Proteomic and Genetic Approach for Lunasin Peptide and Gene Presence Detection in Various Plants †
Abstract
:1. Introduction
2. Materials and Methods
2.1. Biological Material
2.2. Protein and DNA Extraction
2.2.1. Protein Extraction
2.2.2. DNA Extraction
2.3. Analyses of Lunasin Peptide Presence and Detection of Genetic Information for Lunasin
2.3.1. Electrophoretic Separation
2.3.2. Polymerase Chain Reaction (PCR)
2.3.3. Detection System
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mejia, E.; Vásconez, M.; De Lumen, B.; Nelson, R. Lunasin Concentration in Different Soybean Genotypes, Commercial Soy Protein, and Isoflavone Products. J. Agric. Food Chem. 2004, 52, 5882–5887. [Google Scholar] [CrossRef] [PubMed]
- Jeong, H.J.; Jeong, J.B.; Kim, D.S.; De Lumen, B.O. Inhibition of Core Histone Acetylation by the Cancer Preventive Peptide Lunasin. J. Agric. Food Chem. 2007, 55, 632–637. [Google Scholar] [CrossRef]
- Nakurte, I.; Klavins, K.; Kirhnere, I. Discovery of lunasin peptide in triticale (X Triticosecale Wittmack). J. Cereal Sci. 2012, 56, 510–514. [Google Scholar] [CrossRef]
- Wan, X.; Liu, H.; Sun, Y. Lunasin: A promising polypeptide for the prevention and treatment of cancer (Review). Oncol. Lett. 2017, 13, 3997–4001. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsieh, C.H.; Martínez-Villaluenga, C.; De Lumen, B.O. Updating the research on the chemopreventive and therapeutic role of the peptide lunasin: Properties of the peptide lunasin. J. Sci. Food Agric. 2018, 98, 2070–2079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Lumen, B.O. Lunasin: A Cancer-Preventive Soy Peptide. Nutr. Rev. 2005, 63, 16–21. [Google Scholar] [CrossRef] [PubMed]
