Necrotrophic Effectors SnTox from the Stagonospora nodorum (Berk.) Manipulate the Redox Metabolism of the Host Plant to Hijack Its Early Defense Response †
Abstract
1. Introduction
2. Material and Methods
2.1. Research Objects and Seedling Resistance
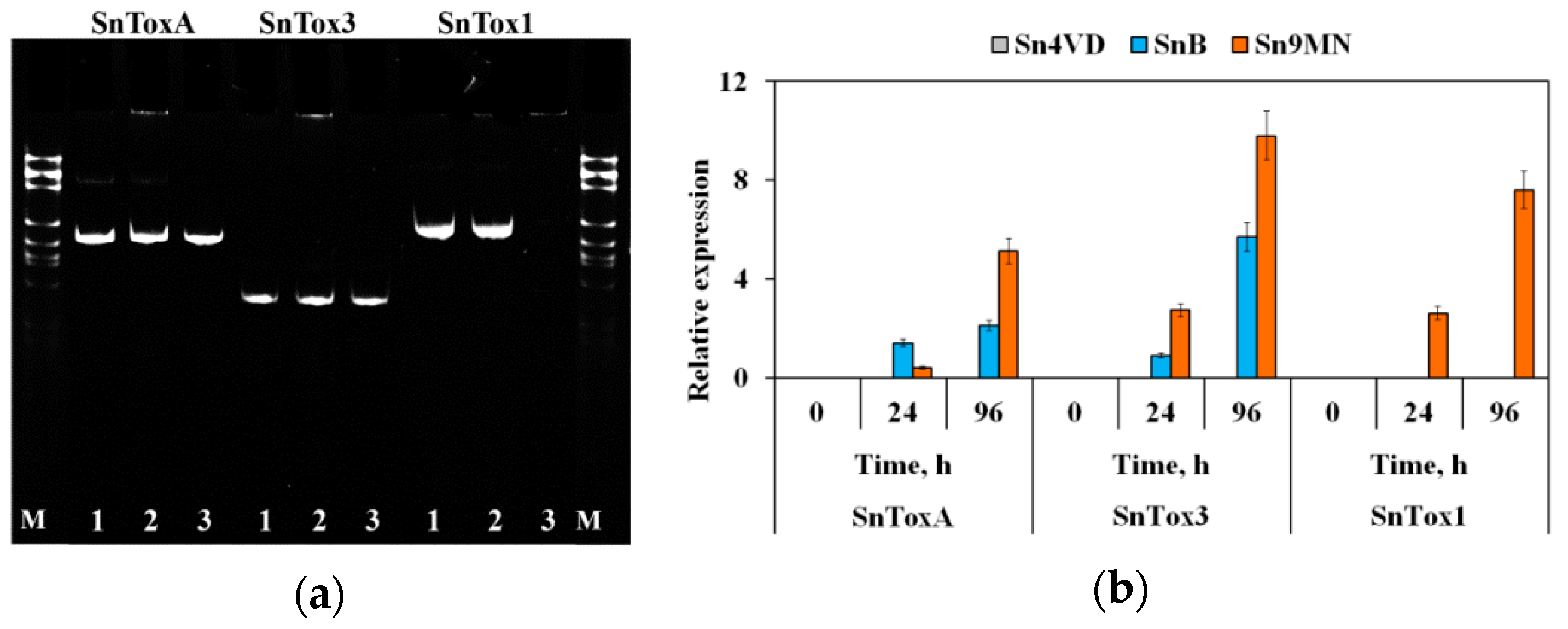
2.2. Isolation of DNA and RNA and Performing the Polymerase Chain Reaction (PCR) and Biochemical Parameters
2.3. Statistical Analysis
3. Results
3.1. The Role of Compatible Interactions in Causing Disease
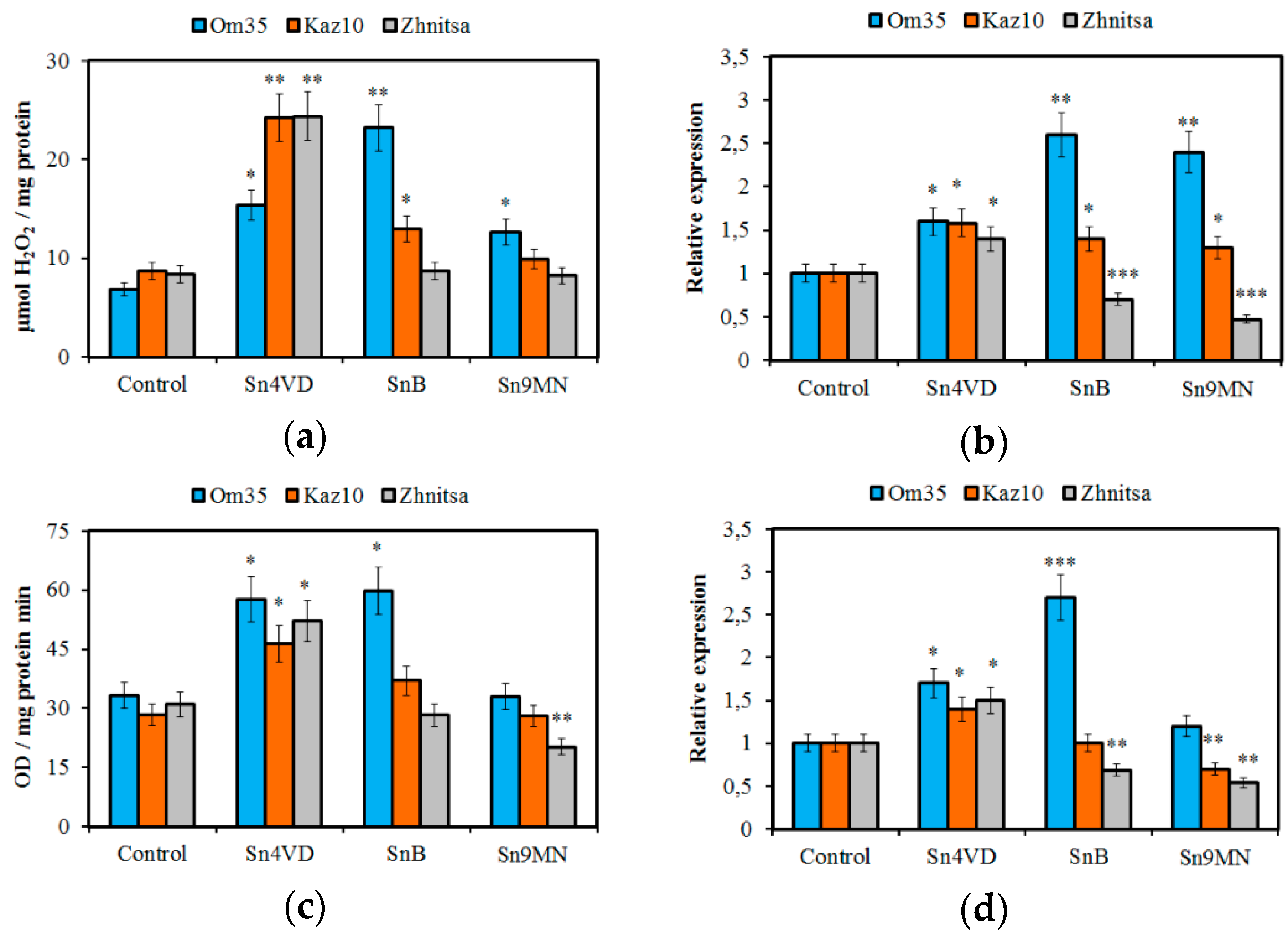
3.2. The Role of Compatible Interactions in the Suppression of ROS Production
4. Discussion
5. Conclusions
Supplementary Materials
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jones, J.D.G.; Dang, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Jwa, N.-S.; Hwang, B.K. Convergent evolution of pathogen effectors toward reactive oxygen species signaling networks in plants. Front. Plant Sci. 2017, 8, 1687. [Google Scholar] [CrossRef] [PubMed]
- Friesen, T.L.; Faris, J.D. Characterization of the wheat-Stagonospora nodorum disease system: What is the molecular basis of this quantitative necrotrophic disease interaction? Can. J. Plant Pathol. 2010, 32, 20–28. [Google Scholar] [CrossRef]
- Shi, G.; Zhang, Z.; Friesen, T.L.; Raats, D.; Fahima, T.; Brueggeman, R.S.; Lu, S.; Trick, H.N.; Liu, Z.; Chao, W.; et al. The hijacking of a receptor kinase–driven pathway by a wheat fungal pathogen leads to disease. Sci. Adv. 2016, 2, e1600822. [Google Scholar] [CrossRef] [PubMed]
