Mathematical Models of Epidemics with Infection Time
Abstract
:1. Introduction
2. Materials and Methods
2.1. SIS Model
2.2. SIRS Model
2.3. Distributed Delay
2.4. Random Delay
2.5. Exposed Population Model
2.6. Fractional Differential Equations
3. Results
3.1. SIS
3.1.1. ODE Model
3.1.2. Discrete Delay Models
3.1.3. Distributed Delay Models
3.1.4. Exposed Population Model
3.2. Sirs
3.2.1. Ode Model
3.2.2. Discrete Delay Models
3.2.3. Distributed Delay Models
3.2.4. Exposed Population Model
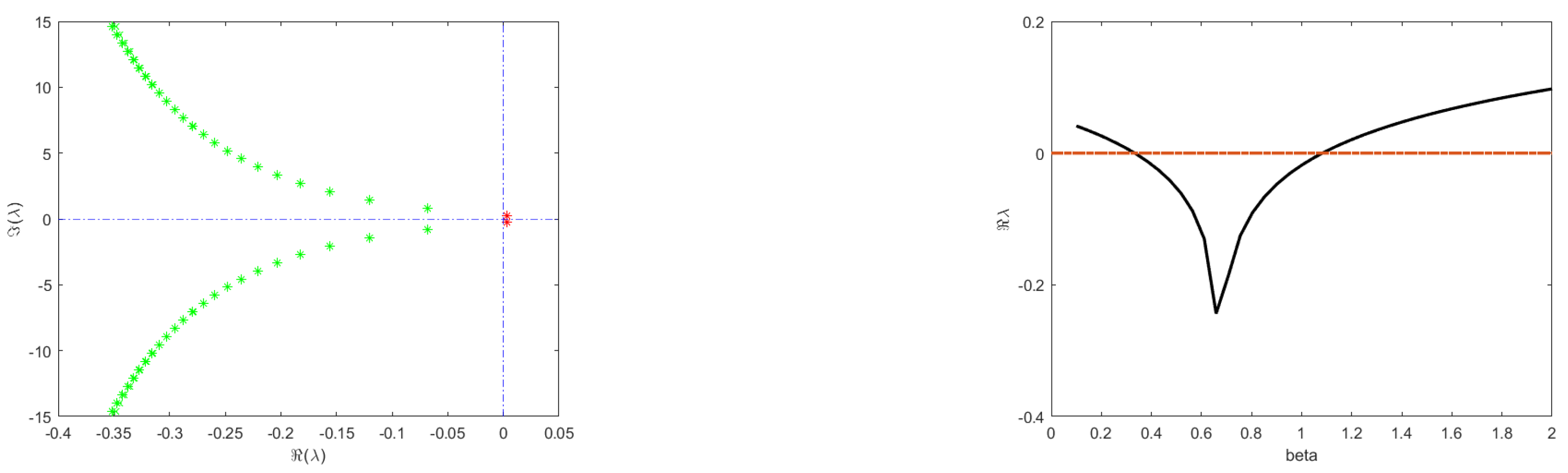
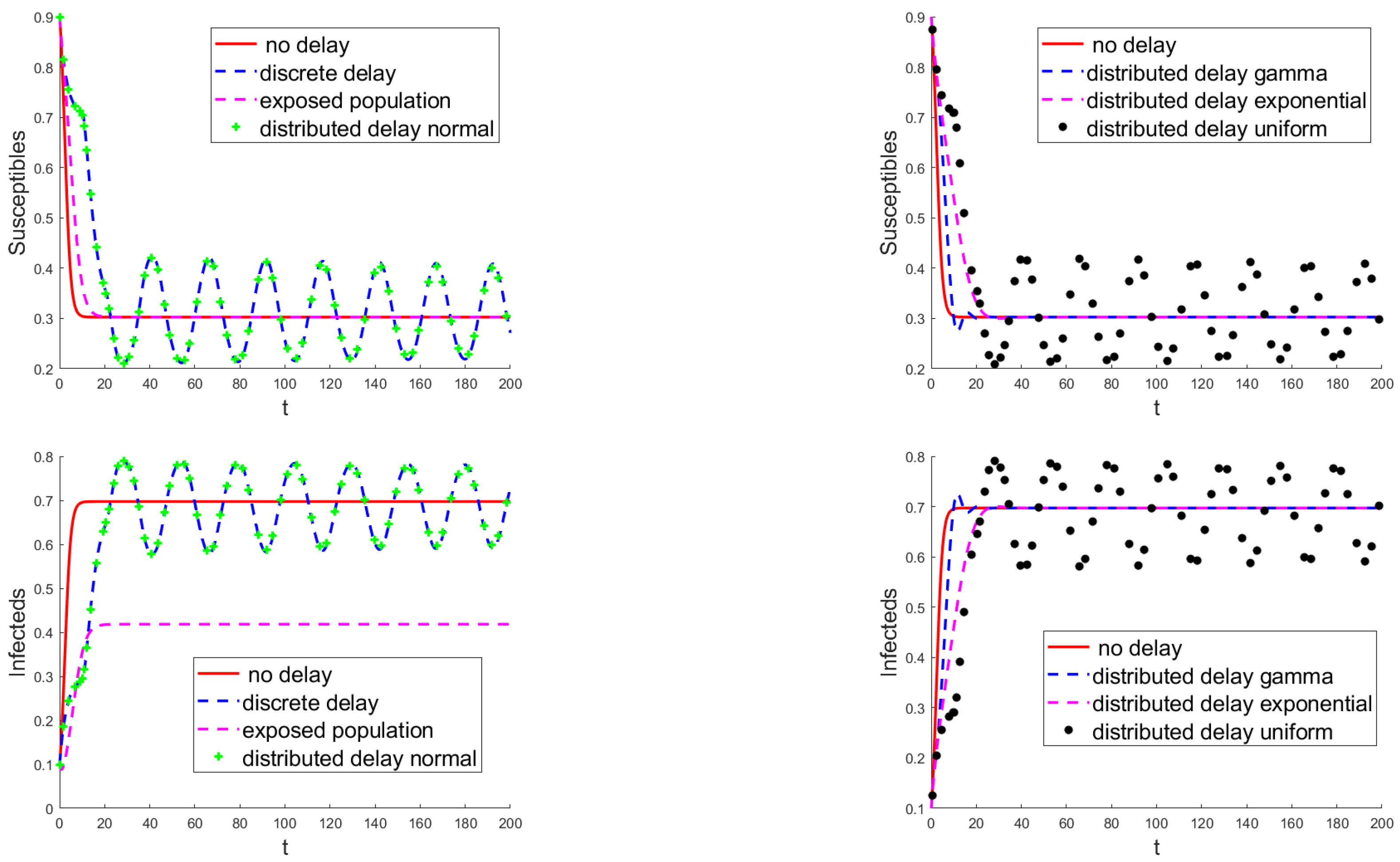
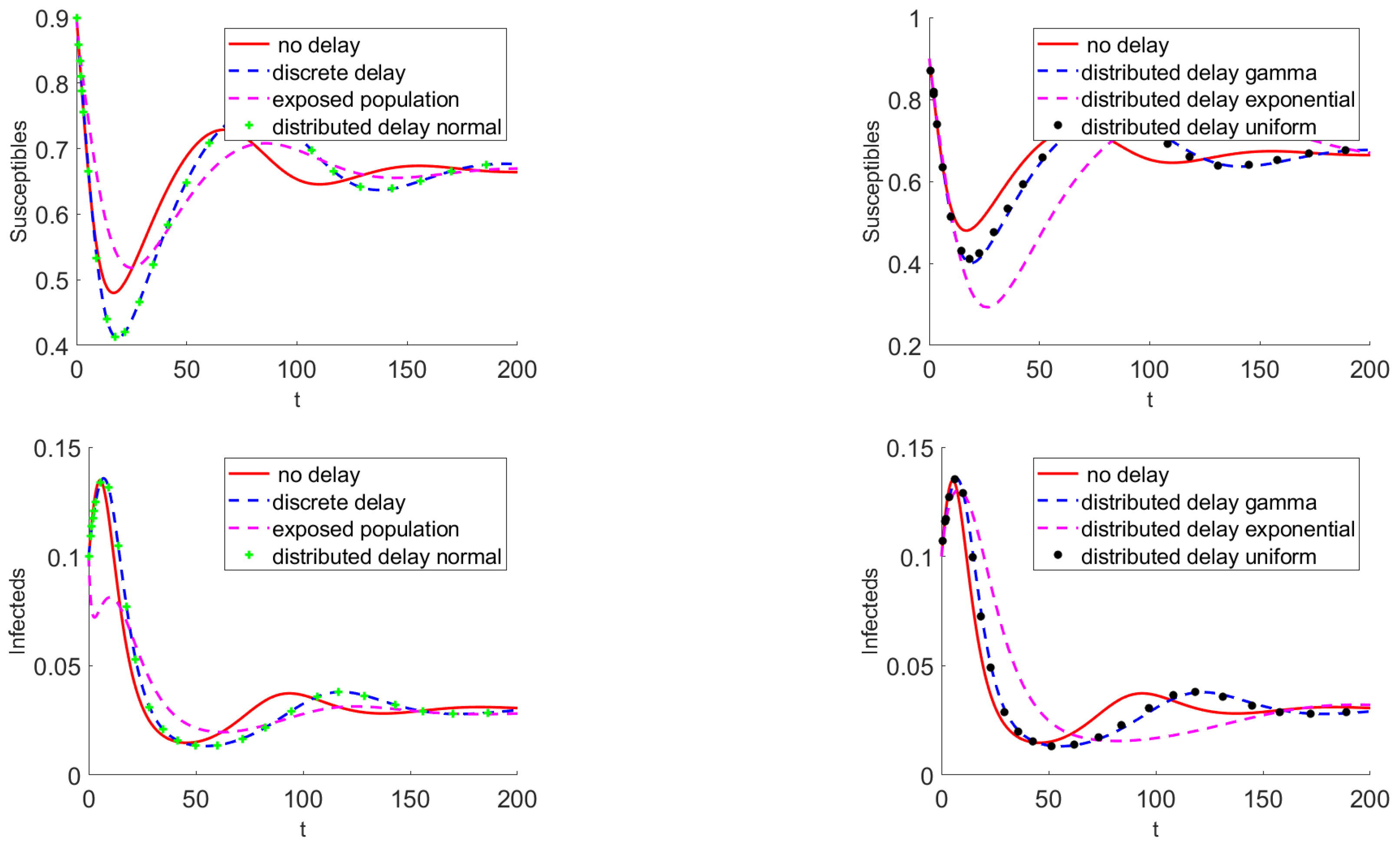
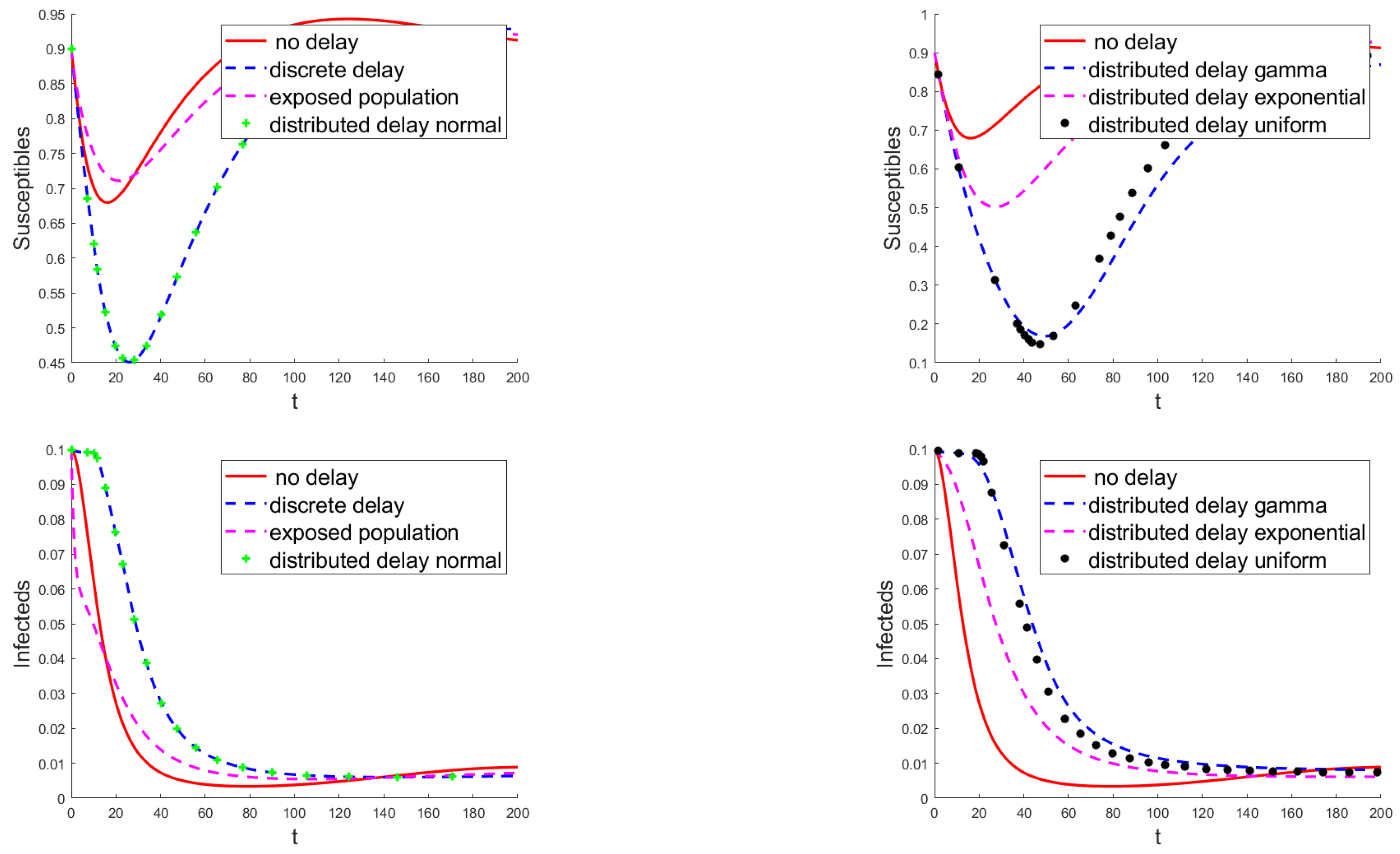
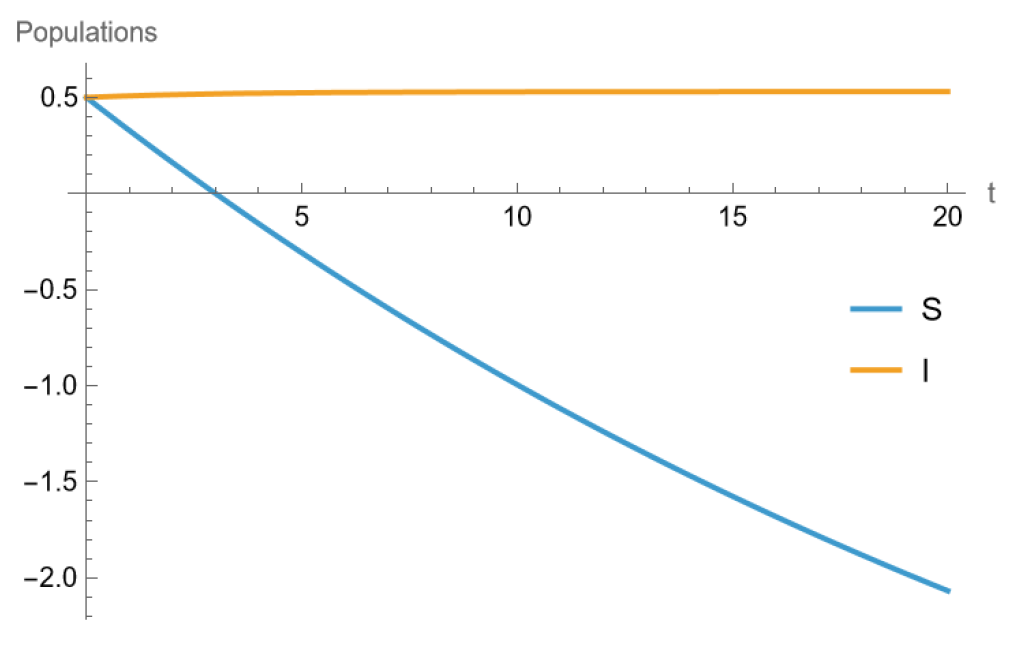
3.3. Numerical Results
4. Discussion
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| ODE | Ordinary differential equation |
| DDE | Delay differential equation |
| SIS | Susceptible–Infective–Susceptible |
| SIRS | Susceptible–Infective–Recovered–Susceptible |
| SEIS | Susceptible–Exposed–Infective–Susceptible |
| SEIRS | Susceptible–Exposed–Infective–Susceptible–Recovered |
| HIV | Human Immunodeficiency Virus |
| COVID | Coronavirus Disease |
| CDC | Centers for Disease Control and Prevention |
References
- Cleveland Clinic Medical. Incubation Period (Incubation Time) of Infections & Diseases—my.clevelandclinic.org. 2024. Available online: https://my.clevelandclinic.org/health/articles/incubation-period (accessed on 2 April 2025).
