Measuring miRNA in Livestock Using Sensor Technologies: Challenges and Potential Approaches †
Abstract
:1. MicroRNAs: Small Molecules, High Impact
2. Role of miRNAs in Livestock Diseases and Conditions
2.1. Immunity
2.2. Mycobacterium Avium ssp. Paratuberculosis
2.3. Foot and Mouth Disease
2.4. Heat Stress in Livestock
2.5. Tumorigenesis and Cancer
2.6. Pregnancy and Lactation
2.7. Endometritis
3. Challenges to MiRNA Detection
4. MiRNA Research: Technological Advancements
4.1. Next-Generation Sequencing (NGS)
4.2. RNA Sequencing (RNA-Seq)
4.3. MinION and GridION
4.4. Ion Torrent Sequencing
4.5. Electrochemical Sensing
4.6. Loop-Mediated Isothermal Amplification (LAMP)
4.7. Surface Plasmon Resonance
5. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Saliminejad, K.; Khorram Khorshid, H.R.; Soleymani Fard, S.; Ghaffari, S.H. An Overview of MicroRNAs: Biology, Functions, Therapeutics, and Analysis Methods. J. Cell. Physiol. 2019, 234, 5451–5465. [Google Scholar] [CrossRef]
- Baptista, B.; Riscado, M.; Queiroz, J.A.; Pichon, C.; Sousa, F. Non-coding RNAs: Emerging from the discovery to therapeutic applications. Biochem. Pharmacol. 2021, 189, 114469. [Google Scholar] [CrossRef] [PubMed]
- Fabian, M.R.; Sonenberg, N.; Filipowicz, W. Regulation of MRNA Translation and Stability by MicroRNAs. Annu. Rev. Biochem. 2010, 79, 351–379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, Z.; Deng, H.; Shen, W.; Ren, Y. A Label-Free Biosensor for Electrochemical Detection of Femtomolar MicroRNAs. Anal. Chem. 2013, 85, 1624–1630. [Google Scholar] [CrossRef] [PubMed]
- Du, W.; Lv, M.; Li, J.; Yu, R.; Jiang, J. A Ligation-Based Loop-Mediated Isothermal Amplification (Ligation-LAMP) Strategy for Highly Selective MicroRNA Detection. Chem. Commun. 2016, 52, 12721–12724. [Google Scholar] [CrossRef] [PubMed]
- Rahaie, M.; Noroozi, S.K. A Nanobiosensor Based on Graphene Oxide and DNA Binding Dye for Multi-MicroRNAs Detection. Biosci. Rep. 2019, 39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.W.; Li, Z.; Moore, P.S.; Monaghan, A.P.; Chang, Y.; Nichols, M.; John, B. A Sensitive Non-Radioactive Northern Blot Method to Detect Small RNAs. Nucleic Acids Res. 2010, 38, e98. [Google Scholar] [CrossRef] [Green Version]
- Liang, G.; Malmuthuge, N.; Guan, Y.; Ren, Y.; Griebel, P.J.; Guan, L.L. Altered MicroRNA Expression and Pre-MRNA Splicing Events Reveal New Mechanisms Associated with Early Stage Mycobacterium Avium Subspecies Paratuberculosis Infection. Sci. Rep. 2016, 6, 24964. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.; Lee, S.; Son, J.; Lim, H.; Kim, E.; Kim, D.; Ha, S.; Hur, T.; Lee, S.; Choi, I. Analysis of Circulating-MicroRNA Expression in Lactating Holstein Cows under Summer Heat Stress. PLoS ONE 2020, 15, e0231125. [Google Scholar] [CrossRef]
- Casas, E.; Cai, G.; Kuehn, L.A.; Register, K.B.; McDaneld, T.G.; Neill, J.D. Association of MicroRNAs with Antibody Response to Mycoplasma Bovis in Beef Cattle. PLoS ONE 2016, 11, e0161651. [Google Scholar] [CrossRef]