- Phan, H.T.T.; Rybak, K.; Furuki, E.; Breen, S.; Solomon, P.S.; Oliver, R.P.; Tan, K.C. Differential effector gene expression underpins epistasis in a plant fungal disease. Plant J. 2016, 87, 343–354. [Google Scholar] [CrossRef] [PubMed]
- McDonald, M.C.; Solomon, P.S. Just the surface: Advances in the discovery and characterization of necrotrophic wheat effectors. Curr. Opin. Microbiol. 2018, 46, 14–18. [Google Scholar] [CrossRef]
- Faris, J.D.; Zhang, Z.; Lu, H.; Lu, S.; Reddy, L.; Cloutier, S.; Fellers, J.P.; Meinhardt, S.W.; Rasmussen, J.B.; Xu, S.S.; et al. A unique wheat disease resistance-like gene governs effector-triggered susceptibility to necrotrophic pathogens. Proc. Natl. Acad. Sci. USA 2010, 107, 13544–13549. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Zhang, Z.; Faris, J.D.; Oliver, R.P.; Syme, R.; McDonald, M.C.; McDonald, B.A.; Solomon, P.S.; Lu, S.; Shelver, W.L.; et al. The cysteine rich necrotrophic effector SnTox1 produced by Stagonospora nodorum triggers susceptibility of wheat lines harboring Snn1. PLoS Pathog. 2012, 8, e1002467. [Google Scholar] [CrossRef] [PubMed]
- Veselova, S.V.; Burkhanova, G.F.; Nuzhnaya, T.V.; Maksimov, I.V. Roles of ethylene and cytokinins in development of defense responses in Triticum aestivum plants infected with Septoria nodorum. Russ. J. Plant Physiol. 2016, 63, 609–619. [Google Scholar] [CrossRef]
- Liu, Z.H.; Friesen, T.L.; Rasmussen, J.B.; Ali, S.; Meinhardt, S.W.; Faris, J.D. Quantitative trait loci analysis and mapping of seedling resistance to Stagonospora nodorum leaf blotch in wheat. Phytopathology 2004, 94, 1061–1067. [Google Scholar] [CrossRef]