- Odani, S.; Koide, T.; Ono, T. Amino acid sequence of a soybean (Glycine max) seed polypeptide having a poly(L-aspartic acid) structure. J. Biol. Chem. 1987, 262, 10502–10505. [Google Scholar] [CrossRef]
- Guha, S.; Majumder, K. Structural-features of food-derived bioactive peptides with anti-inflammatory activity: A brief review. J. Food Biochem. 2019, 43, e12531. [Google Scholar] [CrossRef]
- Alaswad, A.A.; Krishnan, H.B. Immunological Investigation for the Presence of Lunasin, a Chemopreventive Soybean Peptide, in the Seeds of Diverse Plants. J. Agric. Food Chem. 2016, 64, 2901–2909. [Google Scholar] [CrossRef] [PubMed]
- Dia, V.P.; Frankland-Searby, S.; Del Hierro, F.L. Structural property of soybean lunasin and development of a method to quantify lunasin in plasma using an optimized immunoassay protocol. Food Chem. 2013, 138, 334–341. [Google Scholar] [CrossRef]
- Aleksis, R.; Jaudzems, K.; Muceniece, R. Lunasin is a redox sensitive intrinsically disordered peptide with two transiently populated α-helical regions. Peptides 2016, 85, 56–62. [Google Scholar] [CrossRef] [PubMed]
- Wrigley, C.W. Identification of Cereal Varieties by Gel Electrophoresis of the Grain Proteins; Springer: Berlin/Heidelberg, Germany, 1992; pp. 17–41. [Google Scholar]
- Kuťka-Hlozáková, T.; Gregová, E.; Gálová, Z. Genetic diversity of GLU-1 in European wheat gnetic resources and varieties. J. Microbiol. Biotechnol. Food Sci. 2015, 4, 23–25. [Google Scholar] [CrossRef] [Green Version]
