeutic Peptides and Proteins: Technology Landscape of Lipid-Based Nanocarriers. Adv. Drug Deliv. Rev. 2022, 182, 114097. [Google Scholar] [CrossRef] [PubMed]
- Veider, F.; Knoll, P.; Jörgensen, A.M.; Stengel, D.; Bernkop-Schnürch, A. Oral Drug Delivery: Influence of Mucus on Cellular Interactions and Uptake of Lipid-Based Nanocarriers in Caco-2 Cells. Acta Biomater. 2023, 167, 416–424. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Patrone, N.; Liang, J.F. Peptide Self-Assembly on Cell Membranes to Induce Cell Lysis. Biomacromolecules 2012, 13, 3327–3333. [Google Scholar] [CrossRef]
- Pueyo, M.T.; Mutafci, B.A.; Soto-Arriaza, M.A.; Di Mascio, P.; Carmona-Ribeiro, A.M. The Self-Assembly of a Cyclic Lipopeptides Mixture Secreted by a B. Megaterium Strain and Its Implications on Activity against a Sensitive Bacillus Species. PLoS ONE 2014, 9, e97261. [Google Scholar] [CrossRef]
- Pueyo, M.T.; Bloch, C.; Carmona-Ribeiro, A.M.; di Mascio, P. Lipopeptides Produced by a Soil Bacillus Megaterium Strain. Microb. Ecol. 2009, 57, 367–378. [Google Scholar] [CrossRef]
- Lincopan, N.; Espíndola, N.; Vaz, A.; Carmonaribeiro, A. Cationic Supported Lipid Bilayers for Antigen Presentation. Int. J. Pharm. 2007, 340, 216–222. [Google Scholar] [CrossRef]
- Ribeiro, R.T.; Braga, V.H.A.; Carmona-Ribeiro, A.M. Biomimetic Cationic Nanoparticles Based on Silica: Optimizing Bilayer Deposition from Lipid Films. Biomimetics 2017, 2, 20. [Google Scholar] [CrossRef]
- Mathiazzi, B.I.; Carmona-Ribeiro, A.M. Hybrid Nanoparticles of Poly (Methyl Methacrylate) and Antimicrobial Quaternary Ammonium Surfactants. Pharmaceutics 2020, 12, 340. [Google Scholar] [CrossRef]
- Simonson, A.W.; Aronson, M.R.; Medina, S.H. Supramolecular Peptide Assemblies as Antimicrobial Scaffolds. Molecules 2020, 25, 2751. [Google Scholar] [CrossRef]
- Ahmed, E.M. Hydrogel: Preparation, Characterization, and Applications: A Review. J. Adv. Res. 2015, 6, 105–121. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Chen, T.; Dong, C.; Pan, X. Biomedical Applications of Hemicellulose-Based Hydrogels. Curr. Med. Chem. 2020, 27, 4647–4659. [Google Scholar] [CrossRef]
- Yang, Y.; Xu, L.; Wang, J.; Meng, Q.; Zhong, S.; Gao, Y.; Cui, X. Recent Advances in Polysaccharide-Based Self-Healing Hydrogels for Biomedical Applications. Carbohydr. Polym. 2022, 283, 119161. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Cao, Y.; Ma, L.; Sun, J.; Ramos-Mucci, L.; Ma, Y.; Yang, X.; Zhu, Z.; Zhang, J.; Xiao, B. Oral Antimicrobial Peptide-EGCG Nanomedicines for Synergistic Treatment of Ulcerative Colitis. J. Control. Release 2022, 347, 544–560. [Google Scholar] [CrossRef] [PubMed]
- Alves, P.M.; Pereira, R.F.; Costa, B.; Tassi, N.; Teixeira, C.; Leiro, V.; Monteiro, C.; Gomes, P.; Costa, F.; Martins, M.C.L. Thiol–Norbornene Photoclick Chemistry for Grafting Antimicrobial Peptides onto Chitosan to Create Antibacterial Biomaterials. ACS Appl. Polym. Mater. 2022, 4, 5012–5026. [Google Scholar] [CrossRef]
- Cheng, H.; Shi, Z.; Yue, K.; Huang, X.; Xu, Y.; Gao, C.; Yao, Z.; Zhang, Y.S.; Wang, J. Sprayable Hydrogel Dressing Accelerates Wound Healing with Combined Reactive Oxygen Species-Scavenging and Antibacterial Abilities. Acta Biomater. 2021, 124, 219–232. [Google Scholar] [CrossRef] [PubMed]