- Chowell, G.; Miller, M.; Viboud, C. Seasonal influenza in the United States, France, and Australia: Transmission and prospects for control. Epidemiol. Infect. 2008, 136, 852–864. [Google Scholar] [PubMed]
- Deng, Y.; You, C.; Liu, Y.; Qin, J.; Zhou, X.H. Estimation of incubation period and generation time based on observed length-biased epidemic cohort with censoring for COVID-19 outbreak in China. Biometrics 2021, 77, 929–941. [Google Scholar]
- Gressani, O.; Torneri, A.; Hens, N.; Faes, C. Flexible Bayesian estimation of incubation times. Am. J. Epidemiol. 2024, 194, 490–501. [Google Scholar]
- Kermack, W.O.; McKendrick, A.G. Contributions to the mathematical theory of epidemics—I. 1927. Bull. Math. Biol. 1991, 53, 33–55. [Google Scholar]
- Allen, L. An Introduction to Mathematical Biology; Pearson-Prentice Hall: Saddle River, NJ, USA, 2007. [Google Scholar]
- Edelstein-Keshet, L. Mathematical Models in Biology; SIAM: Philadelphia, PA, USA, 2005. [Google Scholar]
- Zhang, X.; Zhao, Y.; Neumann, A.U. Partial immunity and vaccination for influenza. J. Comput. Biol. 2010, 17, 1689–1696. [Google Scholar]
- Muli, F.M.M.; Okelo, B.; Magwanga, R.; Ongati, O. Mathematical Analysis of COVID-19 model with Vaccination and Partial Immunity to Reinfection. J. Math. Anal. Model. 2023, 4, 121–145. [Google Scholar]
- Chen-Charpentier, B. SIRS Epidemic Models with Delays, Partial and Temporary Immunity and Vaccination. AppliedMath 2024, 4, 666–689. [Google Scholar] [CrossRef]
- Arif, F.; Majeed, Z.; Rahman, J.U.; Iqbal, N.; Kafle, J. Mathematical Modeling and Numerical Simulation for the Outbreak of COVID-19 Involving Loss of Immunity and Quarantined Class. Comput. Math. Methods Med. 2022, 2022, 3816492. [Google Scholar] [CrossRef]
- Haq, I.U.; Ullah, N.; Ali, N.; Nisar, K.S. A new mathematical model of COVID-19 with quarantine and vaccination. Mathematics 2022, 11, 142. [Google Scholar] [CrossRef]
- Saha, P.; Mondal, B.; Ghosh, U. Dynamical Behaviors of an Epidemic Model with Partial Immunity Having Nonlinear Incidence and Saturated Treatment in Deterministic and Stochastic Environments. Chaos Solitons Fractals 2023, 174, 113775. [Google Scholar] [CrossRef]
- Li, M.Y.; Muldowney, J.S. Global stability for the SEIR model in epidemiology. Math. Biosci. 1995, 125, 155–164. [Google Scholar] [PubMed]
- He, S.; Peng, Y.; Sun, K. SEIR modeling of the COVID-19 and its dynamics. Nonlinear Dyn. 2020, 101, 1667–1680. [Google Scholar]
- Burke, D.S. Origins of the problematic E in SEIR epidemic models. Infect. Dis. Model. 2024, 9, 673–679. [Google Scholar]
- Kuang, Y. Delay Differential Equations: With Applications in Population Dynamics; Academic press: Cambridge, MA, USA, 1993. [Google Scholar]
- Bellen, A.; Zennaro, M. Numerical Methods for Delay Differential Equations; Oxford University Press: Oxford, UK, 2013. [Google Scholar]
- Cooke, K.L.; Yorke, J.A. Some equations modelling growth processes and gonorrhea epidemics. Math. Biosci. 1973, 16, 75–101. [Google Scholar]
- Khan, Q.J.A.; Krishnan, E.V. An Epidemic Model with a Time Delay in Transmission. Appl. Math. 2003, 48, 193–203. [Google Scholar] [CrossRef]
- Liu, L. A delayed SIR model with general nonlinear incidence rate. Adv. Differ. Equ. 2015, 2015, 329. [Google Scholar] [CrossRef]
- Jackson, M.; Chen-Charpentier, B.M. Modeling plant virus propagation with delays. J. Comput. Appl. Math. 2017, 309, 611–621. [Google Scholar]
- Beretta, E.; Takeuchi, Y. Global stability of an SIR epidemic model with time delays. J. Math. Biol. 1995, 33, 250–260. [Google Scholar]
