Unraveling Phylogenetic Relationships via A. carbonarius and A. tubingensis Sequence Analyses †
Abstract
:1. Introduction
2. Methods
2.1. Organisms, Growth Conditions, and DNA Isolation
2.2. PCR Amplification, Cloning and Sequencing
2.3. Sequence Alignment and Phylogenetic Analyses
3. Results
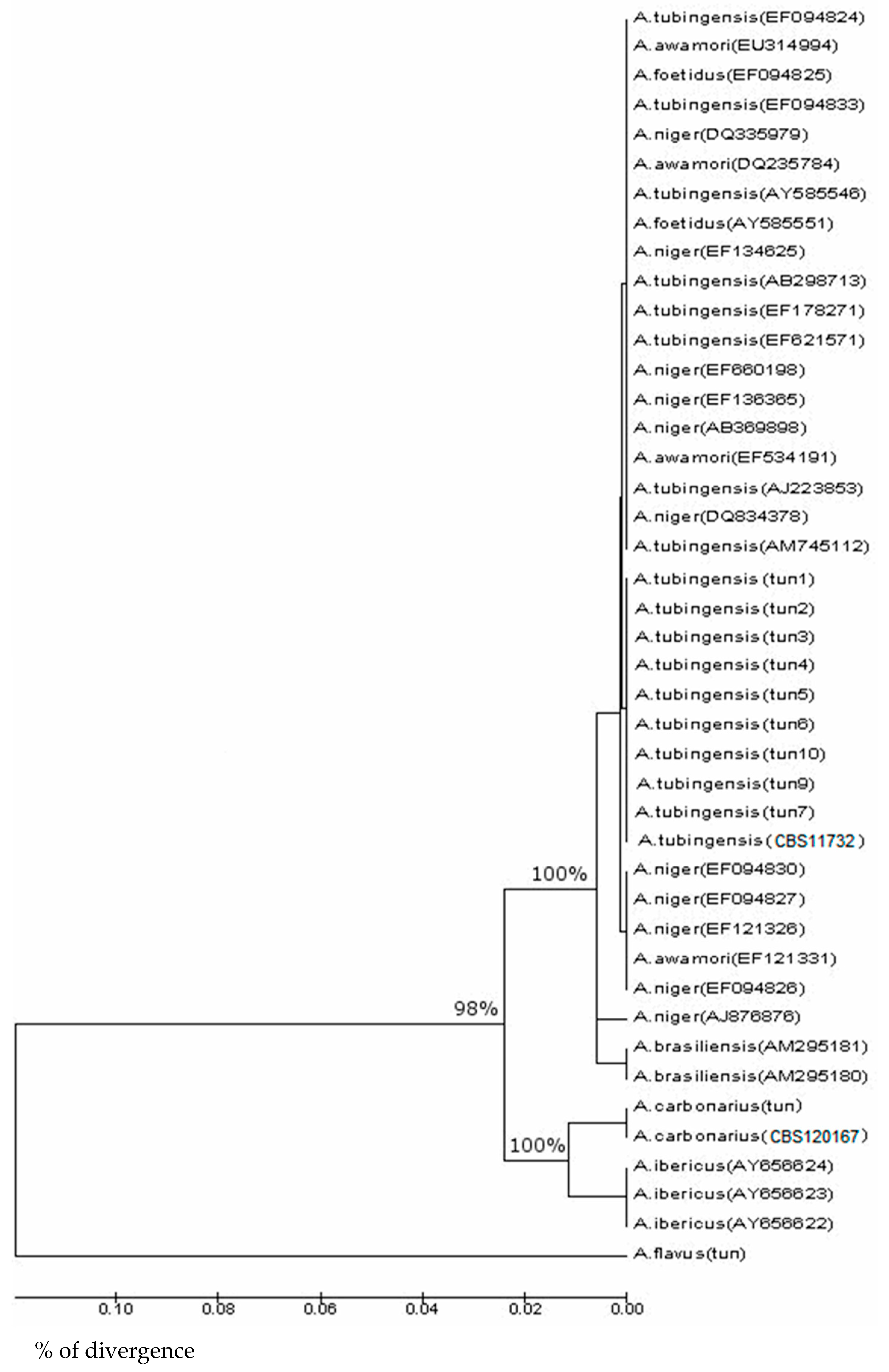
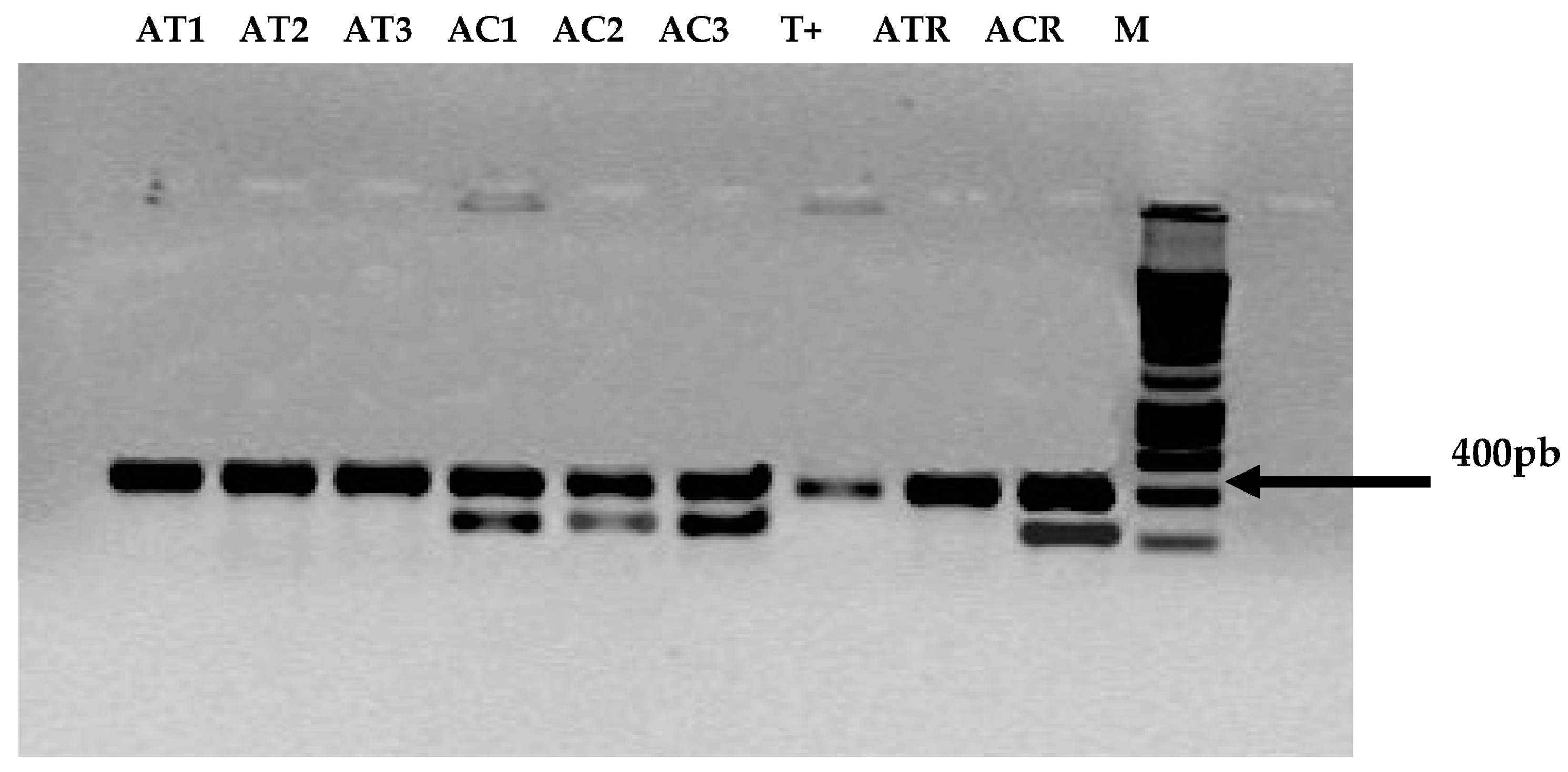
3.1. Screening of ITS-5.8 S rDNA in A. carbonarius and A. tubingensis
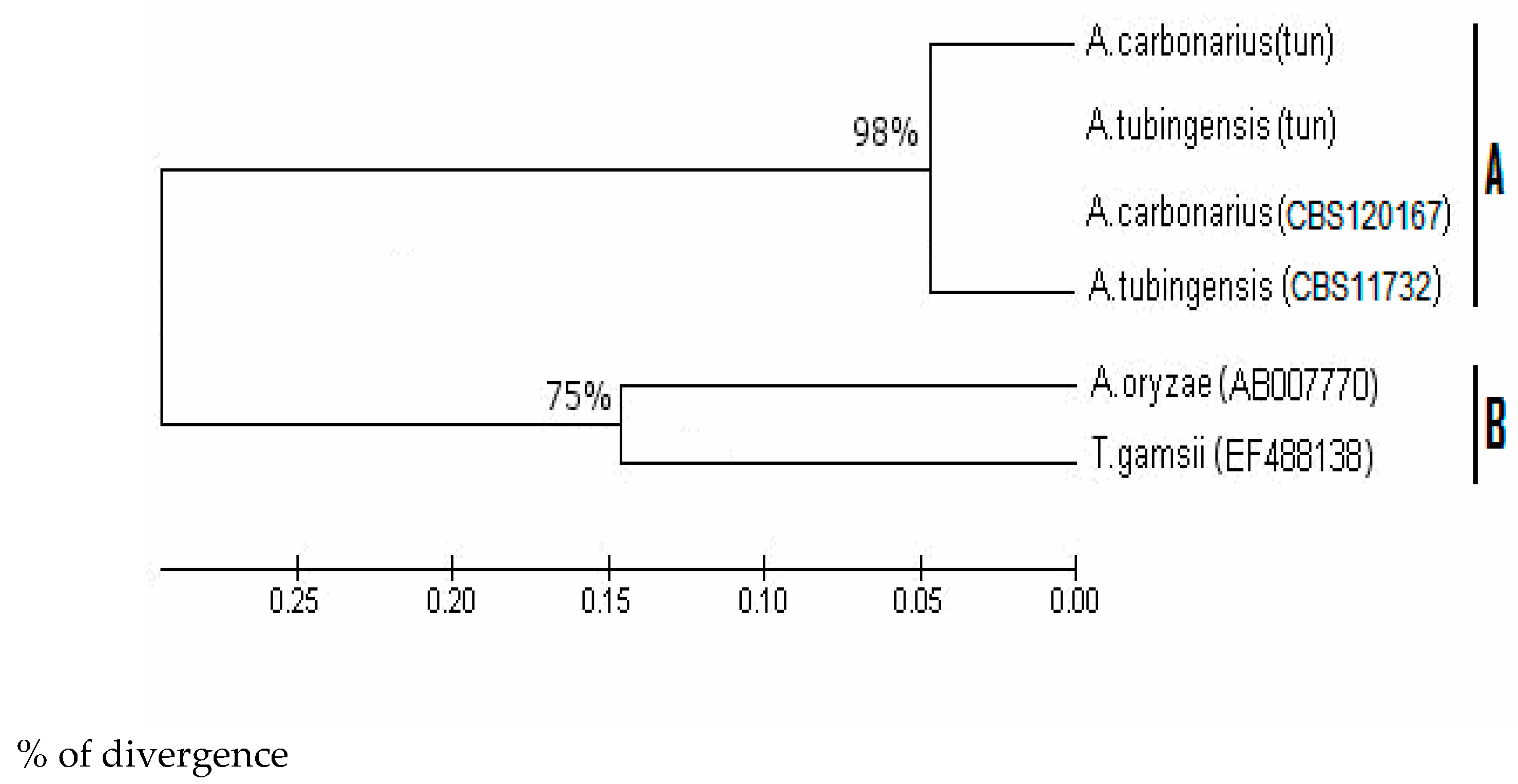
3.2. Screening and Phylogenetic Analysis of (niiA-niaD) Gene in A. carbonarius and A. tubingensis
3.3. Screening and Phylogenetic Analysis of ß-Tubulin Sequences
3.4. Screening and Phylogenetic Analysis of eEF-1 Sequences
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Abarca, M.L.; Bragulat, M.R.; Castella, G.; Cabanes, F.J. Ochratoxin A production by strains of Aspergillus niger var. niger. Appl. Environ. Microbiol. 1994, 60, 2650–2652. [Google Scholar] [CrossRef] [PubMed]
- De Jong, S.N.; Levesque, C.A.; Verkley, G.J.M.; Abeln, E.C.A.; Rahe, J.E.; Braun, P.G. Phylogenetic relationships among Neofabraea species causing tree cankers and bull’s-eye rot of apple based on DNA sequencing of ITS nuclear rDNA, mitochondrial rDNA, and the beta- tubulin gene. Mycol. Res. 2001, 105, 658–669. [Google Scholar] [CrossRef]
- Melki Ben Fredj, S.; Chebil, S.; Mliki, A. Isolation and characterization of ochratoxin A and aflatoxin B1 producing fungi infecting grapevines. Afr. J. Microbiol. Res. 2009, 3, 523–527. [Google Scholar]
