Effects of Delphinidin-3-Sambubiosid on Different Pathways of Human Cells According to a Bioinformatic Analysis †
Abstract
:1. Introduction
2. Methods
2.1. Bioinformatic Analysis
2.2. Literature Search and Data Selection
2.3. Inclusion and Exclusion Criteria
2.4. Results
2.4.1. Enriched Analysis of Gene Ontology and Metabolic Pathways
2.4.2. Protein–Protein Interaction Network
3. Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nguyen, T.T.; Phan-Thi, H.; Pham-Hoang, B.N.; Ho, P.T.; Waché, Y.; Nguyen, T.T. Encapsulation of Hibiscus sabdariffa L. anthocyanins as natural colors in yeast. Food Res Int. 2018, 107, 275–280. [Google Scholar] [CrossRef] [PubMed]
- Kalt, W.; Cassidy, A.; Howard, L.R.; Krikorian, R.; Stull, A.J.; Tremblay, F.; Zamora-Ros, R. Recent Research on the Health Benefits of Blueberries and Their Anthocyanins. Adv. Nutr. 2020, 11, 224–236. [Google Scholar] [CrossRef] [PubMed]
- Reis, J.F.; Monteiro, V.V.S.; De Souza Gomes, R.; Do Carmo, M.M.; Da Costa, G.V.; Ribera, P.C.; Monteiro, M.C. Action mechanism and cardiovascular effect of anthocyanins: A systematic review of animal and human studies. J. Transl. Med. 2016, 14, 316. [Google Scholar] [CrossRef] [PubMed]
- Kartinah, N.T.; Fadilah, F.; Ibrahim, E.I.; Suryati, Y. The Potential of Hibiscus sabdariffa Linn in Inducing Glucagon-Like Peptide-1 via SGLT-1 and GLPR in DM Rats. BioMed. Res. Int. 2019, 2019, 8724824. [Google Scholar] [CrossRef] [PubMed]
- Grosdidier, A.; Zoete, V.; Michielin, O. SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Res. 2011, 39, W270–W277. [Google Scholar] [CrossRef] [PubMed]
- Grosdidier, A.; Zoete, V.; Michielin, O. Fast docking using the CHARMM force field with EADock DSS. J. Comput. Chem. 2011, 32, 2149–2159. [Google Scholar] [CrossRef]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein–protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2022, 51, D638–D646. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The STRING database in 2017: Quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. 2017, 45, D362–D368. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING v10: Protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef] [PubMed]
- Franceschini, A.; Lin, J.; von Mering, C.; Jensen, L.J. SVD-phy: Improved prediction of protein functional associations through singular value decomposition of phylogenetic profiles. Bioinformatics 2016, 32, 1085–1087. [Google Scholar] [CrossRef] [PubMed]