- Ma, W.; Takeuchi, Y.; Hara, T.; Beretta, E. Permanence of an SIR epidemic model with distributed time delays. Tohoku Math. J. Second Ser. 2002, 54, 581–591. [Google Scholar]
- McCluskey, C.C. Complete global stability for an SIR epidemic model with delay—Distributed or discrete. Nonlinear Anal. Real World Appl. 2010, 11, 55–59. [Google Scholar]
- Van den Driessche, P. Reproduction numbers of infectious disease models. Infect. Dis. Model. 2017, 2, 288–303. [Google Scholar] [CrossRef] [PubMed]
- Doldo, P.; Pender, J. A note on the interpretation of distributed delay equations. arXiv 2021, arXiv:2106.11413. [Google Scholar]
- González-Parra, G.; Arenas, A.J.; Chen-Charpentier, B.M. A fractional order epidemic model for the simulation of outbreaks of influenza A (H1N1). Math. Methods Appl. Sci. 2014, 37, 2218–2226. [Google Scholar] [CrossRef]
- Area, I.; Batarfi, H.; Losada, J.; Nieto, J.J.; Shammakh, W.; Torres, Á. On a fractional order Ebola epidemic model. Adv. Differ. Equ. 2015, 2015, 1–12. [Google Scholar] [CrossRef]
- Hamdan, N.I.; Kilicman, A. A fractional order SIR epidemic model for dengue transmission. Chaos Solitons Fractals 2018, 114, 55–62. [Google Scholar] [CrossRef]
- Chatterjee, A.N.; Ahmad, B. A fractional-order differential equation model of COVID-19 infection of epithelial cells. Chaos Solitons Fractals 2021, 147, 110952. [Google Scholar] [CrossRef]
- Chen, Y.; Liu, F.; Yu, Q.; Li, T. Review of fractional epidemic models. Appl. Math. Model. 2021, 97, 281–307. [Google Scholar] [CrossRef]
- Petrás, I. Fractional Derivatives, Fractional Integrals, and Fractional Differential Equations in Matlab; IntechOpen: Rijeka, Croatia, 2011. [Google Scholar]
- Scherer, R.; Kalla, S.L.; Tang, Y.; Huang, J. The Grünwald–Letnikov method for fractional differential equations. Comput. Math. Appl. 2011, 62, 902–917. [Google Scholar] [CrossRef]
- Li, Z.; Liu, L.; Dehghan, S.; Chen, Y.; Xue, D. A review and evaluation of numerical tools for fractional calculus and fractional order controls. Int. J. Control 2017, 90, 1165–1181. [Google Scholar] [CrossRef]
- Garrappa, R. Numerical solution of fractional differential equations: A survey and a software tutorial. Mathematics 2018, 6, 16. [Google Scholar] [CrossRef]
- Siettos, C.I.; Russo, L. Mathematical modeling of infectious disease dynamics. Virulence 2013, 4, 295–306. [Google Scholar] [CrossRef] [PubMed]
- Brauer, F. Mathematical epidemiology: Past, present, and future. Infect. Dis. Model. 2017, 2, 113–127. [Google Scholar] [CrossRef]
- Mata, A.S.; Dourado, S.M. Mathematical modeling applied to epidemics: An overview. Sao Paulo J. Math. Sci. 2021, 15, 1025–1044. [Google Scholar] [CrossRef] [PubMed]
- Pauk, J.N.; Igwe, C.L.; Herwig, C. Workflow for adaptation, analysis and application of mechanistic models for experimental planning of protein refolding processes. In Computer Aided Chemical Engineering; Elsevier: Amsterdam, The Netherlands, 2023; Volume 52, pp. 481–486. [Google Scholar]
- Matsui, S.; Le-Rademacher, J.; Mandrekar, S.J. Statistical models in clinical studies. J. Thorac. Oncol. 2021, 16, 734–739. [Google Scholar] [CrossRef] [PubMed]
- Baker, R.E.; Pena, J.M.; Jayamohan, J.; Jérusalem, A. Mechanistic models versus machine learning, a fight worth fighting for the biological community? Biol. Lett. 2018, 14, 20170660. [Google Scholar] [CrossRef]
- Érdi, P.; Tóth, J. Mathematical Models of Chemical Reactions: Theory and Applications of Deterministic and Stochastic Models; Manchester University Press: Manchester, UK, 1989. [Google Scholar]