- De Zoysa, G.H.; Wang, K.; Lu, J.; Hemar, Y.; Sarojini, V. Covalently Immobilized Battacin Lipopeptide Gels with Activity against Bacterial Biofilms. Molecules 2020, 25, 5945. [Google Scholar] [CrossRef]
- Qian, C.-D.; Wu, X.-C.; Teng, Y.; Zhao, W.-P.; Li, O.; Fang, S.-G.; Huang, Z.-H.; Gao, H.-C. Battacin (Octapeptin B5), a New Cyclic Lipopeptide Antibiotic from Paenibacillus Tianmuensis Active against Multidrug-Resistant Gram-Negative Bacteria. Antimicrob. Agents Chemother. 2012, 56, 1458–1465. [Google Scholar] [CrossRef]
- Xie, S.-X.; Song, L.; Yuca, E.; Boone, K.; Sarikaya, R.; VanOosten, S.K.; Misra, A.; Ye, Q.; Spencer, P.; Tamerler, C. Antimicrobial Peptide–Polymer Conjugates for Dentistry. ACS Appl. Polym. Mater. 2020, 2, 1134–1144. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, X.; Jiang, W.; Wang, K.; Luo, J.; Li, W.; Zhou, X.; Zhang, L. Antimicrobial Peptide GH12 Suppresses Cariogenic Virulence Factors of Streptococcus mutans. J. Oral Microbiol. 2018, 10, 1442089. [Google Scholar] [CrossRef] [PubMed]
- Nordström, R.; Nyström, L.; Andrén, O.C.J.; Malkoch, M.; Umerska, A.; Davoudi, M.; Schmidtchen, A.; Malmsten, M. Membrane Interactions of Microgels as Carriers of Antimicrobial Peptides. J. Colloid Interface Sci. 2018, 513, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Caselli, L.; Malmsten, M. Skin and Wound Delivery Systems for Antimicrobial Peptides. Curr. Opin. Colloid Interface Sci. 2023, 65, 101701. [Google Scholar] [CrossRef]
- Nordström, R.; Andrén, O.C.J.; Singh, S.; Malkoch, M.; Davoudi, M.; Schmidtchen, A.; Malmsten, M. Degradable Dendritic Nanogels as Carriers for Antimicrobial Peptides. J. Colloid Interface Sci. 2019, 554, 592–602. [Google Scholar] [CrossRef] [PubMed]
- Dart, A.; Bhave, M.; Kingshott, P. Antimicrobial Peptide-Based Electrospun Fibers for Wound Healing Applications. Macromol. Biosci. 2019, 19, e1800488. [Google Scholar] [CrossRef]
- Hsu, Y.-H.; Yu, Y.-H.; Chou, Y.-C.; Lu, C.-J.; Lin, Y.-T.; Ueng, S.W.-N.; Liu, S.-J. Sustained Release of Antifungal and Antibacterial Agents from Novel Hybrid Degradable Nanofibers for the Treatment of Polymicrobial Osteomyelitis. Int. J. Mol. Sci. 2023, 24, 3254. [Google Scholar] [CrossRef]
- Li, G.; Lai, Z.; Shan, A. Advances of Antimicrobial Peptide-Based Biomaterials for the Treatment of Bacterial Infections. Adv. Sci. 2023, 10, 2206602. [Google Scholar] [CrossRef]
- Cesaro, A.; Lin, S.; Pardi, N.; de la Fuente-Nunez, C. Advanced Delivery Systems for Peptide Antibiotics. Adv. Drug Deliv. Rev. 2023, 196, 114733. [Google Scholar] [CrossRef]
- Giordano, N.P.; Cian, M.B.; Dalebroux, Z.D. Outer Membrane Lipid Secretion and the Innate Immune Response to Gram-Negative Bacteria. Infect. Immun. 2020, 88, e00920-19. [Google Scholar] [CrossRef]
- Upert, G.; Luther, A.; Obrecht, D.; Ermert, P. Emerging peptide antibiotics with therapeutic potential. Med. Drug Discov. 2021, 9, 100078. [Google Scholar] [CrossRef]
- Cardoso, P.; Glossop, H.; Meikle, T.G.; Aburto-Medina, A.; Conn, C.E.; Sarojini, V.; Valery, C. Molecular engineering of antimicrobial peptides: Microbial targets, peptide motifs and translation opportunities. Biophys. Rev. 2021, 13, 35–69. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, S.; Chatterjee, R.; Chakravortty, D. Evolving and assembling to pierce through: Evolutionary and structural aspects of antimicrobial peptides. Comput. Struct. Biotechnol. J. 2022, 20, 2247–2258. [Google Scholar] [CrossRef] [PubMed]