- Franceschini, A.; Szklarczyk, D.; Frankild, S.; Kuhn, M.; Simonovic, M.; Roth, A.; Lin, J.; Minguez, P.; Bork, P.; von Mering, C.; et al. STRING v9.1: Protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013, 41, D808–D815. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Franceschini, A.; Kuhn, M.; Simonovic, M.; Roth, A.; Minguez, P.; Doerks, T.; Stark, M.; Muller, J.; Bork, P.; et al. The STRING database in 2011: Functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011, 39, D561–D568. [Google Scholar] [CrossRef]
- Jensen, L.J.; Kuhn, M.; Stark, M.; Chaffron, S.; Creevey, C.; Muller, J.; Doerks, T.; Julien, P.; Roth, A.; Simonovic, M.; et al. STRING 8--a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009, 37, D412–D416. [Google Scholar] [CrossRef]
- von Mering, C.; Jensen, L.J.; Kuhn, M.; Chaffron, S.; Doerks, T.; Krüger, B.; Snel, B.; Bork, P. STRING 7--recent developments in the integration and prediction of protein interactions. Nucleic Acids Res. 2007, 35, D358–D362. [Google Scholar] [CrossRef]
- Von Mering, C.; Jensen, L.J.; Snel, B.; Hooper, S.D.; Krupp, M.; Foglierini, M.; Jouffre, N.; Huynen, M.A.; Bork, P. STRING: Known and predicted protein-protein associations, integrated and transferred across organisms. Nucleic Acids Res. 2005, 33, D433–D437. [Google Scholar] [CrossRef]
- Von Mering, C.; Huynen, M.; Jaeggi, D.; Schmidt, S.; Bork, P.; Snel, B. STRING: A database of predicted functional associations between proteins. Nucleic Acids Res. 2003, 31, 258–261. [Google Scholar] [CrossRef]
- Snel, B. STRING: A web-server to retrieve and display the repeatedly occurring neighbourhood of a gene. Nucleic Acids Res. 2000, 28, 3442–3444. [Google Scholar] [CrossRef]
- Syed, D.N.; Afaq, F.; Sarfaraz, S.; Khan, N.; Kedlaya, R.; Setaluri, V.; Mukhtar, H. Delphinidin inhibits cell proliferation and invasion via modulation of Met receptor phosphorylation. Toxicol. Appl. Pharmacol. 2008, 231, 52–60. [Google Scholar] [CrossRef]
- Teller, N.; Thiele, W.; Boettler, U.; Sleeman, J.; Marko, D. Delphinidin inhibits a broad spectrum of receptor tyrosine kinases of the ErbB and VEGFR family. Mol. Nutr. Food Res. 2009, 53, 1075–1083. [Google Scholar] [CrossRef]
- Pal, H.C.; Sharma, S.; Strickland, L.R.; Agarwal, J.; Athar, M.; Elmets, C.A.; Afaq, F. Delphinidin Reduces Cell Proliferation and Induces Apoptosis of Non-Small-Cell Lung Cancer Cells by Targeting EGFR/VEGFR2 Signaling Pathways. PLoS ONE 2013, 8, e77270. [Google Scholar] [CrossRef]