- Schanzenbach, C.I.; Kirchner, B.; Ulbrich, S.E.; Pfaffl, M.W. Can Milk Cell or Skim Milk MiRNAs Be Used as Biomarkers for Early Pregnancy Detection in Cattle? PLoS ONE 2017, 12, e0172220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Otto, T.; Candido, S.V.; Pilarz, M.S.; Sicinska, E.; Bronson, R.T.; Bowden, M.; Lachowicz, I.A.; Mulry, K.; Fassl, A.; Han, R.C.; et al. Cell Cycle-Targeting MicroRNAs Promote Differentiation by Enforcing Cell-Cycle Exit. Proc. Nat. Acad. Sci. USA 2017, 114, 10660–10665. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Yang, C.; Du, J.; Zhang, B.; He, Y.; Hu, Q.; Li, M.; Zhang, Y.; Wang, C.; Zhong, J. Characterization of MiRNA Profiles in the Mammary Tissue of Dairy Cattle in Response to Heat Stress. BMC Genom. 2018, 19, 975. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Correia, C.N.; Nalpas, N.C.; McLoughlin, K.E.; Browne, J.A.; Gordon, S.V.; MacHugh, D.E.; Shaughnessy, R.G. Circulating MicroRNAs as Potential Biomarkers of Infectious Disease. Front. Immunol. 2017, 8, 118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ioannidis, J.; Donadeu, F.X. Circulating MiRNA Signatures of Early Pregnancy in Cattle. BMC Genom. 2016, 17, 184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Liu, C. Coding or Noncoding, the Converging Concepts of RNAs. Front. Genet. 2019, 10, 496. [Google Scholar] [CrossRef]
- Ioannidis, J.; Donadeu, F.X. Comprehensive Analysis of Blood Cells and Plasma Identifies Tissue-Specific MiRNAs as Potential Novel Circulating Biomarkers in Cattle. BMC Genom. 2018, 19, 243. [Google Scholar] [CrossRef] [Green Version]
- Chen, M.; Yao, Y.L.; Yang, Y.; Zhu, M.; Tang, Y.; Liu, S.; Li, K.; Tang, Z. Comprehensive Profiles of MRNAs and MiRNAs Reveal Molecular Characteristics of Multiple Organ Physiologies and Development in Pigs. Front. Genet. 2019, 10, 756. [Google Scholar] [CrossRef] [Green Version]
- Gupta, S.K.; Maclean, P.H.; Ganesh, S.; Shu, D.; Buddle, B.M.; Wedlock, D.N.; Heiser, A. Detection of MicroRNA in Cattle Serum and Their Potential Use to Diagnose Severity of Johne’s Disease. J. Dairy Sci. 2018, 101, 10259–10270. [Google Scholar] [CrossRef]
- Muroya, S.; Shibata, M.; Hayashi, M.; Oe, M.; Ojima, K. Differences in Circulating MicroRNAs between Grazing and Grain-Fed Wagyu Cattle Are Associated with Altered Expression of Intramuscular MicroRNA, the Potential Target PTEN, and Lipogenic Genes. PLoS ONE 2016, 11, e0162496. [Google Scholar] [CrossRef]
- Sengar, G.S.; Deb, R.; Singh, U.; Raja, T.V.; Kant, R.; Sajjanar, B.; Alex, R.; Alyethodi, R.R.; Kumar, A.; Kumar, S.; et al. Differential Expression of MicroRNAs Associated with Thermal Stress in Frieswal (Bos Taurus x Bos Indicus) Crossbred Dairy Cattle. Cell Stress Chaperon. 2018, 23, 155–170. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Zhou, Y.; Zhang, H.; Meng, X.; Ai, S. Electrochemical Determination of MicroRNA-21 Based on Graphene, LNA Integrated Molecular Beacon, AuNPs and Biotin Multifunctional Bio Bar Codes and Enzymatic Assay System. Biosens. Bioelectron. 2012, 33, 247–253. [Google Scholar] [CrossRef] [PubMed]
- Labib, M.; Berezovski, M.V. Electrochemical Sensing of MicroRNAs: Avenues and Paradigms. Biosens. Bioelectron. 2015, 68, 83–94. [Google Scholar] [CrossRef]