- Juhász, T.; Quemé-Peña, M.; Kővágó, B.; Mihály, J.; Ricci, M.; Horváti, K.; Bősze, S.; Zsila, F.; Beke-Somfai, T. Interplay between membrane active host defense peptides and heme modulates their assemblies and in vitro activity. Sci. Rep. 2021, 11, 18328. [Google Scholar] [CrossRef] [PubMed]
- Glossop, H.D.; De Zoysa, G.H.; Hemar, Y.; Cardoso, P.; Wang, K.; Lu, J.; Valéry, C.; Sarojini, V. Battacin-Inspired Ultrashort Peptides: Nanostructure Analysis and Antimicrobial Activity. Biomacromolecules 2019, 20, 2515–2529. [Google Scholar] [CrossRef]
- Claro, B.; Peón, A.; González-Freire, E.; Goormaghtigh, E.; Amorín, M.; Granja, J.R.; Garcia-Fandiño, R.; Bastos, M. Macromolecular assembly and membrane activity of antimicrobial D,L-α-Cyclic peptides. Colloids Surf. B Biointerfaces 2021, 208, 112086. [Google Scholar] [CrossRef]
- Guo, Z.; Wang, Y.; Tan, T.; Ji, Y.; Hu, J.; Zhang, Y. Antimicrobial d-Peptide Hydrogels. ACS Biomater. Sci. Eng. 2021, 7, 1703–1712. [Google Scholar] [CrossRef]
- Dahan, S.; Aibinder, P.; Khalfin, B.; Moran-Gilad, J.; Rapaport, H. Hybrid Hydrogels of FKF-Peptide Assemblies and Gelatin for Sustained Antimicrobial Activity. ACS Biomater. Sci. Eng. 2023, 9, 352–362. [Google Scholar] [CrossRef]
- Tan, A.; Xu, F.; Yokoyama, C.; Yano, S.; Konno, H. Design, synthesis, and evaluation of the self-assembled antimicrobial peptides based on the ovalbumin-derived peptide TK913. J. Pept. Sci. 2022, 28, e3375. [Google Scholar] [CrossRef]
- Baltutis, V.; O’Leary, P.D.; Martin, L.L. Self-Assembly of Linear, Natural Antimicrobial Peptides: An Evolutionary Perspective. ChemPlusChem 2022, 87, e202200240. [Google Scholar] [CrossRef]
- Matthyssen, T.; Li, W.; Holden, J.A.; Lenzo, J.C.; Hadjigol, S.; O’Brien-Simpson, N.M. The Potential of Modified and Multimeric Antimicrobial Peptide Materials as Superbug Killers. Front. Chem. 2022, 9, 795433. [Google Scholar] [CrossRef]
- Fa, K.; Liu, H.; Gong, H.; Zhang, L.; Liao, M.; Hu, X.; Ciumac, D.; Li, P.; Webster, J.; Petkov, J.; et al. In-Membrane Nanostructuring of Cationic Amphiphiles Affects Their Antimicrobial Efficacy and Cytotoxicity: A Comparison Study between a De Novo Antimicrobial Lipopeptide and Traditional Biocides. Langmuir 2022, 38, 6623–6637. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Liu, X.; Nie, J.; Zhao, H.; Li, W. Nano-Antimicrobial Peptides Based on Constitutional Isomerism-Dictated Self-Assembly. Biomacromolecules 2022, 23, 1302–1313. [Google Scholar] [CrossRef] [PubMed]
- Hurtado-Rios, J.J.; Carrasco-Navarro, U.; Almanza-Pérez, J.C.; Ponce-Alquicira, E. Ribosomes: The New Role of Ribosomal Proteins as Natural Antimicrobials. Int. J. Mol. Sci. 2022, 23, 9123. [Google Scholar] [CrossRef] [PubMed]
- Caselli, L.; Rodrigues, G.R.; Franco, O.L.; Malmsten, M. Pulmonary delivery systems for antimicrobial peptides. Crit. Rev. Biotechnol. 2023, 1–18. [Google Scholar] [CrossRef]
- Townsend, J.A.; Marty, M.T. What’s the defect? Using mass defects to study oligomerization of membrane proteins and peptides in nanodiscs with native mass spectrometry. Methods 2023, 218, 1–13. [Google Scholar] [CrossRef]