- Fridrich, D.; Teller, N.; Esselen, M.; Pahlke, G.; Marko, D. Comparison of delphinidin, quercetin and (–)-epigallocatechin-3-gallate as inhibitors of the EGFR and the ErbB2 receptor phosphorylation. Mol. Nutr. Food Res. 2008, 52, 815–822. [Google Scholar] [CrossRef]
- Haberthür, U.; Caflisch, A. FACTS: Fast analytical continuum treatment of solvation. J. Comput. Chem. 2008, 29, 701–715. [Google Scholar] [CrossRef]
- Yan, L.; Zhang, Z.; Liu, Y.; Ren, S.; Zhu, Z.; Wei, L.; Feng, J.; Duan, T.; Sun, X.; Xie, T.; et al. Anticancer Activity of Erianin: Cancer-Specific Target Prediction Based on Network Pharmacology. Front. Mol. Biosci. 2022, 9, 862932. [Google Scholar] [CrossRef]
- Pawitan, Y.; Michiels, S.; Koscielny, S.; Gusnanto, A.; Ploner, A. False discovery rate, sensitivity and sample size for microarray studies. Bioinformatics 2005, 21, 3017–3024. [Google Scholar] [CrossRef]
- Kiefer, F.N.; Neysari, S.; Humar, R.; Li, W.; Munk, V.C.; Battegay, E.J. Hypertension and Angiogenesis. Curr. Pharm. Des. 2003, 9, 1733–1744. [Google Scholar] [CrossRef]
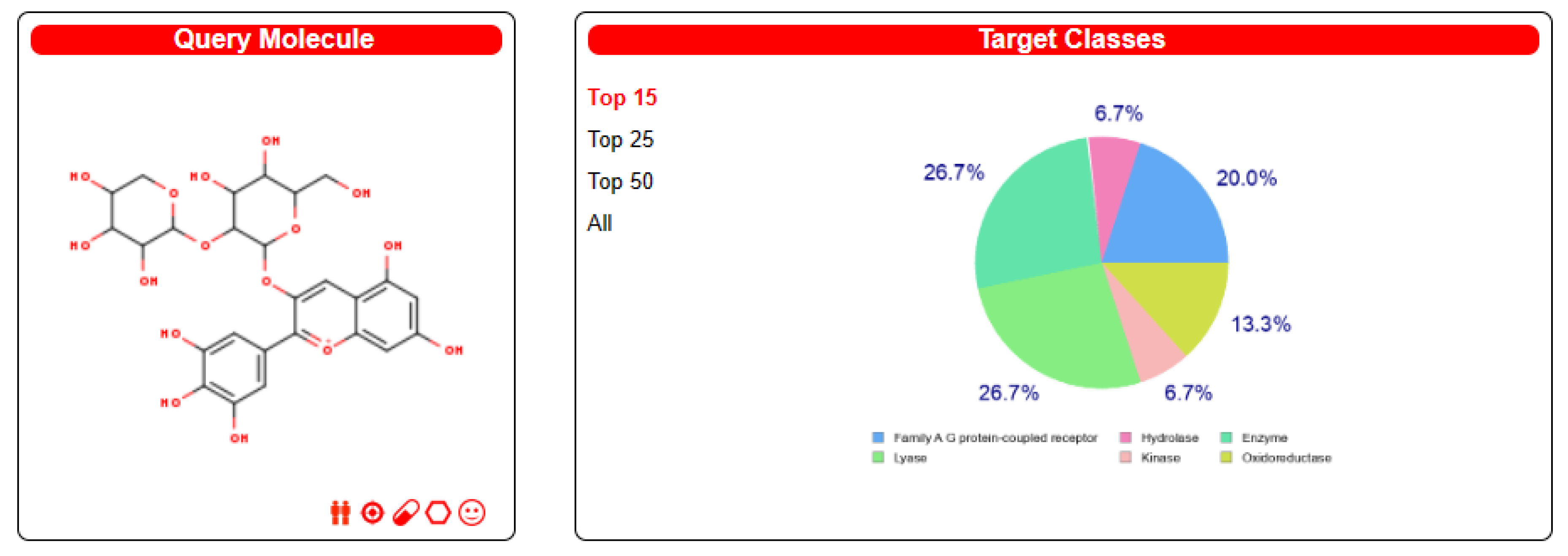
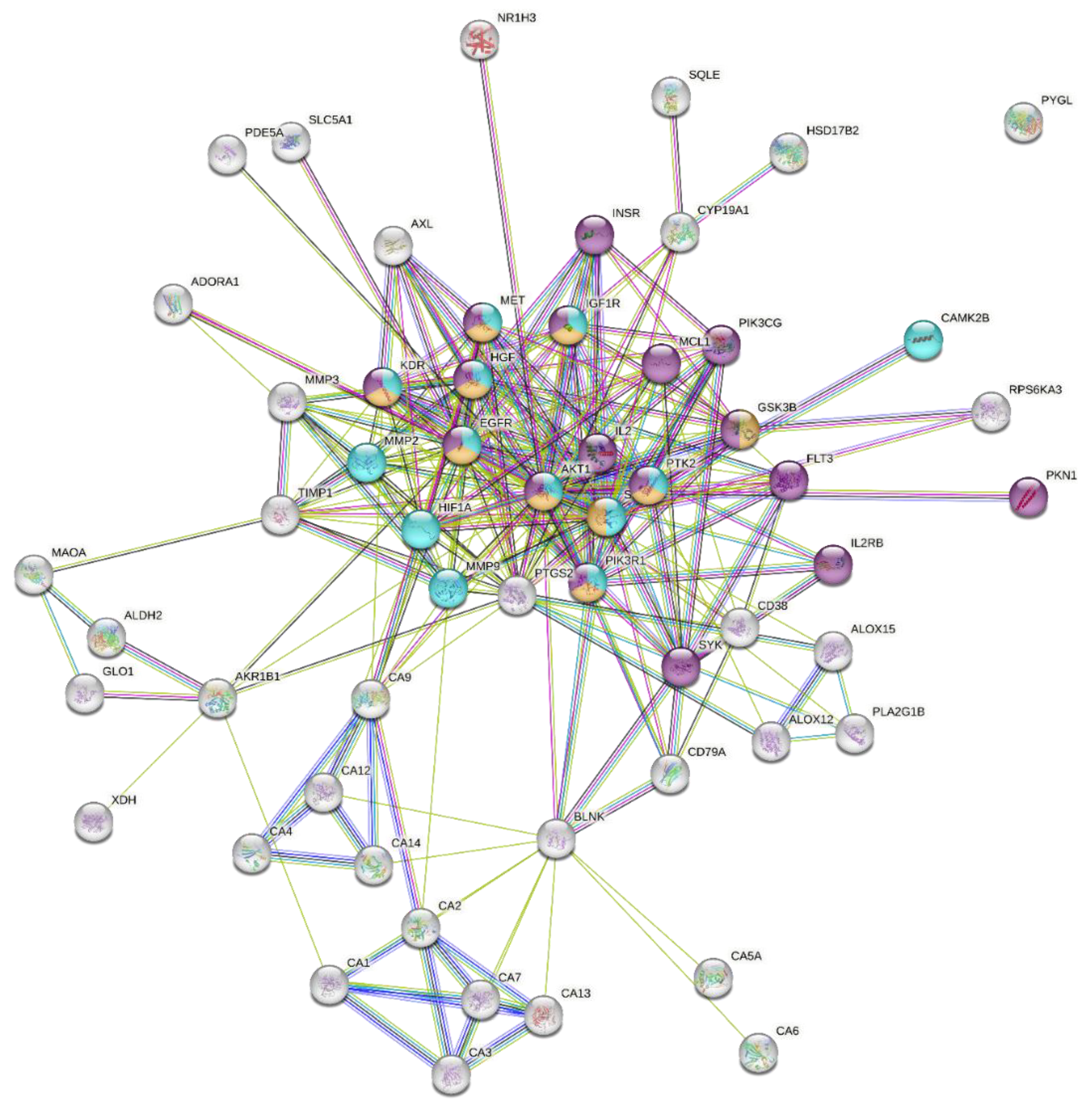
| Enrichment FDR | Genes | Pathway Genes | Fold Enrichment | Pathway | Genes |
|---|---|---|---|---|---|
| 4.20 × 10−18 | 10 | 17 | 141.151703 | Nitrogen metabolism | CA2, CA9, CA14, CA6, CA1, CA3, CA4, CA7, CA5A CA13 |
| 1.49 × 10−9 | 15 | 354 | 10.1677074 | PI3K-Akt signaling pathway | GSK3B, PIK3CG, MET, IL2, FLT3, PKN1, KDR, IGF1R, AKT1, MCL1, PIK3R1, EGFR, SYK, PTK2, INSR |
| 2.21 × 10−9 | 9 | 79 | 27.3369754 | EGFR tyrosine kinase inhibitor resistance | GSK3B, MET, KDR, IGF1R, AKT1, PIK3R1, EGFR, AXL, SRC |
| 2.73 × 10−9 | 27 | 1527 | 4.24287044 | Metabolic pathways | CD38, PTGS2, CA12, AKR1B1, HSD17B2, PYGL, CA2, SQLE, PIK3CG, CA9, ALOX12, ALDH2, CA14, GLO1, CA6, CA1, CYP19A1, PDE5A, XDH, ALOX15, CA3, CA4, CA7, PLA2G1B, CA5A, CA13, MAOA |
| 1.98 × 10−7 | 8 | 95 | 20.2069806 | Endocrine resistance | MMP2, MMP9, IGF1R, AKT1, PIK3R1, EGFR, PTK2, SRC |
| 3.94 × 10−6 | 7 | 108 | 15.5528265 | Insulin resistance | NR1H3, GSK3B, PYGL, AKT1, PIK3R1, INSR, RPS6KA3 |
| 4.89 × 10−6 | 6 | 70 | 20.5678196 | Central carbon metabolism in cancer | HIF1A, MET, FLT3, AKT1, PIK3R1, EGFR |
| 2.28 × 10−5 | 8 | 214 | 8.97038859 | Lipid and atherosclerosis | CAMK2B, GSK3B, MMP9, AKT1, PIK3R1, MMP3, PTK2, SRC |