- Veselova, S.V.; Burkhanova, G.F.; Nuzhnaya, T.V.; Rumyantsev, S.D.; Maksimov, I.V. Effect of the host-specific toxin SnTOX3 from Stagonospora nodorum on ethylene signaling pathway regulation and redox-state in common wheat. Vavilovskii Zhurnal Genet. i Selektsii Vavilov J. Genet. Breed. 2019, 23, 856–864. [Google Scholar] [CrossRef]
- Winterberg, B.; Du Fall, L.A.; Song, X.M.; Pascovici, D.; Care, N.; Molloy, M.; Ohms, S.; Solomon, P.S. The necrotrophic effector protein SnTox3 re-programs metabolism and elicits a strong defence response in susceptible wheat leaves. BMC Plant Biol. 2014, 14, 215. [Google Scholar] [CrossRef]
- Podgórska, A.; Burian, M.; Szal, B. Extra-cellular but extra-ordinarily important for cells: Apoplastic reactive oxygen species metabolism. Front. Plant Sci. 2017, 8, 1353. [Google Scholar] [CrossRef]
- Qi, J.; Wang, J.; Gong, Z.; Zhou, J.M. Apoplastic ROS signaling in plant immunity. Curr. Opin. Plant Biol. 2017, 38, 92–100. [Google Scholar] [CrossRef] [PubMed]

| Variety | Reaction to Damage | Isolate S. nodorum | ||
|---|---|---|---|---|
| Sn4VD (toxa/tox3/tox1) | SnB (ToxA/Tox3/tox1) | Sn9MN (ToxA/Tox3/Tox1) | ||
| Omskaya 35 (tsn1/snn3/Snn1) | Necrosis, % | 0.05 ± 0.002 | 5 ± 0.7 | 23 ± 2 |
| Chlorosis, % | 0 | 3 ± 0.5 | 0 | |
| Damage zone, % | 0.05 ± 0.001 | 8 ± 1 | 23 ± 2 | |
| Damage score | 1 | 2 | 3 | |
| Resistance group * | RR | R | M | |
| Kazahstanskaya 10 (tsn1/Snn3/Snn1) | Necrosis, % | 0.05 ± 0.002 | 16 ± 2 | 31 ± 3 |
| Chlorosis, % | 0 | 35 ± 3 | 25 ± 2 | |
| Damage zone, % | 0.05 ± 0.001 | 51 ± 5 | 56 ± 4 | |
| Damage score | 1 | 4 | 4 | |
| Resistance group * | RR | S | S | |