- Jeong, H.J.; Jeong, J.B.; Hsieh, C.C. Lunasin Is Prevalent in Barley and Is Bioavailable and Bioactive in In Vivo and In Vitro Studies. Nutr. Cancer 2010, 62, 1113–1119. [Google Scholar] [CrossRef] [PubMed]
- Jeong, H.J.; Lee, J.R.; Jeong, J.B. The Cancer Preventive Seed Peptide Lunasin From Rye Is Bioavailable and Bioactive. Nutr. Cancer 2009, 61, 680–686. [Google Scholar] [CrossRef] [PubMed]
- Nakurte, I.; Kirhnere, I.; Namniece, J. Detection of the lunasin peptide in oats (Avena sativa L). J. Cereal Sci. 2013, 57, 319–324. [Google Scholar] [CrossRef]
- Silva-Sánchez, C.; de la Rosa, A.P.; León-Galván, M.F.; de Lumen, B.O.; de León-Rodríguez, A.; de Mejía, E.G. Bioactive peptides in amaranth (Amaranthus hypochondriacus) seed. J. Agric. Food Chem. 2008, 56, 1233–1240. [Google Scholar] [CrossRef] [PubMed]
- Ren, G.; Zhu, Y.; Shi, Z. Detection of lunasin in quinoa (Chenopodium quinoa Willd.) and the in vitro evaluation of its antioxidant and anti-inflammatory activities. J. Sci. Food Agric. 2017, 97, 4110–4116. [Google Scholar] [CrossRef] [PubMed]
- Dinelli, G.; Bregola, V.; Bosi, S.; Fiori, J.; Gotti, R.; Simonetti, E.; Trozzi, C.; Leoncini, E.; Prata, C.; Massaccesi, L.; et al. Lunasin in wheat: A chemical and molecular study on its presence or absence. Food Chem. 2014, 151, 520–525. [Google Scholar] [CrossRef] [PubMed]
- Michell, R.; Lovergrove, A.; Shewry, P. Lunasin in cereal seeds: What is the origin? J. Cereal Sci. 2013, 57, 267–269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
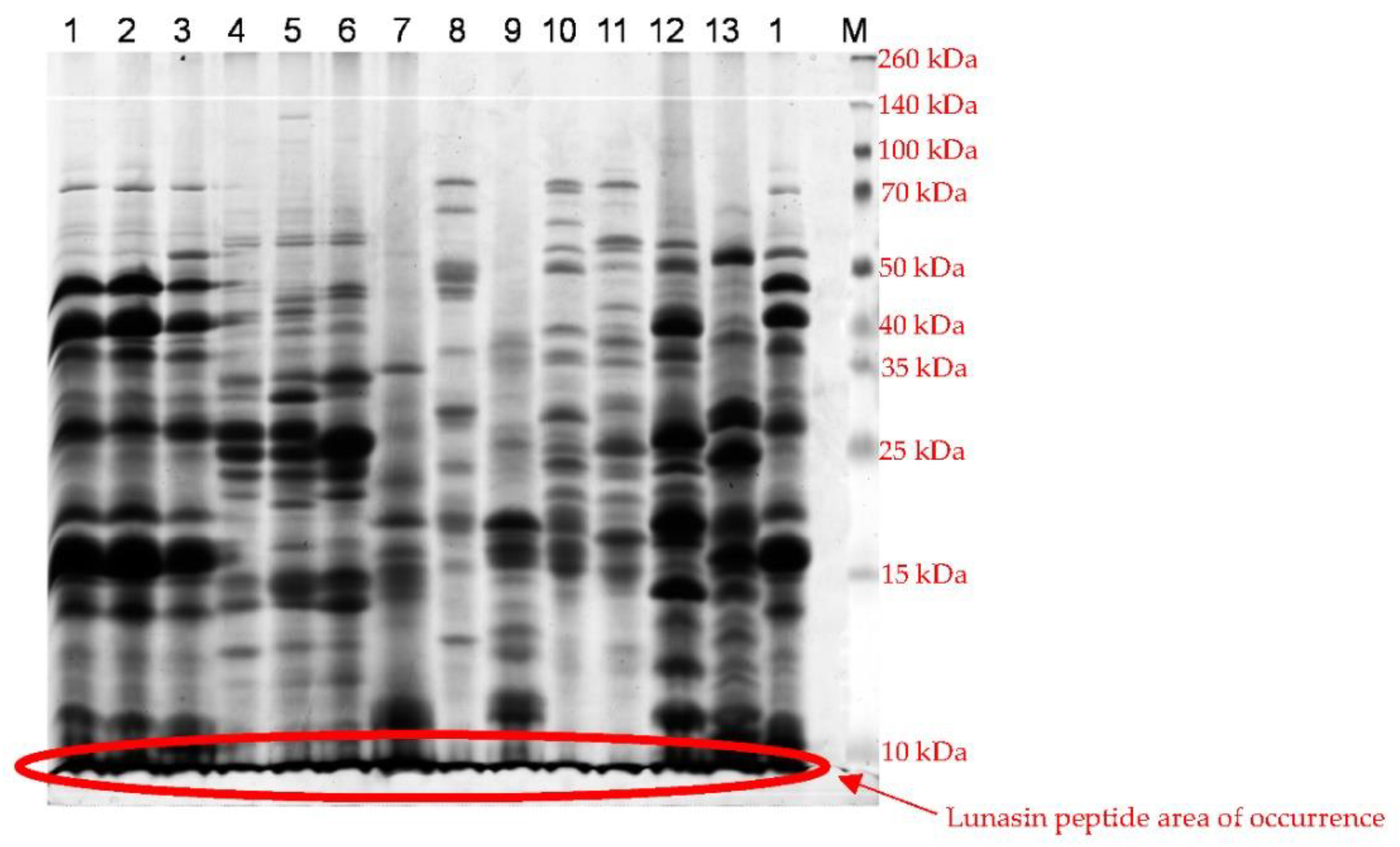