- Gao, A.; Lu, N.; Wang, Y.; Dai, P.; Li, T.; Gao, X.; Wang, Y.; Fan, C. Enhanced Sensing of Nucleic Acids with Silicon Nanowire Field Effect Transistor Biosensors. Nano Lett. 2012, 12, 5262–5268. [Google Scholar] [CrossRef] [PubMed]
- Gu, J.; Qiao, Z.; He, X.; Yu, Y.; Lei, Y.; Tang, J.; Shi, H.; He, D.; Wang, K. Enzyme-Free Amplified Detection of MiRNA Based on Target-Catalyzed Hairpin Assembly and DNA-Stabilized Fluorescent Silver Nanoclusters. Analyst 2020, 145, 5194–5199. [Google Scholar] [CrossRef]
- Zhang, J.; Li, S.; Li, L.; Li, M.; Guo, C.; Yao, J.; Mi, S. Exosome and Exosomal MicroRNA: Trafficking, Sorting, and Function. Genom. Proteom. Bioinform. 2015, 13, 17–24. [Google Scholar] [CrossRef] [Green Version]
- Shi, C.; Liu, Q.; Ma, C.; Zhong, W. Exponential Strand-Displacement Amplification for Detection of MicroRNAs. Anal. Chem. 2014, 86, 336–339. [Google Scholar] [CrossRef] [Green Version]
- Cammaerts, S.; Strazisar, M.; De Rijk, P.; Del Favero, J. Genetic Variants in MicroRNA Genes: Impact on MicroRNA Expression, Function, and Disease. Front. Genet. 2015, 6. [Google Scholar] [CrossRef] [Green Version]
- Horikawa, A.; Ogasawara, H.; Okada, K.; Kobayashi, M.; Muroya, S.; Hojito, M. Grazing-Induced Changes in Muscle MicroRNA-206 and -208b Expression in Association with Myogenic Gene Expression in Cattle. Anim. Sci. J. 2015, 86, 952–960. [Google Scholar] [CrossRef]
- Basagoudanavar, S.H.; Hosamani, M.; Tamil Selvan, R.P.; Sreenivasa, B.P.; Sanyal, A.; Venkataramanan, R. Host Serum MicroRNA Profiling during the Early Stage of Foot-and-Mouth Disease Virus Infection. Arch. Virol. 2018, 163, 2055–2063. [Google Scholar] [CrossRef]
- Sengar, G.S.; Deb, R.; Singh, U.; Junghare, V.; Hazra, S.; Raja, T.V.; Alex, R.; Kumar, A.; Alyethodi, R.R.; Kant, R.; et al. Identification of Differentially Expressed MicroRNAs in Sahiwal (Bos Indicus) Breed of Cattle during Thermal Stress. Cell Stress Chaperon. 2018, 23, 1019–1032. [Google Scholar] [CrossRef] [PubMed]
- Deng, R.; Zhang, K.; Li, J. Isothermal Amplification for MicroRNA Detection: From the Test Tube to the Cell. Accounts Chem. Res. 2017, 50, 1059–1068. [Google Scholar] [CrossRef] [PubMed]
- Gines, G.; Menezes, R.; Nara, K.; Kirstetter, A.-S.; Taly, V.; Rondelez, Y. Isothermal Digital Detection of MicroRNAs Using Background-Free Molecular Circuit. Sci. Adv. 2020, 6, eaay5952. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, D.; Xia, L.; Zhou, Q.; Wang, L.; Chen, D.; Gao, X.; Li, Y. Label-Free Detection of MiRNA Using Surface-Enhanced Raman Spectroscopy. Anal. Chem. 2020, 92, 12769–12773. [Google Scholar] [CrossRef]
- Persano, S.; Guevara, M.L.; Wolfram, J.; Blanco, E.; Shen, H.; Ferrari, M.; Pompa, P.P. Label-Free Isothermal Amplification Assay for Specific and Highly Sensitive Colorimetric MiRNA Detection. ACS Omega 2016, 1, 448–455. [Google Scholar] [CrossRef]
- Rosa, A.; Ballarino, M. Long Noncoding RNA Regulation of Pluripotency. Stem Cells Int. 2016, 2016, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Yao, S. MicroRNA Biogenesis and Their Functions in Regulating Stem Cell Potency and Differentiation. Biol. Proced. Online 2016, 18, 8. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Ruan, K. MicroRNA Detection by Microarray. Anal. Bioanal. Chem. 2009, 394, 1117–1124. [Google Scholar] [CrossRef]