- Schutze, J.; Krasko, A.; Custodio, M.R.; Efremova, S.M.; Muller, I.M.; Muller, W.E.G. Evolutionary relationships of Metazoa within the eukaryotes based on molecular data from Porifera. Proc. R. Soc. Lond. 1999, 266, 63–73. [Google Scholar] [CrossRef] [PubMed]
- Melki Ben Fredj, S.; Gautier, A.; Brygoo, Y.; Mliki, A. Molecular strategy to discriminate between two ochratoxin A producing fungal species in Aspergillus niger aggregate group isolated from fresh and dried grapes. Ann. Microbiol. 2009, 59, 635–641. [Google Scholar] [CrossRef]
- Myburg, H.; Gryzenhout, M.; Heath, R.; Roux, J.; Wingfield, B.; Wingfield, M. Cryphonectria canker on Tibouchina in South Africa. Mycol. Res. 2002, 106, 1299–1306. [Google Scholar] [CrossRef]
- Keeling, P.J.; Luker, M.A.; Palmer, J.D. Evidence from beta-tubulin phylogeny that microsporidia evolved from within the fungi. Mol. Biol. Evol. 2000, 17, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Raper, K.B.; Fennell, D.I. The Genus Aspergillus; Williams & Wilkins: Baltimore, MD, USA, 1965. [Google Scholar] [CrossRef]
- Teren, J.; Varga, J.; Hamari, Z.; Rinyu, E.; Kevei, F. Immunochemical detection of ochratoxin A in black Aspergillus strains. Mycopathologia 1996, 134, 171–176. [Google Scholar] [CrossRef] [PubMed]
- Gams, W.; Christensen, M.; Onions, A.H.S.; Pitt, J.I.; Samson, R.A. Infrageneric taxa of Aspergillus. In Advances in Penicillium and Aspergillus Systematics; Samson, R.A., Pitt, J.I., Eds.; Plenum Press: New York, NY, USA, 1985; pp. 55–61. [Google Scholar] [CrossRef]
- Hawksworth, D.L. Fungi and biodiversity: International incentives. Microbiologia 1997, 13, 221–226. [Google Scholar] [PubMed]