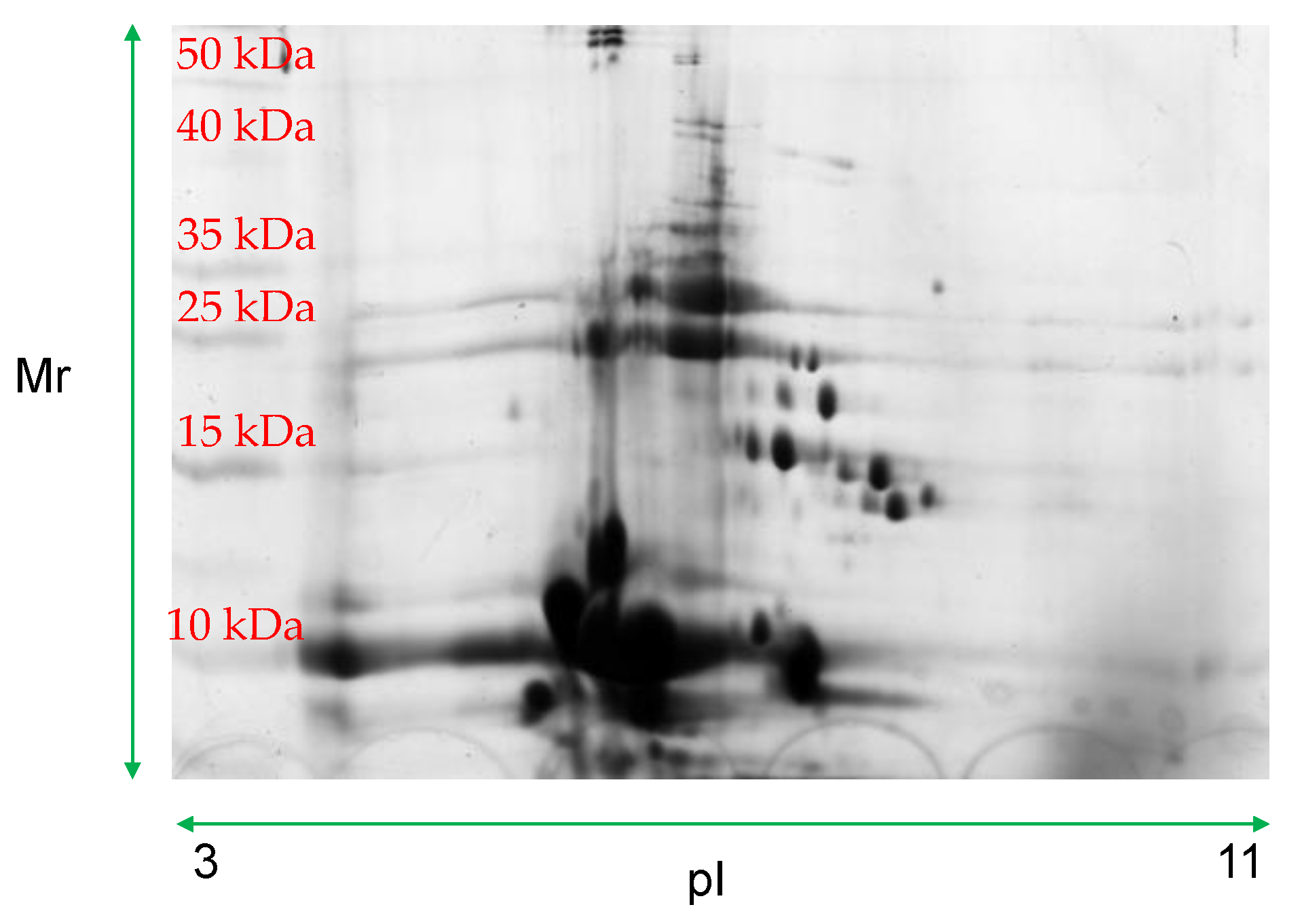

| Spot Number | Molecular Weight | Isoelectric Point |
|---|---|---|
| 3 | 5.93 | 3.47 |
| 3001 | 5.99 | 5.82 |
| 4001 | 5.74 | 6.11 |
| 6002 | 5.97 | 7.04 |
| 7002 | 5.87 | 7.41 |
| 7003 | 5.96 | 7.67 |
| 8001 | 5.72 | 9.51 |
| 9001 | 5.89 | 10.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chňapek, M.; Rajnincová, D.; Balážová, Ž.; Ražná, K.; Vivodík, M.; Hromadová, Z.; Mikolášová, L.; Gálová, Z. Proteomic and Genetic Approach for Lunasin Peptide and Gene Presence Detection in Various Plants. Biol. Life Sci. Forum 2022, 11, 86. https://doi.org/10.3390/IECPS2021-12004
Chňapek M, Rajnincová D, Balážová Ž, Ražná K, Vivodík M, Hromadová Z, Mikolášová L, Gálová Z. Proteomic and Genetic Approach for Lunasin Peptide and Gene Presence Detection in Various Plants. Biology and Life Sciences Forum. 2022; 11(1):86. https://doi.org/10.3390/IECPS2021-12004
Chicago/Turabian StyleChňapek, Milan, Dana Rajnincová, Želmíra Balážová, Katarína Ražná, Martin Vivodík, Zuzana Hromadová, Lucia Mikolášová, and Zdenka Gálová. 2022. "Proteomic and Genetic Approach for Lunasin Peptide and Gene Presence Detection in Various Plants" Biology and Life Sciences Forum 11, no. 1: 86. https://doi.org/10.3390/IECPS2021-12004
APA StyleChňapek, M., Rajnincová, D., Balážová, Ž., Ražná, K., Vivodík, M., Hromadová, Z., Mikolášová, L., & Gálová, Z. (2022). Proteomic and Genetic Approach for Lunasin Peptide and Gene Presence Detection in Various Plants. Biology and Life Sciences Forum, 11(1), 86. https://doi.org/10.3390/IECPS2021-12004