| 2.52 × 10−5 | 5 | 56 | 21.424812 | Regulation of lipolysis in adipocytes | PTGS2, AKT1, PIK3R1, ADORA1, INSR |
| 0.0001554 | 8 | 294 | 6.52946652 | MAPK signaling pathway | MET, FLT3, KDR, IGF1R, AKT1, EGFR, INSR, RPS6KA3 |
| 0.00042661 | 5 | 112 | 10.712406 | TNF signaling pathway | PTGS2, MMP9, AKT1, PIK3R1, MMP3 |
| 0.0008788 | 5 | 137 | 8.7575874 | Insulin signaling pathway | GSK3B, PYGL, AKT1, PIK3R1, INSR |
| 0.00134156 | 5 | 155 | 7.74057725 | Non-alcoholic fatty liver disease | NR1H3, GSK3B, AKT1, PIK3R1, INSR |
| 0.0024404 | 3 | 47 | 15.3164614 | Carbohydrate digestion and absorption | SLC5A1, AKT1, PIK3R1 |
| 0.00373995 | 4 | 120 | 7.99859649 | AMPK signaling pathway | IGF1R, AKT1, PIK3R1, INSR |
| 0.01950816 | 3 | 107 | 6.72779144 | Glucagon signaling pathway | CAMK2B, PYGL, AKT1 |
| 0.02834824 | 2 | 46 | 10.432952 | Type II diabetes mellitus | PIK3R1, INSR |
| 0.02929805 | 2 | 47 | 10.2109742 | Pyruvate metabolism | ALDH2, GLO1 |
| Gene Symbol | Protein Name | Protein Function |
|---|---|---|
| AKT1 | RAC-alpha serine/threonine-protein kinase | Regulates many processes, including metabolism, proliferation, cell survival, growth, and angiogenesis |
| PTK2 | Focal adhesion Kinase 1 | Related to the increase in glucose uptake and glycogen synthesis in insulin-sensitive tissues. |
| IL2 | Interleukin-2 | Required for T-cell proliferation and other cells of the immune system |
| PIK3R1 | Phosphoinositide-3-kinase regulatory subunit alpha/beta/delta | Necessary for the insulin-stimulated increase in glucose uptake and glycogen synthesis |
| SYK | Spleen-associated tyrosine kinase | Regulates biological processes including immunity, cell adhesion, vascular development, and others |
| PTGS2 | Prostaglandin G/H synthase 2 | Plays a role in the production of inflammatory prostaglandins |
| MMP9 | Matrix metalloproteinase-9 | Key role in local proteolysis of the extracellular matrix and leukocyte migration |
| HIF1A | Hypoxia-inducible factor 1-alpha | Master transcriptional regulator in response to hypoxia |
| MMP2 | Matrix metalloproteinase-2 (gelatinase a) | Involved in angiogenesis, tissue repair, tumor invasion, inflammation, and atherosclerotic plaque rupture |
| KDR | Vascular endothelial growth factor receptor 2 | Essential in the regulation of angiogenesis, promotes the proliferation, survival, and migration of endothelial cells |