- Inui, T.; Takeda, Y.; Iizuka, H. Taxonomical studies on genus Rhizopus. J. Gen. Appl. Microbiol. 1965, 11, 1–121. [Google Scholar] [CrossRef]
- Edgcomb, V.P.; Roger, A.J.; Simpson, A.G.B.; Kysela, D.T.; Sogin, M.L. Evolutionary relationships among “jakobid” flagellates as indicated by alpha- and beta-tubulin phylogenies. Mol. Biol. Evol. 2001, 18, 514–522. [Google Scholar] [CrossRef] [PubMed]
- Keller, N.P. Fungal secondary metabolism: Regulation, function and drug discovery. Nat. Rev. Microbiol. 2019, 17, 167–180. [Google Scholar] [CrossRef] [PubMed]
- Hawksworth, D.L. The fungal dimension of biodiversity: Magnitude, significance, and conservation. Mycol. Res. 1991, 95, 641–655. [Google Scholar] [CrossRef]
- Peterson, S.W. Phylogenetic relationships in Aspergillus based on rDNA sequence analysis. In Integration of Modern Taxonomic Methods for Penicillium and Aspergillus Classification; Samson, R.A., Pitt, J.I., Eds.; Harwood Academic Publishers: Amsterdam, The Netherlands, 2000; pp. 323–355. [Google Scholar] [CrossRef]
- Baldauf, S.L.; Roger, A.J.; Wenk-Siefert, I.; Doolittle, W.F. A kingdom-level phylogeny of eukaryotes based on combined protein data. Science 2000, 290, 972–977. [Google Scholar] [CrossRef] [PubMed]
- Geiser, D.M.; Frisvad, J.; Taylor, J. Evolutionary relationships in Aspergillus section Fumigati inferred from partial ß-tubulin and hydrophobin DNA sequences. Mycologia 1998, 90, 831–845. [Google Scholar] [CrossRef]