- Ouyang, T.; Liu, Z.; Han, Z.; Ge, Q. MicroRNA Detection Specificity: Recent Advances and Future Perspective. Anal. Chem. 2019, 91, 3179–3186. [Google Scholar] [CrossRef] [Green Version]
- Ye, D.; Shen, Z.; Zhou, S. Function of microRNA-145 and mechanisms underlying its role in malignant tumor diagnosis and treatment. Cancer Manag. Res. 2019, 11, 969–979. [Google Scholar] [CrossRef]
- Taxis, T.M. MicroRNA Expression and Implications for Infectious Diseases in Livestock. CAB Rev. Perspect. Agric. Veter. Sci. Nutr. Nat. Resour. 2017, 2017, 12. [Google Scholar] [CrossRef]
- Wang, M.; Moisá, S.; Khan, M.J.; Wang, J.; Bu, D.; Loor, J.J. MicroRNA Expression Patterns in the Bovine Mammary Gland Are Affected by Stage of Lactation. J. Dairy Sci. 2012, 95, 6529–6535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lenkala, D.; LaCroix, B.; Gamazon, E.R.; Geeleher, P.; Im, H.K.; Huang, R.S. The Impact of MicroRNA Expression on Cellular Proliferation. Hum. Genet. 2014, 133, 931–938. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, H.-Z.; Chen, Y.; Guan, L.L. MicroRNA Expression Profiles across Blood and Different Tissues in Cattle. Sci. Data 2019, 6, 190013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Timoneda, O.; Núñez-Hernández, F.; Balcells, I.; Muñoz, M.; Castelló, A.; Vera, G.; Pérez, L.J.; Egea, R.; Mir, G.; Córdoba, S.; et al. The Role of Viral and Host MicroRNAs in the Aujeszky’s Disease Virus during the Infection Process. PLoS ONE 2014, 9, e86965. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Cheng, Z.; Wang, L.; Jiao, B.; Yang, H.; Wang, X. MiR-21-3p Centric Regulatory Network in Dairy Cow Mammary Epithelial Cell Proliferation. J. Agric. Food Chem. 2019, 67, 11137–11147. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, M.; Sharma, N.; Singh, A.K.; Gera, M.; Pulicherla, K.K.; Jeong, D.K. Transformation of Animal Genomics by Next-Generation Sequencing Technologies: A Decade of Challenges and Their Impact on Genetic Architecture. Criti. Rev. Biotechnol. 2018, 38, 1157–1175. [Google Scholar] [CrossRef] [PubMed]
- Schnall-Levin, M.; Rissland, O.S.; Johnston, W.K.; Perrimon, N.; Bartel, D.P.; Berger, B. Unusually Effective MicroRNA Targeting within Repeat-Rich Coding Regions of Mammalian MRNAs. Genome Res. 2011, 21, 1395–1403. [Google Scholar] [CrossRef] [Green Version]
- Sun, J.; Aswath, K.; Schroeder, S.G.; Lippolis, J.D.; Reinhardt, T.A.; Sonstegard, T.S. MicroRNA Expression Profiles of Bovine Milk Exosomes in Response to Staphylococcus Aureus Infection. BMC Genom. 2015, 16, 806. [Google Scholar] [CrossRef] [Green Version]
- Barbu, M.G.; Condrat, C.E.; Thompson, D.C.; Bugnar, O.L.; Cretoiu, D.; Toader, O.D.; Suciu, N.; Voinea, S.C. MicroRNA Involvement in Signaling Pathways During Viral Infection. Front. Cell Dev. Biol. 2020, 8, 143. [Google Scholar] [CrossRef] [PubMed]