- Lan, X.; Zhong, J.; Huang, R.; Liu, Y.; Ma, X.; Li, X.; Zhao, D.; Qing, G.; Zhang, Y.; Liu, L.; et al. Conformation Dependent Architectures of Assembled Antimicrobial Peptides with Enhanced Antimicrobial Ability. Adv. Healthc. Mater. 2023, 4, e2301688. [Google Scholar] [CrossRef]
- Ramos-Martín, F.; D’Amelio, N. Biomembrane lipids: When physics and chemistry join to shape biological activity. Biochimie 2022, 203, 118–138. [Google Scholar] [CrossRef]
- Akshaya, S.; Rowlo, P.K.; Dukle, A.; Nathanael, A.J. Antibacterial Coatings for Titanium Implants: Recent Trends and Future Perspectives. Antibiotics 2022, 11, 1719. [Google Scholar] [CrossRef]
- Yang, W.; Seo, J.; Kim, J.H. Protein-mimetic peptoid nanoarchitectures for pathogen recognition and neutralization. Nanoscale 2023, 15, 975–986. [Google Scholar] [CrossRef]
- Rosetti, B.; Scarel, E.; Colomina-Alfaro, L.; Adorinni, S.; Pierri, G.; Bellotto, O.; Mamprin, K.; Polentarutti, M.; Bandiera, A.; Tedesco, C.; et al. Self-Assembly of Homo- and Hetero-Chiral Cyclodipeptides into Supramolecular Polymers towards Antimicrobial Gels. Polymers 2022, 14, 4554. [Google Scholar] [CrossRef]
- Serrano-Aroca, Á.; Cano-Vicent, A.; Sabater I Serra, R.; El-Tanani, M.; Aljabali, A.; Tambuwala, M.M.; Mishra, Y.K. Scaffolds in the microbial resistant era: Fabrication, materials, properties and tissue engineering applications. Mater. Today Bio 2022, 16, 100412. [Google Scholar] [CrossRef] [PubMed]
- Liao, M.; Gong, H.; Quan, X.; Wang, Z.; Hu, X.; Chen, Z.; Li, Z.; Liu, H.; Zhang, L.; McBain, A.J.; et al. Intramembrane Nanoaggregates of Antimicrobial Peptides Play a Vital Role in Bacterial Killing. Small 2023, 19, e2204428. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Wu, Z.; Zhou, J.; Wang, Z.; Yang, C.; Wang, P.; Fareed, M.S.; He, Y.; Su, J.; Cha, R.; et al. The Antimicrobial, Hemostatic, and Anti-Adhesion Effects of a Peptide Hydrogel Constructed by the All-d-Enantiomer of Antimicrobial Peptide Jelleine-1. Adv. Healthc. Mater. 2023, 8, e2301612. [Google Scholar] [CrossRef]
- Fuertes Llanos, M.; Gómara, M.J.; Haro, I.; Sánchez López, E. Peptide Amphiphiles for Pharmaceutical Applications. Curr. Med. Chem. 2023. [Google Scholar] [CrossRef]
- Cardoso, P.; Appiah Danso, S.; Hung, A.; Dekiwadia, C.; Pradhan, N.; Strachan, J.; McDonald, B.; Firipis, K.; White, J.F.; Aburto-Medina, A.; et al. Rational design of potent ultrashort antimicrobial peptides with programmable assembly into nanostructured hydrogels. Front. Chem. 2023, 10, 1009468. [Google Scholar] [CrossRef] [PubMed]
- Blomstrand, E.; Rajasekharan, A.K.; Atefyekta, S.; Andersson, M. Cross-linked lyotropic liquid crystal particles functionalized with antimicrobial peptides. Int. J. Pharm. 2022, 627, 122215. [Google Scholar] [CrossRef]
- Sharma, R.; Lalhall, A.; Puri, S.; Wangoo, N. Design of Fmoc-Phenylalanine Nanofibrillar Hydrogel and Mechanistic Studies of Its Antimicrobial Action against Both Gram-Positive and Gram-Negative Bacteria. ACS Appl. Bio Mater. 2023, 6, 494–506. [Google Scholar] [CrossRef]
- Hamley, I.W. Self-Assembly, Bioactivity, and Nanomaterials Applications of Peptide Conjugates with Bulky Aromatic Terminal Groups. ACS Appl. Bio Mater. 2023, 6, 384–409. [Google Scholar] [CrossRef]