- O’Donnell, K.; Cigelnik, E.; Casper, H.H. Molecular phylogenetic, morphological and mycotoxin data support reidentification of the Quorn mycoprotein fungus as Fusarium venenatum. Fungal Genet. Biol. 1998, 23, 57–67. [Google Scholar] [CrossRef]
- Eichler, E.E.; Sankoff, D. Structural dynamics of eukaryotic chromosome evolution. Science 2003, 301, 793–797. [Google Scholar] [CrossRef] [PubMed]
- Keeling, P.J.; Doolittle, W.F. Alphα-tubulin from early-diverging eukaryotic lineages and the evolution of the tubulin family. Mol. Biol. Evol. 1996, 13, 1297–1305. [Google Scholar] [CrossRef] [PubMed]
- Ayliffe, M.A.; Dodds, P.N.; Lawrence, G.J. Characterisation of a beta-tubulin gene from Melampsora lini and comparison of fungal beta-tubulin genes. Mycol. Res. 2001, 105, 818–826. [Google Scholar] [CrossRef]
- Mages, W.; Cresnar, B.; Harper, J.F.; Brüderlein, M.; Schmitt, R. Volvox carteri alpha-2- tubulin-encoding and beta-2-tubulin-encoding genes: Regulatory signals and transcription. Gene 1995, 160, 47–54. [Google Scholar] [CrossRef]
- Serra, R.; Lourenço, A.; Aliipio, P.; Venâncio, A. Influence of the region of origin on the mycobiota of grapes with emphasis on Aspergillus and Penicillium species. Mycol. Res. 2006, 110, 971–978. [Google Scholar] [CrossRef]
- Aoki, T.; O’Donnell, K. Morphological and molecular characterization of Fusarium pseudograminearum sp. nov., formerly recognised as the Group 1 population of F. graminearum. Mycologia 1999, 91, 597–609. [Google Scholar] [CrossRef]
- Hawksworth, D.L. The magnitude of fungal diversity: The 1.5 million species estimate revisited. Mycol. Res. 2001, 105, 1422–1432. [Google Scholar] [CrossRef]