| Zhnitsa (Tsn1/Snn3/Snn1) | Necrosis, % | 1 ± 0.1 | 18 ± 2 | 27 ± 2 |
| Chlorosis, % | 2 ± 0.2 | 55 ± 4 | 57 ± 5 | |
| Damage zone, % | 3 ± 0.3 | 73 ± 6 | 84 ± 6 | |
| Damage score | 1 | 5 | 5 | |
| Resistance group * | RR | SS | SS | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Veselova, S.; Nuzhnaya, T.; Burkhanova, G.; Rumyantsev, S.; Maksimov, I. Necrotrophic Effectors SnTox from the Stagonospora nodorum (Berk.) Manipulate the Redox Metabolism of the Host Plant to Hijack Its Early Defense Response. Biol. Life Sci. Forum 2021, 4, 106. https://doi.org/10.3390/IECPS2020-08765
Veselova S, Nuzhnaya T, Burkhanova G, Rumyantsev S, Maksimov I. Necrotrophic Effectors SnTox from the Stagonospora nodorum (Berk.) Manipulate the Redox Metabolism of the Host Plant to Hijack Its Early Defense Response. Biology and Life Sciences Forum. 2021; 4(1):106. https://doi.org/10.3390/IECPS2020-08765
Chicago/Turabian StyleVeselova, Svetlana, Tatyana Nuzhnaya, Guzel Burkhanova, Sergey Rumyantsev, and Igor Maksimov. 2021. "Necrotrophic Effectors SnTox from the Stagonospora nodorum (Berk.) Manipulate the Redox Metabolism of the Host Plant to Hijack Its Early Defense Response" Biology and Life Sciences Forum 4, no. 1: 106. https://doi.org/10.3390/IECPS2020-08765
APA StyleVeselova, S., Nuzhnaya, T., Burkhanova, G., Rumyantsev, S., & Maksimov, I. (2021). Necrotrophic Effectors SnTox from the Stagonospora nodorum (Berk.) Manipulate the Redox Metabolism of the Host Plant to Hijack Its Early Defense Response. Biology and Life Sciences Forum, 4(1), 106. https://doi.org/10.3390/IECPS2020-08765