- Kuosmanen, S.M.; Kansanen, E.; Sihvola, V.; Levonen, A.-L. MicroRNA Profiling Reveals Distinct Profiles for Tissue-Derived and Cultured Endothelial Cells. Sci. Rep. 2017, 7, 10943. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lawless, N.; Reinhardt, T.A.; Bryan, K.; Baker, M.; Pesch, B.; Zimmerman, D.; Zuelke, K.; Sonstegard, T.; O’Farrelly, C.; Lippolis, J.D.; et al. MicroRNA Regulation of Bovine Monocyte Inflammatory and Metabolic Networks in an in Vivo Infection Model. G3 Genes|Genomes|Genetics 2014, 4, 957–971. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oladejo, A.O.; Li, Y.; Wu, X.; Imam, B.H.; Shen, W.; Ding, X.Z.; Wang, S.; Yan, Z. MicroRNAome: Potential and Veritable Immunomolecular Therapeutic and Diagnostic Baseline for Lingering Bovine Endometritis. Front. Veter. Sci. 2020, 7, 614054. [Google Scholar] [CrossRef]
- Sato, F.; Tsuchiya, S.; Meltzer, S.J.; Shimizu, K. MicroRNAs and Epigenetics. FEBS J. 2011, 278, 1598–1609. [Google Scholar] [CrossRef]
- Miretti, S.; Lecchi, C.; Ceciliani, F.; Baratta, M. MicroRNAs as Biomarkers for Animal Health and Welfare in Livestock. Front. Veter. Sci. 2020, 7, 578193. [Google Scholar] [CrossRef]
- Friedman, R.C.; Farh, K.K.-H.; Burge, C.B.; Bartel, D.P. Most Mammalian MRNAs Are Conserved Targets of MicroRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef] [Green Version]
- Jia, L.; Jiang, M.; Wu, K.; Hu, J.; Wang, Y.; Quan, W.; Hao, M.; Liu, H.; Wei, H.; Fan, W.; et al. Nanopore Sequencing of African Swine Fever Virus. Sci. China Life Sci. 2020, 63, 160–164. [Google Scholar] [CrossRef]
- Deiuliis, J.A. MicroRNAs as Regulators of Metabolic Disease: Pathophysiologic Significance and Emerging Role as Biomarkers and Therapeutics. Int. J. Obes. 2016, 40, 88–101. [Google Scholar] [CrossRef] [Green Version]
- Cortez, M.A.; Bueso-Ramos, C.; Ferdin, J.; Lopez-Berestein, G.; Sood, A.K.; Calin, G.A. MicroRNAs in Body Fluids—The Mix of Hormones and Biomarkers. Nat. Rev. Clin. Oncol. 2011, 8, 467–477. [Google Scholar] [CrossRef] [Green Version]
- Shirjang, S.; Mansoori, B.; Asghari, S.; Duijf, P.H.G.; Mohammadi, A.; Gjerstorff, M.; Baradaran, B. MicroRNAs in Cancer Cell Death Pathways: Apoptosis and Necroptosis. Free Radic. Biol. Med. 2019, 139, 1–15. [Google Scholar] [CrossRef]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. MiRBase: From MicroRNA Sequences to Function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef] [PubMed]
- Leti, F.; DiStefano, J.K. MiRNA Quantification Method Using Quantitative Polymerase Chain Reaction in Conjunction with C q Method. In Methods in Molecular Biology; DiStefano, J., Ed.; Humana Press: New York, NY, USA, 2018; Volume 1706, pp. 257–265. [Google Scholar]
- Zheng, C.Y.; Zou, X.; Lin, H.J.; Zhao, B.C.; Zhang, M.L.; Luo, C.H.; Fu, S.X. MiRNA-185 Regulates the VEGFA Signaling Pathway in Dairy Cows with Retained Fetal Membranes. Theriogenology 2018, 110, 116–121. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.-X.; Guo, T.; Lu, H.; Yue, L.; Li, Y.; Jin, D.; Zhang, G.-J.; Yang, F. Nanostructuring Synergetic Base-Stacking Effect: An Enhanced Versatile Sandwich Sensor Enables Ultrasensitive Detection of MicroRNAs in Blood. ACS Sens. 2020, 5, 2514–2522. [Google Scholar] [CrossRef]