- Dissanayake, S.; He, J.; Yang, S.H.; Brimble, M.A.; Harris, P.W.R.; Cameron, A.J. Flow-Based Fmoc-SPPS Preparation and SAR Study of Cathelicidin-PY Reveals Selective Antimicrobial Activity. Molecules 2023, 28, 1993. [Google Scholar] [CrossRef]
- Cole, T.J.; Parker, J.K.; Feller, A.L.; Wilke, C.O.; Davies, B.W. Evidence for Widespread Class II Microcins in Enterobacterales Genomes. Appl. Environ. Microbiol. 2022, 88, e0148622. [Google Scholar] [CrossRef]
- George, N.L.; Orlando, B.J. Architecture of a complete Bce-type antimicrobial peptide resistance module. Nat. Commun. 2023, 14, 3896. [Google Scholar] [CrossRef]
- Prasad, A.K.; Martin, L.L.; Panwar, A.S. Helical intermediate formation and its role in amyloids of an amphibian antimicrobial peptide. Phys. Chem. Chem. Phys. 2023, 25, 12134–12147. [Google Scholar] [CrossRef]
- Makowska, M.; Kosikowska-Adamus, P.; Zdrowowicz, M.; Wyrzykowski, D.; Prahl, A.; Sikorska, E. Lipidation of Naturally Occurring α-Helical Antimicrobial Peptides as a Promising Strategy for Drug Design. Int. J. Mol. Sci. 2023, 24, 3951. [Google Scholar] [CrossRef] [PubMed]
- Niu, M.; Gu, X.; Yang, J.; Cui, H.; Hou, X.; Ma, Y.; Wang, C.; Wei, G. Dual-Mechanism Glycolipidpeptide with High Antimicrobial Activity, Immunomodulatory Activity, and Potential Application for Combined Antibacterial Therapy. ACS Nano 2023, 17, 6292–6316. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Zeng, S.; Wang, Y.; Yin, X.; Zhang, B.; Zhang, B.; Xu, S.; Zhang, Y.; Zheng, J.; Fan, J.; et al. Metal coordinating-induced self-assembly of cyclic lipopeptides into high-performance antimicrobial supramolecules. Food Chem. 2023, 422, 136203. [Google Scholar] [CrossRef] [PubMed]
- Xiao, X.; Ji, J.; Wang, H.; Nangia, S.; Wang, H.; Libera, M. Self-Defensive Antimicrobial Surfaces Using Polymyxin-Loaded Poly(styrene sulfonate) Microgels. ACS Biomater. Sci. Eng. 2022, 8, 4827–4837. [Google Scholar] [CrossRef] [PubMed]
- Thankappan, B.; Thomas, A.; Sakthivadivel, A.; Shanmuganathan, N.; Angayarkanni, J. In vitro and in vivo antimicrobial activity of self-assembled melittin nanoparticles: A comparative study with melittin peptide. Colloids Surf. B Biointerfaces 2023, 226, 113331. [Google Scholar] [CrossRef]
- Yadav, N.; Kumar, U.; Chauhan, V.S. Conformationally restricted, dipeptide-based, self-assembled nanoparticles for efficient vancomycin delivery. Nanomedicine 2022, 17, 2023–2035. [Google Scholar] [CrossRef]
- Yu, W.; Sun, Y.; Li, W.; Guo, X.; Liu, X.; Wu, W.; Yu, W.; Wang, J.; Shan, A. Self-Assembly of Antimicrobial Peptide-Based Micelles Breaks the Limitation of Trypsin. ACS Appl. Mater. Interfaces 2023, 15, 494–510. [Google Scholar] [CrossRef]
- Rashid, R.; Nair, Z.J.; Chia, D.M.H.; Chong, K.K.L.; Cazenave Gassiot, A.; Morley, S.A.; Allen, D.K.; Chen, S.L.; Chng, S.S.; Wenk, M.R.; et al. Depleting Cationic Lipids Involved in Antimicrobial Resistance Drives Adaptive Lipid Remodeling in Enterococcus faecalis. mBio 2023, 14, e0307322. [Google Scholar] [CrossRef]
- Romero-Calle, D.X.; Pedrosa-Silva, F.; Ribeiro Tomé, L.M.; Fonseca, V.; Guimarães Benevides, R.; de Oliveira Santos, L.T.S.; de Oliveira, T.; da Costa, M.M.; Alcantara, L.C.J.; de Carvalho Azevedo, V.A.; et al. Molecular Characterization of Salmonella Phage Wara Isolated from River Water in Brazil. Microorganisms 2023, 11, 1837. [Google Scholar] [CrossRef] [PubMed]