- Maddison, W.P.; Maddison, D.R. MacClade: Analysis of Phylogeny and Character Evolution; Version 3; Sinauer Associates: Sunderland, MA, USA, 1992. [Google Scholar] [CrossRef]
- Melki Ben Fredj, S.; Chebil, S.; Lebrihi, A.; Lasram, S.; Ghorbel, A.; Mliki, A. Occurrence of pathogenic fungal species in Tunisian vineyards. Int. J. Food Microbiol. 2007, 113, 245–250. [Google Scholar] [CrossRef]

| Primer Code | Primer Sequence (5′ to 3′) | Annealing Temperature (X) °C |
|---|---|---|
| PN1 PN34 | AGTAAAAGTCGTAACAAGG TTGCCGCTTCACTCGCCGTT | 55 |
| PU PR | CGTTGTAAAACGACGGCCAGT GTACCAGTATCGACAAAGGAG | 52 |
| Nit-462F Nit-873R | GTTACGGAAGCAAAGAGGTA GTTATTAGGGGCTATGGCAC | 54 |
| ß-tubF ß-tubR | CTCGAGCGTAGTAACGTCTAC AAACCCTGGAGGCAGTCGC | 55 |
| eEF1-526F eEF1-1567R | GTCGTYGTYATYGGHCAYGT ACHGT RCC RATACCACC RAT CTT | 58 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Melki, S.B.F.; Omoregie, E.S.; Mliki, A. Unraveling Phylogenetic Relationships via A. carbonarius and A. tubingensis Sequence Analyses. Biol. Life Sci. Forum 2024, 31, 31. https://doi.org/10.3390/ECM2023-16445
Melki SBF, Omoregie ES, Mliki A. Unraveling Phylogenetic Relationships via A. carbonarius and A. tubingensis Sequence Analyses. Biology and Life Sciences Forum. 2024; 31(1):31. https://doi.org/10.3390/ECM2023-16445
Chicago/Turabian StyleMelki, Sabah Ben Fredj, Ehi Sheena Omoregie, and Ahmed Mliki. 2024. "Unraveling Phylogenetic Relationships via A. carbonarius and A. tubingensis Sequence Analyses" Biology and Life Sciences Forum 31, no. 1: 31. https://doi.org/10.3390/ECM2023-16445