- Lawless, N.; Foroushani, A.B.K.; McCabe, M.S.; O’Farrelly, C.; Lynn, D.J. Next Generation Sequencing Reveals the Expression of a Unique MiRNA Profile in Response to a Gram-Positive Bacterial Infection. PLoS ONE 2013, 8, e57543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, X.; Nie, Q.; Zhang, X.; Nolan, L.K.; Lamont, S.J. Novel MicroRNA Involved in Host Response to Avian Pathogenic Escherichia Coli Identified by Deep Sequencing and Integration Analysis. Infect. Immun. 2017, 85, e00688-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, H.; Giordano, F.; Ning, Z. Oxford Nanopore MinION Sequencing and Genome Assembly. Genom. Proteom. Bioinform. 2016, 14, 265–279. [Google Scholar] [CrossRef] [Green Version]
- Stenfeldt, C.; Arzt, J.; Smoliga, G.; LaRocco, M.; Gutkoska, J.; Lawrence, P. Proof-of-Concept Study: Profile of Circulating MicroRNAs in Bovine Serum Harvested during Acute and Persistent FMDV Infection. Virol. J. 2017, 14, 71. [Google Scholar] [CrossRef] [Green Version]
- Okotrub, K.A.; Surovtsev, N.V.; Semeshin, V.F.; Omelyanchuk, L.V. Raman Spectroscopy for DNA Quantification in Cell Nucleus. Cytom. Part A 2015, 87, 68–73. [Google Scholar] [CrossRef]
- Wen, Q.; Liang, X.; Pan, H.; Li, J.; Zhang, Y.; Zhu, W.; Long, Z. Rapid and Sensitive Electrochemical Detection of MicroRNAs by Gold Nanoparticle-Catalyzed Silver Enhancement. Analyst 2020, 145, 7893–7897. [Google Scholar] [CrossRef]
- Gebert, L.F.R.; MacRae, I.J. Regulation of MicroRNA Function in Animals. Nat. Rev. Mol. Cell Biol. 2019, 20, 21–37. [Google Scholar] [CrossRef]
- Malvisi, M.; Palazzo, F.; Morandi, N.; Lazzari, B.; Williams, J.L.; Pagnacco, G.; Minozzi, G. Responses of Bovine Innate Immunity to Mycobacterium Avium Subsp. Paratuberculosis Infection Revealed by Changes in Gene Expression and Levels of MicroRNA. PLoS ONE 2016, 11, e0164461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xue, T.; Liang, W.; Li, Y.; Sun, Y.; Xiang, Y.; Zhang, Y.; Dai, Z.; Duo, Y.; Wu, L.; Qi, K.; et al. Ultrasensitive Detection of MiRNA with an Antimonene-Based Surface Plasmon Resonance Sensor. Nat. Commun. 2019, 10, 28. [Google Scholar] [CrossRef] [PubMed]
- Lecchi, C.; Zamarian, V.; Gini, C.; Avanzini, C.; Polloni, A.; Rota Nodari, S.; Ceciliani, F. Salivary MicroRNAs Are Potential Biomarkers for the Accurate and Precise Identification of Inflammatory Response after Tail Docking and Castration in Piglets. J. Anim. Sci. 2020, 98, skaa153. [Google Scholar] [CrossRef]
- Hu, F.; Xu, J.; Chen, Y. Surface Plasmon Resonance Imaging Detection of Sub-Femtomolar MicroRNA. Anal. Chem. 2017, 89, 10071–10077. [Google Scholar] [CrossRef]
- Sohel, M.M.H. Macronutrient modulation of mRNA and microRNA function in animals: A review. Anim. Nutr. 2020, 6, 258–268. [Google Scholar] [CrossRef]
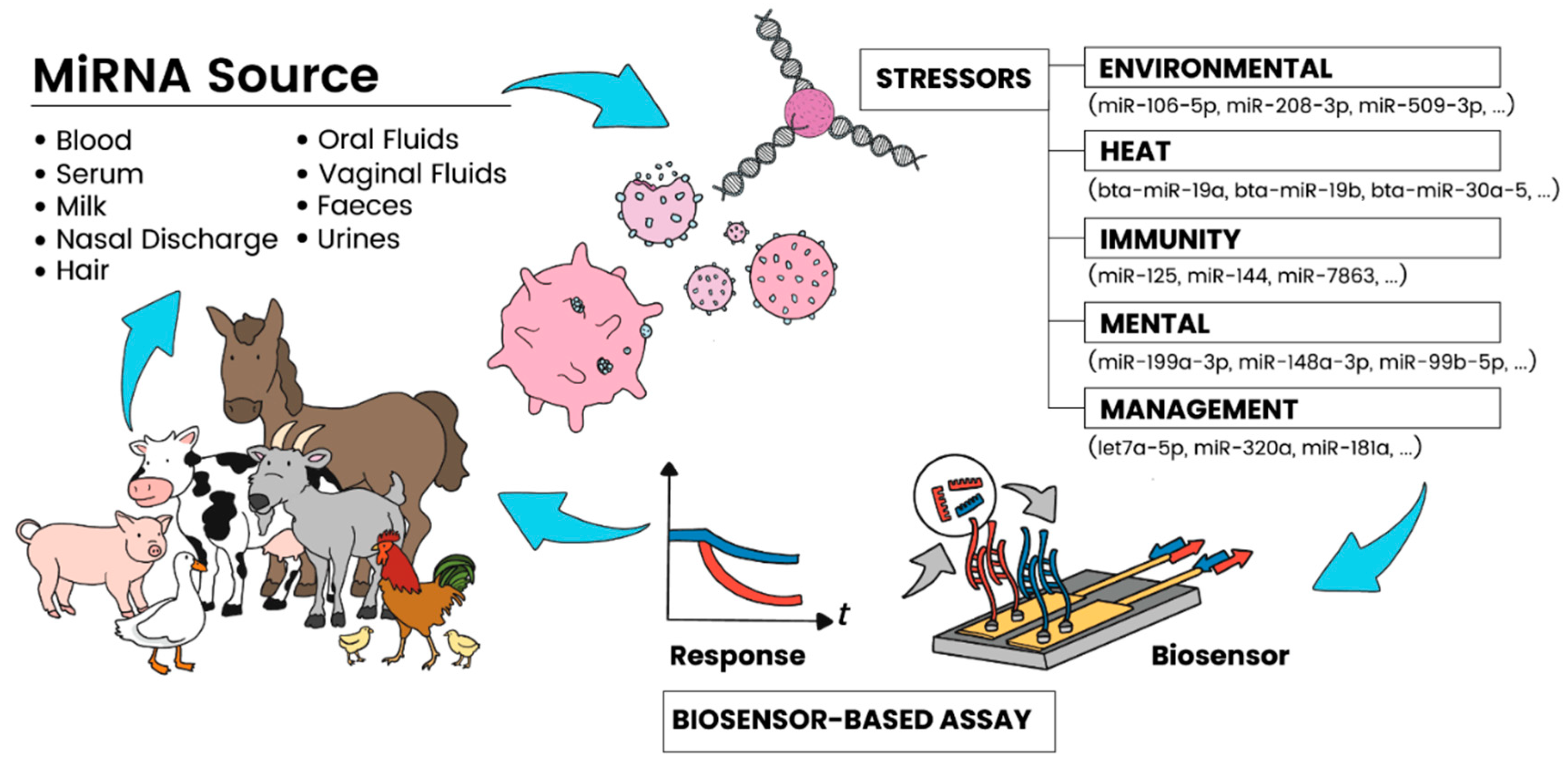

| Livestock Disease/Condition | Analyte Source | miRNAs Involved | miRNA Characteristics | Detection Method | References |
|---|---|---|---|---|---|
| Pregnancy | |||||
| Pregnancy diagnosis | Plasma samples | miR-99b, miR-152, miR-101, miR-103 | C19MC cluster miRNAs, miR-516-5p, miR-518b, miR-520a and miR-525 | Small-RNA sequencing, RT-qPCR | [40] |