- Holling, C.S. The components of predation as revealed by a study of small-mammal predation of the European Pine Sawfly1. Can. Entomol. 1959, 91, 293–320. [Google Scholar] [CrossRef]
- DeAngelis, D.L.; Goldstein, R.; O’Neill, R.V. A model for tropic interaction. Ecology 1975, 56, 881–892. [Google Scholar] [CrossRef]
- Beddington, J.R. Mutual interference between parasites or predators and its effect on searching efficiency. J. Anim. Ecol. 1975, 40, 331–340. [Google Scholar] [CrossRef]
- Paulhus, C.; Wang, X.S. Global stability analysis of a delayed susceptible–infected–susceptible epidemic model. J. Biol. Dyn. 2015, 9, 45–50. [Google Scholar] [CrossRef]
- Takeuchi, Y.; Ma, W.; Beretta, E. Global asymptotic properties of a delay SIR epidemic model with finite incubation times. Nonlinear Anal. Theory Methods Appl. 2000, 42, 931–947. [Google Scholar] [CrossRef]
- Diekmann, O.; Gyllenberg, M.; Metz, J. Finite dimensional state representation of linear and nonlinear delay systems. J. Dyn. Differ. Equations 2018, 30, 1439–1467. [Google Scholar] [CrossRef]
- Cassidy, T.; Gillich, P.; Humphries, A.R.; van Dorp, C.H. Numerical methods and hypoexponential approximations for gamma distributed delay differential equations. IMA J. Appl. Math. 2022, 87, 1043–1089. [Google Scholar] [CrossRef]
- Zhao, W.; Zhang, T.; Chang, Z.; Meng, X.; Liu, Y. Dynamical analysis of SIR epidemic models with distributed delay. J. Appl. Math. 2013, 2013, 154387. [Google Scholar] [CrossRef]
- Caraballo, T.; Fatini, M.E.; Khalifi, M.E.; Gerlach, R.; Pettersson, R. Analysis of a Stochastic Distributed Delay Epidemic Model with Relapse and Gamma Distribution Kernel. Chaos Solitons Fractals 2020, 133, 109643. [Google Scholar] [CrossRef]
- Nevermann, D.H.; Gros, C. Mapping dynamical systems with distributed time delays to sets of ordinary differential equations. J. Phys. A Math. Theor. 2023, 56, 345702. [Google Scholar] [CrossRef]
- Podlubny, I. Fractional Differential Equations: An Introduction to Fractional Derivatives, Fractional Differential Equations, to Methods of Their Solution and Some of Their Applications; Elsevier: Amsterdam, The Netherlands, 1998. [Google Scholar]
- Kilbas, A.A.; Srivastava, H.M.; Trujillo, J.J. Theory and Applications of Fractional Differential Equations; Elsevier: Amsterdam, The Netherlands, 2006; Volume 204. [Google Scholar]
- Li, C.; Zhang, F. A survey on the stability of fractional differential equations: Dedicated to Prof. YS Chen on the Occasion of his 80th Birthday. Eur. Phys. J. Spec. Top. 2011, 193, 27–47. [Google Scholar] [CrossRef]
- Jin, B. Fractional Differential Equations; Springer: Berlin/Heidelberg, Germany, 2021. [Google Scholar]
- Chen-Charpentier, B. On Population Models with Delays and Dependence on Past Values. Axioms 2024, 13, 206. [Google Scholar] [CrossRef]
- Diekmann, O.; Heesterbeek, J.A.P.; Metz, J.A.J. On the definition and the computation of the basic reproduction ratio R 0 in models for infectious diseases in heterogeneous populations. J. Math. Biol. 1990, 28, 365–382. [Google Scholar] [CrossRef]
- Vargas-De-León, C. On the global stability of SIS, SIR and SIRS epidemic models with standard incidence. Chaos Solitons Fractals 2011, 44, 1106–1110. [Google Scholar] [CrossRef]
- Korobeinikov, A.; Wake, G.C. Lyapunov functions and global stability for SIR, SIRS, and SIS epidemiological models. Appl. Math. Lett. 2002, 15, 955–960. [Google Scholar] [CrossRef]
- Wei, H.M.; Li, X.Z.; Martcheva, M. An Epidemic Model of a Vector-Borne Disease with Direct Transmission and Time Delay. J. Math. Anal. Appl. 2008, 342, 895–908. [Google Scholar] [CrossRef]