| MET | Hepatocyte growth factor receptor | Regulates processes like proliferation, scattering, morphogenesis, and survival |
| HGF | Hepatocyte growth factor | Growth factor for a broad spectrum of tissues and cell types |
| EGFR | Epidermal growth factor receptor | Converts extracellular cues into appropriate cellular responses |
| IGF1R | Insulin-like growth factor 1 receptor | Involved in cell growth and survival control |
| CA9 | Carbonic anhydrase 9 | Involved in pH regulation |
| BLNK | B-cell linker protein | Important for the activation of NF-kappa-B and NFAT |
| Genes | Results at the Gene Expression Level | Results at the Protein Level | Results of Pathway Impact |
|---|---|---|---|
| MET | Syed, D. N. 2008: Suppress the phosphorylation of the protein [21] | ||
| IGF1R | Teller et al., 2009: Inhibition of its kinase activity [22] | ||
| EGFR | Harish Chandra Pal, et al., 2013: Reduction in the expression of the gen [23] | Fredrich D, Et all, 2008: Suppress phosphorylation of the protein [24] | Harish Chandra Pal, et al., 2013: Inhibition of the PI3K-Akt pathway [23] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zúñiga-Hernández, S.R.; García-Iglesias, T.; Macías-Carballo, M.; Perez-Larios, A.; Rodríguez-Razón, C.M. Effects of Delphinidin-3-Sambubiosid on Different Pathways of Human Cells According to a Bioinformatic Analysis. Biol. Life Sci. Forum 2023, 29, 13. https://doi.org/10.3390/IECN2023-15797
Zúñiga-Hernández SR, García-Iglesias T, Macías-Carballo M, Perez-Larios A, Rodríguez-Razón CM. Effects of Delphinidin-3-Sambubiosid on Different Pathways of Human Cells According to a Bioinformatic Analysis. Biology and Life Sciences Forum. 2023; 29(1):13. https://doi.org/10.3390/IECN2023-15797
Chicago/Turabian StyleZúñiga-Hernández, Sergio R., Trinidad García-Iglesias, Monserrat Macías-Carballo, Alejandro Perez-Larios, and Christian Martin Rodríguez-Razón. 2023. "Effects of Delphinidin-3-Sambubiosid on Different Pathways of Human Cells According to a Bioinformatic Analysis" Biology and Life Sciences Forum 29, no. 1: 13. https://doi.org/10.3390/IECN2023-15797
APA StyleZúñiga-Hernández, S. R., García-Iglesias, T., Macías-Carballo, M., Perez-Larios, A., & Rodríguez-Razón, C. M. (2023). Effects of Delphinidin-3-Sambubiosid on Different Pathways of Human Cells According to a Bioinformatic Analysis. Biology and Life Sciences Forum, 29(1), 13. https://doi.org/10.3390/IECN2023-15797