| Pregnancy and lactation in Holstein cows | Mammary gland tissue, serum | Group 1. miR-10a, miR-15b, miR-16, miR-21, miR-31, miR-33b, miR-145, miR-146b, miR-155, miR-181a, miR-205, miR-221, and miR-223 | Group 1. All miRNAs except miR-31 showed increases in expression between -30 days prepartum and 7 days postpartum miR-31, a known inhibitor of cyclin gene expression Expression of miR-221 was shown to further increase 30 days postpartum, which corresponded to early lactation. A role for miR-221 in the control of endothelial cell proliferation and/or angiogenesis | RT-qPCR | [36] |
| Retained fetal membrane syndrome | Serum | miR-185 | miRNA-185 regulates the VEGFA signaling pathway in dairy cows with retained fetal membranes | qPCR and Western blotting | [41] |
| Endometritis | Bovine mammary gland epithelial cells | miR-21-3p | Plays an important role in promoting the viability and proliferation of the epithelial tissue in the mammary glands of dairy cows | MTT assay, flow cytometry analysis, Dual luciferase assay, RT-qPCR, and Western blot | [42] |
| Liver function and post-partum | Blood cells, plasma | miR-802 | Insulin sensitivity and lipid metabolism | Small RNA sequencing, RT-qPCR | [38] |
| Heat stress in pregnancy | Serum | Group 1. bta-miR-19a, bta-miR-19b, bta-miR-30a-5p, bta-miR-2284 Group 2. bta-miR-146b, bta-miR-20b, bta-miR-29d-3p, bta-miR-1246 | Group 1. miRNAs were differentially expressed in both pregnant and non-pregnant cows under heat stress conditions. Group 2. Targeted progesterone biosynthesis (StAR) and the function of corpus luteum-related genes (CCL11, XCL), | [31] | |
| Heat stress | |||||
| Heat stress | Serum | bta-miR-21-5p, bta-miR-99a-5p, bta-miR-146b, bta-miR-145, bta-miR-2285, bta-miR-133a, bta-miR-29c bta-miR-423-5p | miRNAs may act as dominant regulators during heat stress | Deep RNA sequencing, stem-loop qPCR | [32] |
| Diseases | |||||
| Paratuberculosis | Serum | 1. miR-1976, miR-873-3p, miR-520f-3p, and miR-126-3p 2. Increase in miR-6517, miR-7857, miR-24-1, miR-24- 2, miR-378c 3. Decrease in miR-19b, miR-19b-2, miR-1271, miR100, miR-301a, miR-32a | miRNA expression can distinguish moderate and severely infected animals from noninfected animals. | NanoString nCounter technology | [20] |
| Foot and mouth disease | Serum | bta-miR-21-5p, bta-miR-101, bta-miR-126-3p, bta-miR-145, bta-miR-197, bta-miR-223 | Compared to prior to infection, on day 2 post-infection, 119 miRNAs were upregulated, of which 39 were significantly upregulated. Serum miRNA upregulation before the appearance of clinical signs. These circulating miRNAs are released by lysed cells or secreted from cells in a paracrine manner | Microarray, RT-qPCR | [21] |
| Foot and mouth disease | Serum | Group 1. bta-miR-23b-5p, let-7 g, bta-miR-22-5p, bta-miR-1224, bta-miR-144, bta-miR-497, bta-miR-455-3p, bta-miR-154a, bta-miR-369-3p, bta-miR-26b, bta-miR-34a, bta-miR-205, bta-miR-181b, bta-miR-146a, bta-miR-17-5p, bta-miR-31 Group 2. bta-miR-26b, bta-miR-34a, bta-miR-205, bta-miR-181b, bta-miR-146a, bta-miR-17-5p, bta-miR-31, bta-miR-150, bta-miR-147 3. miR-1281, bta-miR-17-5p, bta-mir-31 | Group 1. Cellular proliferation or apoptosis Group 2. Immune modulatory function Group 3. Tumor suppressors | miRNA PCR array plates | [43] |
| Bovine mastitis | Milk-isolated monocytes | bta-miR-615, bta-miR-451, bta-miR-451, bta-miR-146b, bta-miR-411a, miR-149 | Downregulated miRNAs are highly enriched for roles in innate immunity; upregulated miRNAs preferentially target genes involved in metabolism | Next-generation sequencing | [25] |