- Al Basir, F.; Takeuchi, Y.; Ray, S. Dynamics of a Delayed Plant Disease Model with Beddington-DeAngelis Disease Transmission. Math. Biosci. Eng. 2021, 18, 583–599. [Google Scholar] [CrossRef]
- Huang, G.; Takeuchi, Y. Global analysis on delay epidemiological dynamic models with nonlinear incidence. J. Math. Biol. 2011, 63, 125–139. [Google Scholar] [CrossRef]
- Enatsu, Y. Lyapunov functional techniques on the global stability of equilibria of SIS epidemic models with delays (Analysis on non-equilibria and nonlinear phenomena: From the evolution equations point of view). Kyoto Univ. Res. Inf. Repos. 2012, 1792, 118–130. [Google Scholar]
- Erneux, T. Applied Delay Differential Equations; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2009; Volume 3. [Google Scholar]
- Marsden, J.E.; McCracken, M. The Hopf Bifurcation and Its Applications; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2012; Volume 19. [Google Scholar]
- Colizza, V.; Barrat, A.; Barthelemy, M.; Valleron, A.J.; Vespignani, A. Modeling the Worldwide Spread of Pandemic Influenza: Baseline Case and Containment Interventions. PLoS Med. 2007, 4, e13. [Google Scholar] [CrossRef]
- Centers for Disease Control. Influenza (Flu). 2024. Available online: https://www.cdc.gov/flu/about/?CDC_AAref_Val=https://www.cdc.gov/flu/about/keyfacts.htm (accessed on 8 April 2025).
- Kanyiri, C.W.; Mark, K.; Luboobi, L. Mathematical Analysis of Influenza A Dynamics in the Emergence of Drug Resistance. Comput. Math. Methods Med. 2018, 2018, e2434560. [Google Scholar] [CrossRef]
- MATLAB. Matlab, Version R2024a; The MathWorks Inc.: Natick, MA, USA, 2024. [Google Scholar]
- Shampine, L.F.; Thompson, S.; Kierzenka, J. Solving Delay Differential Equations with dde23. 2000. Available online: https://www.researchgate.net/publication/228818478_Solving_delay_differential_equations_with_dde23 (accessed on 5 September 2024).
- Burden, R.L.; J Douglas, F.; Burden, A.M. Numerical Analysis; Cengage: Boston, MA, USA, 2016. [Google Scholar]
- Engelborghs, K.; Luzyanina, T.; Roose, D. Numerical bifurcation analysis of delay differential equations using DDE-BIFTOOL. ACM Trans. Math. Softw. (TOMS) 2002, 28, 1–21. [Google Scholar] [CrossRef]
- Flint, S.J.; Racaniello, V.R.; Rall, G.F.; Hatziioannou, T.; Skalka, A.M. Principles of Virology, Volume 2: Pathogenesis and Control; John Wiley & Sons: Hoboken, NJ, USA, 2020; Volume 2. [Google Scholar]
- CDC. Symptoms of COVID-19. 2025. Available online: https://www.cdc.gov/covid/signs-symptoms/index.html (accessed on 1 February 2025).








| Symbol | Parameter Description | Value |
|---|---|---|
| Infection rate | 0.3663 /d | |
| Recovery rate | 1/3 /d | |
| Loss-of-immunity rate | 1/30 /d | |
| Delay in time of infection | 1.9 d | |
| 1/(time of infection) | /d | |
| N | Total population | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen-Charpentier, B. Mathematical Models of Epidemics with Infection Time. AppliedMath 2025, 5, 47. https://doi.org/10.3390/appliedmath5020047
Chen-Charpentier B. Mathematical Models of Epidemics with Infection Time. AppliedMath. 2025; 5(2):47. https://doi.org/10.3390/appliedmath5020047
Chicago/Turabian StyleChen-Charpentier, Benito. 2025. "Mathematical Models of Epidemics with Infection Time" AppliedMath 5, no. 2: 47. https://doi.org/10.3390/appliedmath5020047
APA StyleChen-Charpentier, B. (2025). Mathematical Models of Epidemics with Infection Time. AppliedMath, 5(2), 47. https://doi.org/10.3390/appliedmath5020047