- Sousa, A.; Borøy, V.; Bæverud, A.; Julin, K.; Bayer, A.; Strøm, M.; Johannessen, M.; Škalko-Basnet, N.; Obuobi, S. Polymyxin B stabilized DNA micelles for sustained antibacterial and antibiofilm activity against P. aeruginosa. J. Mater. Chem. B. 2023, 11, 7972–7985. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Wu, Y.; Zhou, Z.; Liu, S.; Mao, S.; Li, G. Co-delivery of EGCG and melittin with self-assembled fluoro-nanoparticles for enhanced cancer therapy. Aging 2023, 15, 4875–4888. [Google Scholar] [CrossRef] [PubMed]
- Manioglu, S.; Modaresi, S.M.; Ritzmann, N.; Thoma, J.; Overall, S.A.; Harms, A.; Upert, G.; Luther, A.; Barnes, A.B.; Obrecht, D.; et al. Antibiotic polymyxin arranges lipopolysaccharide into crystalline structures to solidify the bacterial membrane. Nat. Commun. 2022, 13, 6195. [Google Scholar] [CrossRef]
- Stachurski, O.; Neubauer, D.; Walewska, A.; Iłowska, E.; Bauer, M.; Bartoszewska, S.; Sikora, K.; Hać, A.; Wyrzykowski, D.; Prahl, A.; et al. Understanding the Role of Self-Assembly and Interaction with Biological Membranes of Short Cationic Lipopeptides in the Effective Design of New Antibiotics. Antibiotics 2022, 11, 1491. [Google Scholar] [CrossRef]
- Zhan, W.; Gao, G.; Liu, Z.; Liu, X.; Xu, L.; Wang, M.; Xu, H.D.; Tang, R.; Cao, J.; Sun, X.; et al. Enzymatic Self-Assembly of Adamantane-Peptide Conjugate for Combating Staphylococcus aureus Infection. Adv. Healthc. Mater. 2023, 12, e2203283. [Google Scholar] [CrossRef]
- Dong, L.; Huang, C.; Zhao, B.; Hu, G.; Huang, Y.; Zhang, X.; Hu, X.; Wang, Y.; Sun, X.; Qian, W.; et al. A pH/enzyme dual responsive PMB spatiotemporal release hydrogel promoting chronic wound repair. J. Nanobiotechnol. 2023, 21, 213. [Google Scholar] [CrossRef]
- Zhao, H.; Zhong, L.L.; Yang, C.; Tang, N.; He, Y.; He, W.; Zhao, Z.; Wu, C.; Yuan, P.; Yang, Y.Y.; et al. Antibiotic-Polymer Self-Assembled Nanocomplex to Reverse Phenotypic Resistance of Bacteria toward Last-Resort Antibiotic Colistin. ACS Nano 2023, 17, 15411–15423. [Google Scholar] [CrossRef]
- Zhang, P.; Ouyang, Q.; Zhai, T.; Sun, J.; Wu, J.; Qin, F.; Zhang, N.; Yue, S.; Yang, X.; Zhang, H.; et al. An inflammation-targeted nanoparticle with bacteria forced release of polymyxin B for pneumonia therapy. Nanoscale 2022, 14, 15291–15304. [Google Scholar] [CrossRef]
- Coya, J.M.; Fraile-Ágreda, V.; de Tapia, L.; García-Fojeda, B.; Sáenz, A.; Bengoechea, J.A.; Kronqvist, N.; Johansson, J.; Casals, C. Cooperative action of SP-A and its trimeric recombinant fragment with polymyxins against Gram-negative respiratory bacteria. Front. Immunol. 2022, 13, 927017. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carmona-Ribeiro, A.M. Antimicrobial Peptides and Their Assemblies. Future Pharmacol. 2023, 3, 763-788. https://doi.org/10.3390/futurepharmacol3040047
Carmona-Ribeiro AM. Antimicrobial Peptides and Their Assemblies. Future Pharmacology. 2023; 3(4):763-788. https://doi.org/10.3390/futurepharmacol3040047
Chicago/Turabian StyleCarmona-Ribeiro, Ana Maria. 2023. "Antimicrobial Peptides and Their Assemblies" Future Pharmacology 3, no. 4: 763-788. https://doi.org/10.3390/futurepharmacol3040047
APA StyleCarmona-Ribeiro, A. M. (2023). Antimicrobial Peptides and Their Assemblies. Future Pharmacology, 3(4), 763-788. https://doi.org/10.3390/futurepharmacol3040047