| Mycoplasma bovis | Serum | 1. bta-let-7b, bta-miR- 24-3p, bta-miR- 92a, bta-miR-423-5p 2. Increase miR-155. miR-146a, miR-146b-5p, miR-886-5p 3. Decrease miR-20a, miR-191, miR-378, miR-30c, miR-423-5p. miR-374a, miR-185, miR768-5p, miR-18 | These microRNAs have been recognized as playing a role in the host defense against bacteria. | Next-generation sequencing | [26] |
| Bovine viral diarrhea virus | Serum | Bta-miR-423-5p, bta-miR-151-3p | miRNA expression involved in host immune response | Next-generation sequencing | [27] |
| Staphylococcus aureus | Bovine milk exosomes | bta-miR-142-5p, bta-miR-223 | Potential biomarkers for the early detection of bacterial infection of the mammary gland. | Next-generation sequencing | [28] |
| Aujeszky’s disease virus [ADV] (also known as suid herpesvirus type 1 [SuHV-1]) infection | Tissue samples (Olfactory bulb (OB) and trigeminal ganglia (TG) | miR-206, miR-133a, miR-133b, miR-378 | Pathways related to viral infection processes and immune response | Ion Torrent sequencing, RT-qPCR | [44] |
| Metabolic and other disorders | |||||
| Insulin resistance | miR-1281 ction. Stenfeldt et al., reported that was significantly decreased during both the acute and the chronic stages of the disease Conversely, miR-17-5p was expressed at the highest level during the acute stage. The level of bta-miR-31 was also significantly increased during the persistent stage of the disease. | [22] | |||
| Heat stress | Serum | bta-miR-21-5p, bta-miR-99a-5p, bta-miR-146b, bta-miR-145, bta-miR-2285, bta-miR-133a, bta-miR-29c bta-miR-423-5p | miRNAs may act as dominant regulators during heat stress | Deep RNA sequencing, stem-loop qPCR | [32] |
| Acute pain, 4-d-old piglets | Saliva | miR-19b, miR-27b-3p, miR-215, miR-22-3p, miR-155-5p, hsa-miR-365-5p, hsa-miR-204 | Focal adhesion pathways and cytokines expression | RT-qPCR | [45] |
| Respiratory diseases | Serum, milk | bta-miR-423-5p, bta-miR-151-3p | Avoidance of host immune response | NGS | [27] |
| Skeletal muscle development | Biceps femoris muscle | miR-206, mi-208b | Regulation of muscle gene expression during skeletal muscle adaptation to grazing. | RT-PCR | [46,47] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Neethirajan, S. Measuring miRNA in Livestock Using Sensor Technologies: Challenges and Potential Approaches. Biol. Life Sci. Forum 2022, 10, 3. https://doi.org/10.3390/blsf2022010003
Neethirajan S. Measuring miRNA in Livestock Using Sensor Technologies: Challenges and Potential Approaches. Biology and Life Sciences Forum. 2022; 10(1):3. https://doi.org/10.3390/blsf2022010003
Chicago/Turabian StyleNeethirajan, Suresh. 2022. "Measuring miRNA in Livestock Using Sensor Technologies: Challenges and Potential Approaches" Biology and Life Sciences Forum 10, no. 1: 3. https://doi.org/10.3390/blsf2022010003
APA StyleNeethirajan, S. (2022). Measuring miRNA in Livestock Using Sensor Technologies: Challenges and Potential Approaches. Biology and Life Sciences Forum, 10(1), 3. https://doi.org/10.3390/blsf2022010003






